Abstract
Cryptosporidium spp. are diarrheagenic intestinal parasites with multiple hosts worldwide. A total of 1252 fresh fecal samples of sheep were collected from 10 large-scale farms in southern Xinjiang. Based on the small subunit ribosomal (SSU rRNA) gene of Cryptosporidium, 100 Cryptosporidium-positive samples (8.0%, 100/1252) were detected by PCR. Nine out of 10 farms were positive for Cryptosporidium, with the highest infection rate being 18.4% (23/125) on farm 9 in Qira. The infection rates of Cryptosporidium in pre-weaned lambs, weaned lambs, fattening sheep, and adult sheep were 20.3% (61/301), 10.3% (34/329), 0.9% (3/327), and 0.7% (2/295), respectively. Three Cryptosporidium species were identified, namely, C. xiaoi (n = 61), C. parvum (n = 22), and C. ubiquitum (n = 17). Of them, C. xiaoi was detected on all positive farms and in different age groups of sheep. The subtypes of C. parvum and C. ubiquitum were identified by PCR at the 60 kDa glycoprotein (gp60) gene. Two C. parvum subtypes were identified: IIdA19G1 (n = 21) and IIdA15G1 (n = 1). One C. ubiquitum subtype was identified with XIIa (n = 17). These results indicated the common transmission and genetic diversity of Cryptosporidium in sheep in southern Xinjiang, and further investigations are needed on the zoonotic potential of C. parvum and C. ubiquitum in this region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium spp. are apicomplexan parasites commonly found across many host species, including humans, livestock, companion animals, and wild animals (Menon et al. 2022). As an important reservoir and susceptible host of Cryptosporidium, sheep may suffer from diarrhea and other symptoms, as well as limited growth and performance (Chen et al. 2022; Yang et al. 2021). Cryptosporidiosis in sheep is worthy of attention because of its zoonotic potential and cross-host transmission ability.
Of the over 44 established Cryptosporidium species, the ones that dominantly infect sheep are C. parvum, C. ubiquitum, and C. xiaoi (Chen et al. 2022). C. parvum is the dominant species in European and Australian sheep, whereas C. xiaoi predominates in Asian and African sheep. However, C. ubiquitum appears to be more common in Asia and America (Guo et al. 2021). Subtyping of C. parvum from sheep has identified almost exclusively IIa and IId subtypes with differences across geographic locations or host ages. Host adaptation to C. ubiquitum subtypes is apparent, with subtypes in XIIa found in sheep (Santin 2020).
Cryptosporidium infection is prevalent in sheep worldwide. In China, studies on the epidemiology of Cryptosporidium in sheep were mainly concentrated in the central and eastern regions (Lang et al. 2023; Li et al. 2016; Li et al. 2019b; Mi et al. 2018; Ye et al. 2013; Zhang et al. 2020). Xinjiang Uygur Autonomous Region (hereinafter referred to as Xinjiang) is an important hub connecting the interior of eastern China and Central Asia, with a unique ecological environment, unique climatic conditions, and special dietary habits. Sheep farming has always been an important part of the economy and people’s livelihood in the region. In comparison with surveys of cattle in Xinjiang, epidemiological data of Cryptosporidium from large-scale sheep farms in this area are quite scarce. Therefore, this study aimed to investigate the infection and molecular genetic characteristics of Cryptosporidium on concentrated sheep farms in southern Xinjiang.
Materials and methods
Ethics statement
The protocol in this study was not required to be reviewed and approved by the Animal Ethical Committee.
Fecal sample collection
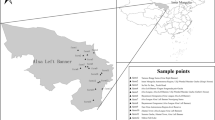
A total of 1252 fresh fecal samples were randomly collected from 10 large-scale sheep farms in southern Xinjiang from May 2021 to August 2022. The sampling locations were distributed in nine cities (Kuqa, Wensu, Wushi, Alaer, Kalpin, Marabishi, Shufu, Qira, and Yutian) in southern Xinjiang (Fig. 1). All the samples were divided into four groups according to host age, including pre-weaned lambs at <3 months old (n = 301), weaned lambs at 3–6 months old (n = 329), fattening sheep at 6–12 months old (n = 327), and adult sheep older than 12 months old (n = 295) (Fig. 1). Most of the stool samples were collected from a single rectum with disposable clean gloves or fresh excreta from individual sheep (especially to avoid contact with the ground) weighing 5–30 g. The marked samples were placed in sealed sampling bags, sent to the laboratory, and stored at a low temperature of 4 °C until detection.
Nucleic acid extraction and PCR amplification
Approximately 200 μg of each fecal sample was selected for nucleic acid extraction using a fecal whole genome extraction kit (E.Z.N.A.® D4015-02, OMEGA Bio-Tek) following the product procedures. The extracted nucleic acid samples were stored at −20 °C.
The small subunit ribosomal (SSU rRNA) gene and the glycoprotein 60 (gp60) gene were detected in Cryptosporidium species and subtypes and were amplified by nested PCR. The primer sequences required for PCR amplification were obtained from others and have been evaluated by peers and were synthesized by Anshengda (Suzhou) Biotechnology Co., Ltd. (Table 1).
For accurate quality control in the assay, positive and negative controls were used for each PCR amplification. The positive controls were derived from the nucleic acids of C. andersoni and C. parvum (subtype IIdA20G1) from cattle. All the controls were identified and stored at the Veterinary Parasitology Laboratory of Tarim University, and the negative control samples were sterilized in double-distilled water. Five microliters of the second PCR amplification products was placed in a 1.5% agarose gel for electrophoresis and then transferred to a gel imaging system for visualization.
Sequencing identification and phylogenetic analysis
A DNA sequencing instrument (ABI PRISMTM3730 XL DNA Analyzer) was used to identify the positive samples, and the sequencing process was commissioned by Ansengda (Suzhou) Biotechnology Co., Ltd. To ensure the accuracy of the sequences, amplicons of positive DNA samples were identified by forward and reverse sequencing. The calibrated sequences were assembled and calibrated by ChromasPro version 1.5 (http://technelysium.com.au/wp/chromaspro/). The SSU rRNA gene sequences of Cryptosporidium obtained in this study were compared with the known sequences in the GenBank database by BLAST (Basic Local Alignment Search Tool) to determine the species or genotype. The gp60 gene sequences obtained from C. parvum and C. ubiquitum were also subjected to the same procedure to determine their subtypes.
The obtained sequences and reference sequences were analyzed by ClustalX 2.1 software (http://www.clustal.org), and a molecular phylogenetic tree was constructed by the general time-reversible model based on the maximum likelihood method in MEGA 7.0 (https://megasoftware.net). The reliability of the phylogenetic tree was tested by bootstrap analysis with 1000 replicates.
Statistical analysis
SPSS (Statistical Product Service Solutions) version 26.0 (IBM Corp.) was used for all statistical analyses. The infection rates and their 95% confidence intervals (CIs) were calculated with the Wald method. Differences in Cryptosporidium infection rates were evaluated with the chi-squared test, and differences were considered significant at P < 0.05.
Results
Cryptosporidium infection of sheep at different farms
Cryptosporidium was identified in 9 out of the 10 sheep farms, with an overall infection rate of 8.0% (100/1252). The infection rate of Cryptosporidium in different farms ranged from 0% (0/120) to 18.40% (23/125), with the highest infection rate being 18.4% (23/125) on farm 9 in Qira (Table 2). In general, the infection rate of Cryptosporidium was significantly different among the farms (χ2 = 45.994, df = 9, P = 0.000).
Cryptosporidium infection of sheep of different ages
The highest infection rate of Cryptosporidium was found in pre-weaned lambs (20.3%, 61/301), followed by weaned lambs (10.3%, 34/329), fattening sheep (0.92%, 3/327), and adult sheep (0.68%, 2/295). The infection rates of Cryptosporidium were significantly different between the four age groups of sheep (χ2 = 107.897, df = 3, P = 0.000) (Table 3).
Distribution of Cryptosporidium species and subtypes
Sequence alignment analysis based on the SSU rRNA gene showed that C. xiaoi (n = 61), C. parvum (n = 22), and C. ubiquitum (n = 17) were identified in the 100 positive samples. Further analysis based on the gp60 gene revealed that C. parvum contained two subtypes, IIdA19G1 (n = 21) and IIdA15G1 (n = 1), while all detected C. ubiquitum belonged to subtype XIIa (n = 17) (Tables 2 and 3).
C. xiaoi was present on all positive farms, but C. ubiquitum and C. parvum were found only on three and four farms, respectively. C. xiaoi was found in all age groups, especially in pre-weaned (n = 29) and weaned (n = 28) lambs, with three positives in fattening sheep and one positive in adult sheep. C. ubiquitum was entirely concentrated in pre-weaned (n = 11) and weaned (n = 6) lambs. C. parvum mainly infected pre-weaned lambs (n = 21), except for one positive occurrence in the adult flock. Notably, the dominant subtype of IIdA19G1 was present in pre-weaned lambs, while one IIdA15G1 was detected in the adult flock (Table 3).
Molecular characterization of Cryptosporidium species and subtypes
Cryptosporidium parvum contained two haplotype sequences, which showed 100% identity with the reference sequence from Camelus dromedarius (MK491509) from Egypt (n = 1) and yak (KP334136) from China (n = 21). Similarly, the two haplotypes contained in C. ubiquitum showed 100% identity with the reference sequence from a bovine (MT044136) from India (n = 16) and 99.79% identity with the reference sequence from a goat (KM199749) from China (n = 1). The dominant haplotype of C. xiaoi showed 100% identity with that from a sheep in Algeria (LC414392) (n = 59), and the other two haplotype sequences were consistent with two goat reference sequences in China (KT235703 and KT235699) (Table 4).
In terms of subtypes, only the C. parvum IIdA15G1 subtype shared the same identity with the reference sequence (MH794167) of sheep from Xinjiang, China. In addition, one isolate of subtype IIdA19G1 had 99.62% identity with the reference sequence of sheep from Shaanxi (KT235713), China, and the others shared 99.09–100% identity with the reference sequence of sheep from Anhui (MH049734), China. All C. ubiquitum XIIa subtypes maintained high identity with reference sequences (MH049733) from sheep from Jiangsu, China (Table 4).
Phylogenetic analysis based on the SSU rRNA gene showed that most isolates of the three Cryptosporidium species identified in this study were clustered together with the ruminant isolates (cattle, sheep, and goat) from China in the genetic evolutionary tree, except for one C. parvum isolate on the same branch as the Egyptian camelid isolate (Fig. 2). Phylogenetic analysis based on the gp60 gene revealed high genetic diversity of C. parvum isolates. There were a variety of haplotypes based on the single nucleotide polymorphisms (SNPs) within subtype IIdA19G1 in this study, and most of them were on independent branches of the genetic evolutionary tree (Fig. 3).
Discussion
The results of this study confirmed that the overall infection rate of Cryptosporidium was 8.0% (100/1252), much higher than that in grazing adult sheep (0.9, 3/318) and lower than that in captive sheep (36.4%, 36/99) in Xinjiang, according to the reports mentioned above (Mi et al. 2018; Qi et al. 2019). It is slightly lower than the total infection rate of sheep in China reported so far (9.6%, 571/5946), and according to the specific analysis of administrative divisions, the infection rate in this study is lower than that reported in East China (11.1%, 134/1215), Northwest China (14.4%, 171/1184), and North China (16.5%, 16.5%). In contrast, it was higher than that reported in South China (4.8%, 82/1701) and Northeast China (4.5%, 25/559) (Guo et al. 2021; Mi et al. 2018; Wang et al. 2022; Yang et al. 2021). Globally, the prevalence of Cryptosporidium infection in sheep was 18.9% (7836/47585), with some heterogeneity between 6 continents, namely, Asia (14.8%), Europe (20.2%), Africa (21.7%), North America (29.8%), South America (20.3%), and Oceania (19.1%) (Chen et al. 2022; Santin 2020). Several factors, including sampling area, sampling time, age distribution of hosts, and detection methods, may explain the differences between the results of this study and those of other reports.
In the current study, the infection rate of Cryptosporidium was negatively and significantly related to sheep age, which was similar to most previous findings for sheep indicating a higher prevalence of Cryptosporidium in lambs than in adult sheep worldwide. A relevant global meta-analysis showed that sheep aged <3 months had a significantly higher prevalence (27.8%, 3284/11938) than those aged 3–12 and >12 months (Chen et al. 2022). Domestically, a survey of Cryptosporidium in sheep from several provinces in China showed that the infection rate of lambs (31.2%, 224/718) was higher than that of adult sheep (22.4%, 71/317) (Mi et al. 2018). The result was consistent with one report in Inner Mongolia that the prevalence in 15- to 16-week-old lambs (weaned) was higher than that in 3- to 4-week-old lambs (pre-weaned) (Ye et al. 2013). It may be that, compared with newborn lambs, older lambs before and after weaning may be more susceptible to Cryptosporidium oocysts secreted by adult sheep or existing in the environment due to lower maternal antibodies, weakened immunity, and weaning stress.
Cryptosporidium xiaoi, C. ubiquitum, and C. parvum were detected in this study, which have been generally recognized as the dominant species infecting sheep (Guo et al. 2021; Santin 2020). Although the majority of sheep studies worldwide have shown the predominance of C. ubiquitum, C. xiaoi, and C. parvum and reported to be more prevalent in European countries (Dessi et al. 2020; Kaupke et al. 2017; Mammeri et al. 2019). In a small-scale study in Poland, C. parvum was found in lambs <4 weeks of age, whereas older hosts were infected with only C. xiaoi (Kaupke et al. 2017). In a longitudinal study conducted in Australia, of the four Cryptosporidium species identified in sheep, C. parvum was detected in lambs of various ages, C. ubiquitum was mostly identified in lambs <2 months of age, C. andersoni was identified in lambs older than 3 months, and C. xiaoi was the only species identified in ewes (Sweeny et al. 2011a). In contrast, the results of this study were somewhat unique and complex, in which C. xiaoi was distributed in sheep of all ages, C. parvum was mostly restricted to pre-weaned lambs, with only one isolate occurring in adult lambs, and C. ubiquitum was found in lambs before and after weaning.
In the current study, C. xiaoi was distributed on all nine positive farms, but C. parvum and C. ubiquitum, which are generally considered to have a wide range of hosts and strong diarrheagenic properties when infecting ruminants and humans, were found only in four and three of the farms, respectively. This was similar to the results of two previous related surveys in Xinjiang (Qi et al. 2019; Mi et al. 2018). The results from Qi et al. 2019 showed that only one of the 15 sheep grazing areas was C. parvum positive. Similarly, in the survey results of Mi et al. in 2018 from two large-scale sheep farms in Xinjiang, only C. xiaoi and C. ubiquitum were found to have high infection rates, while no cases of C. parvum infection were observed. Once colonized on large-scale farms, the species is often not easily eradicated, causing the animals and environment of the positive farms to act as sources for a long time (Cai et al. 2017; Li et al. 2019a; Santin et al. 2008; Sweeny et al. 2011b; Yang et al. 2014). In particular, a 1-year longitudinal survey of newborn calves on two farms in Xinjiang showed that C. parvum infection was basically the same and at a high level all year round, accompanied by the occurrence of herd diarrhoea in calves, suggesting its strong survival and pathogenic ability (Zhang et al. 2022).
All C. ubiquitum isolates in this study were subtyped as XIIa, which was consistent with previous results in sheep from multiple provinces (Qinghai, Beijing, Inner Mongolia, Jilin, Ningxia, Shanghai, and Xinjiang) in China (Li et al. 2019b; Mi et al. 2018) and has also been reported in sheep from Australia, Ghana, and Spain (Ramo et al. 2016; Squire et al. 2017; Yang et al. 2014), suggesting that subtype XIIa of C. ubiquitum is the most common subtype in sheep worldwide. C. parvum subtypes IIdA15G1 (n = 1) and IIdA19G1 (n = 21) were identified in this study which was reported in previous studies to cause large-scale outbreaks of diarrheal disease in newborn calves, resulting in mass mortality in China (Guo et al. 2021; Li et al. 2019a). To date, only one case related to IIdA15G1 has been identified in sheep in China, and several IIdA15G1 isolates from sheep were found in Greece (Papanikolopoulou et al. 2018; Qi et al. 2019). Subtype IIdA19G1 was found only in a small number of sheep in eastern China (Shandong and Shanghai) and has not been reported abroad, although it has previously appeared in humans and other animals such as goats and cattle (Guo et al. 2021; Santin 2020). This study also found that the IIdA19G1 subtype could be divided into several haplotypes based on SNPs, indicating that it had intra-subtype genetic diversity.
In conclusion, this is the first large-scale survey of Cryptosporidium in sheep in southern Xinjiang, which confirmed that Cryptosporidium infection was common in sheep on local intensive farms. The infection rate of Cryptosporidium decreased with the age of the host, including the zoonotic subtypes IIdA19G1 and IIdA15G1 of C. parvum and the subtype XIIa of C. ubiquitum.
Data availability
The nucleotide sequences reported in this study have been deposited in the GenBank database at the National Center for Biotechnology Information under accession numbers OR361825-OR361831.
References
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41(6):2744–2747. https://doi.org/10.1128/JCM.41.6.2744-2747.2003
Cai M, Guo Y, Pan B, Li N, Wang X, Tang C, Feng Y, Xiao L (2017) Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol 241:14–19. https://doi.org/10.1016/j.vetpar.2017.05.005
Chen Y, Qin H, Huang J, Li J, Zhang L (2022) The global prevalence of Cryptosporidium in sheep: a systematic review and meta-analysis. Parasitology 149(12):1652–1665. https://doi.org/10.1017/S0031182022001196
Dessi G, Tamponi C, Varcasia A, Sanna G, Pipia AP, Carta S, Salis F, Diaz P, Scala A (2020) Cryptosporidium infections in sheep farms from Italy. Parasitol Res 119(12):4211–4218. https://doi.org/10.1007/s00436-020-06947-2
Guo Y, Li N, Ryan U, Feng Y, Xiao L (2021) Small ruminants and zoonotic cryptosporidiosis. Parasitol Res 120(12):4189–4198. https://doi.org/10.1007/s00436-021-07116-9
Kaupke A, Michalski MM, Rzezutka A (2017) Diversity of Cryptosporidium species occurring in sheep and goat breeds reared in Poland. Parasitol Res 116(3):871–879. https://doi.org/10.1007/s00436-016-5360-3
Lang J, Han H, Dong H, Qin Z, Fu Y, Qin H, Zhang J, Zhao J, Li X, Zhao G, Li J, Zhang L (2023) Molecular characterization and prevalence of Cryptosporidium spp. in sheep and goats in western Inner Mongolia, China. Parasitol Res 122(2):537–545. https://doi.org/10.1007/s00436-022-07756-5
Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L (2019a) Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol 49(7):569–577. https://doi.org/10.1016/j.ijpara.2019.02.006
Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y (2014) Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis 20(2):217–224. https://doi.org/10.3201/eid2002.121797
Li P, Cai J, Cai M, Wu W, Li C, Lei M, Xu H, Feng L, Ma J, Feng Y, Xiao L (2016) Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet Parasitol 215:58–62. https://doi.org/10.1016/j.vetpar.2015.11.009
Li W, Wang K, Tang L, Chen M, Li H, Kan Z, Gu Y (2019b) Molecular characterization of Cryptosporidium species in sheep and goats in Anhui Province and neighboring provinces. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 31(5):474–478. https://doi.org/10.16250/j.32.1374.2018043
Mammeri M, Cartou L, Chevillot A, Thomas M, Julien C, Vallee I, Polack B, Follet J, Adjou KT (2019) First identification of Cryptosporidium parvum zoonotic subtype IIaA15G2R1 in diarrheal lambs in France. Vet Parasitol Reg Stud Reports 18:100355. https://doi.org/10.1016/j.vprsr.2019.100355
Menon VK, Okhuysen PC, Chappell CL, Mahmoud M, Mahmoud M, Meng Q, Doddapaneni H, Vee V, Han Y, Salvi S, Bhamidipati S, Kottapalli K, Weissenberger G, Shen H, Ross MC, Hoffman KL, Cregeen SJ, Muzny DM, Metcalf GA et al (2022) Fully resolved assembly of Cryptosporidium parvum. Gigascience 11. https://doi.org/10.1093/gigascience/giac010
Mi R, Wang X, Huang Y, Mu G, Zhang Y, Jia H, Zhang X, Yang H, Wang X, Han X, Chen Z (2018) Sheep as a potential source of zoonotic cryptosporidiosis in China. Appl Environ Microbiol 84(18). https://doi.org/10.1128/AEM.00868-18
Papanikolopoulou V, Baroudi D, Guo Y, Wang Y, Papadopoulos E, Lafi SQ, Abd El-Tawab MM, Diakou A, Giadinis ND, Feng Y, Xiao L (2018) Genotypes and subtypes of Cryptosporidium spp. in diarrheic lambs and goat kids in northern Greece. Parasitol Int 67(4):472–475. https://doi.org/10.1016/j.parint.2018.04.007
Qi M, Zhang Z, Zhao A, Jing B, Guan G, Luo J, Zhang L (2019) Distribution and molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi amongst grazing adult sheep in Xinjiang, China. Parasitol Int 71:80–86. https://doi.org/10.1016/j.parint.2019.04.006
Ramo A, Monteagudo LV, Del Cacho E, Sanchez-Acedo C, Quilez J (2016) Intra-species genetic diversity and clonal structure of Cryptosporidium parvum in sheep farms in a confined geographical area in northeastern Spain. PLoS One 11(5):e0155336. https://doi.org/10.1371/journal.pone.0155336
Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlasek I (2003) Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microbiol 69(7):4302–4307. https://doi.org/10.1128/AEM.69.7.4302-4307.2003
Santin M (2020) Cryptosporidium and Giardia in Ruminants. Vet Clin North Am Food Anim Pract 36(1):223–238. https://doi.org/10.1016/j.cvfa.2019.11.005
Santin M, Trout JM, Fayer R (2008) A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol 155(1-2):15–23. https://doi.org/10.1016/j.vetpar.2008.04.018
Squire SA, Yang R, Robertson I, Ayi I, Ryan U (2017) Molecular characterization of Cryptosporidium and Giardia in farmers and their ruminant livestock from the Coastal Savannah zone of Ghana. Infect Genet Evol 55:236–243. https://doi.org/10.1016/j.meegid.2017.09.025
Sweeny JP, Ryan UM, Robertson ID, Jacobson C (2011a) Cryptosporidium and Giardia associated with reduced lamb carcase productivity. Vet Parasitol 182(2-4):127–139. https://doi.org/10.1016/j.vetpar.2011.05.050
Sweeny JP, Ryan UM, Robertson ID, Yang R, Bell K, Jacobson C (2011b) Longitudinal investigation of protozoan parasites in meat lamb farms in southern Western Australia. Prev Vet Med 101(3-4):192–203. https://doi.org/10.1016/j.prevetmed.2011.05.016
Wang P, Zheng L, Liu L, Yu F, Jian Y, Wang R, Zhang S, Zhang L, Ning C, Jian F (2022) Genotyping of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from sheep and goats in China. BMC Vet Res 18(1):361. https://doi.org/10.1186/s12917-022-03447-6
Yang R, Jacobson C, Gardner G, Carmichael I, Campbell AJ, Ng-Hublin J, Ryan U (2014) Longitudinal prevalence, oocyst shedding and molecular characterisation of Cryptosporidium species in sheep across four states in Australia. Vet Parasitol 200(1-2):50–58. https://doi.org/10.1016/j.vetpar.2013.11.014
Yang X, Gong Q, Zhao B, Cai Y, Zhao Q (2021) Prevalence of Cryptosporidium infection in sheep and goat flocks in China during 2010-2019: a systematic review and meta-analysis. Vector Borne Zoonotic Dis 21(9):692–706. https://doi.org/10.1089/vbz.2020.2713
Ye J, Xiao L, Wang Y, Wang L, Amer S, Roellig DM, Guo Y, Feng Y (2013) Periparturient transmission of Cryptosporidium xiaoi from ewes to lambs. Vet Parasitol 197(3-4):627–633. https://doi.org/10.1016/j.vetpar.2013.07.021
Zhang K, Wu Y, Jing B, Xu C, Chen Y, Yu F, Wei Z, Zhang Y, Cui Z, Qi M, Zhang L (2022) Seasonal monitoring of Cryptosporidium species and their genetic diversity in neonatal calves on two large-scale farms in Xinjiang, China. J Eukaryot Microbiol 69(2):e12878. https://doi.org/10.1111/jeu.12878
Zhang Z, Chen D, Zou Y, Hou J, Sun L, Li Z, Yang J, Zou F, Zhu X (2020) First report of Cryptosporidium spp. infection and risk factors in black-boned goats and black-boned sheep in China. Parasitol Res 119(9):2813–2819. https://doi.org/10.1007/s00436-020-06781-6
Funding
This work was supported by the Program for Young and Middle-aged Leading Science, Technology, and Innovation of the Xinjiang Production & Construction Group (2018CB034) and the Open Project Fund State Key Laboratory of Sheep Genetic Improvement and Healthy Production (MYSKLKF202004).
Author information
Authors and Affiliations
Contributions
Zhengrong Wang, Xia Peng, Xinwen Bo and Bowen Zhang collected the faecal samples. Zhengrong Wang, Yanyan Zhang, Fuchang Yu and Aiyun Zhao carried out the PCR assays and sequence analyses. Zhenjie Zhang and Meng Qi designed the study and drafted the current manuscript.
Corresponding authors
Ethics declarations
Consent to participate
Not applicable.
Consent for publication
All the authors consent to publication of this article.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Lihua Xiao
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Peng, X., Bo, X. et al. Molecular evaluation of Cryptosporidium spp. in sheep in southern Xinjiang, China. Parasitol Res 122, 2989–2997 (2023). https://doi.org/10.1007/s00436-023-07988-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07988-z