Abstract
Cryptosporidium spp. are zoonotic intestinal parasites that infect fish, birds, reptiles and mammals. Cryptosporidium spp. are common cause of diarrhea. In this study, a total of 1032 fecal samples were collected from the rectums of sheep and goats. The samples were analyzed using nested polymerase chain reaction (nested PCR) based on the small subunit ribosomal RNA (SSU rRNA) gene of Cryptosporidium spp. The average infection rate of Cryptosporidium spp. was 2.23% (n = 23), and three Cryptosporidium species were identified, namely Cryptosporidium ubiquitum (8/23), Cryptosporidium andersoni (5/23) and Cryptosporidium xiaoi (10/23). Subtyping of C. ubiquitum and C. xiaoi was carried out by DNA sequence analysis of the 60-kDa glycoprotein (gp60) gene. Eight C. ubiquitum isolates were identified as zoonotic subtype XIIa. Nine C. xiaoi isolates were identified as subtypes XXIIIc (n = 1), XXIIIf (n = 3) and XXIIIg (n = 5). Subtype XXIIIg was first found in Chinese sheep. C. ubiquitum subtype XIIa was found in both sheep and goats, suggesting that sheep and goats are important sources of C. ubiquitum infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium spp. are zoonotic intestinal parasites that infect fish (Zahedi et al. 2021), birds (Holubová et al. 2019), reptiles (Jezkova et al. 2016), and mammals (Ryan et al. 2021), it spreads through fecal–oral transmission or by ingestion of contaminated food or water, and Cryptosporidium spp. are common cause of diarrhea in sheep and goats (Santín 2013). A total of 44 Cryptosporidium species and more than 120 genotypes have been described in various animals (Ryan et al. 2021).
Cryptosporidium parvum is one of the most widely transmitted zoonotic species, and it has major public health importance. Sheep and goats are important hosts of C. parvum, and they can also be infected by Cryptosporidium ubiquitum, Cryptosporidium hominis, Cryptosporidium xiaoi, Cryptosporidium andersoni, Cryptosporidium scrofarum and more than a dozen of other Cryptosporidium species (Fiuza et al. 2011; Zhang et al. 2020). Cryptosporidium ubiquitum, C. xiaoi, and C. parvum were commonly found in sheep and goats. However, in different countries or regions, the dominant species or distributions of Cryptosporidium spp. may vary. For example, in Kuwait (Majeed et al. 2018), Spain (Díaz et al. 2015), and Italy (Dessì et al. 2020), the infection rate of C. parvum was higher than C. ubiquitum and C. xiaoi, which were the most common Cryptosporidium species in Ningxia (Yang 2018), Anhui (Li et al. 2019) Xinjiang, Beijing (Mi et al. 2018) and other regions of China. In addition, C. hominis, C. parvum and C. scrofarum have also been detected in sheep and goats within Papua New Guinea (Koinari et al. 2014).
Inner Mongolia is located on the northern frontier of the People’s Republic of China, with an average altitude of about 1000 m. The region encompasses 1,177,500 sq. km with varied vegetation patterns. Sheep farming is an important component of its economic development. Cryptosporidium is an important cause of lamb diarrhea (Fan et al. 2021), and it has caused economic losses to the sheep industry (Scallan et al. 2011). There are few studies of Cryptosporidium infection in sheep and goats within Inner Mongolia. Therefore, we studied the species and distribution of Cryptosporidium in Alxa League, Inner Mongolia, to better understand the zoonotic potential of Cryptosporidium in sheep and goats, and to help reduce the economic losses caused by Cryptosporidium infection.
Materials and methods
Sample collection
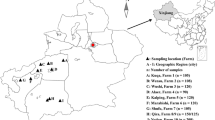
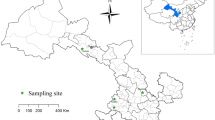
A total of 1032 fecal samples were collected between October 2019 and December 2020 from 16 sheep and goats farm in Alxa, Inner Mongolia (Fig. 1). Rectal sampling was performed directly with a sterile swab, samples were put in clean sampling bags. Sample number, sampling time and place, animal species (sheep or goats), feeding models, age patterns, animal health status (diarrhea or not, the sampled animals were mostly healthy and no obvious phenomenon of diarrhea) and other important information were recorded. The samples were stored in an ice chest and returned to the laboratory of veterinary parasitology, at Henan Agricultural University. We took 0.5–1 g of sample in a 2 ml centrifuge tube, and used this for fecal DNA extraction. We then added feces to 2 ml centrifuge tubes, added 2.5% potassium dichromate, and stored the processed samples at 4℃ in refrigerators.
Map of the sampling locations in Inner Mongolia, China. The figure was originally designed using ArcGIS 10.2 software. The original vector diagram imported in ArcGIS was adapted from Natural Earth (http://www.naturalearthdata.com)
DNA extraction and PCR amplification
The genomic DNA was extracted from the fecal pellets with the E.Z.N.A Stool DNA Kit (Omefa Biotek Inc, Norcross, GA, USA), according to manufacturer’s instructions. The extracted DNA was kept at − 20 °C before it was used for molecular analysis.
Cryptosporidium spp. were identified by nested PCR based on the small subunit (SSU) rRNA gene and gp60 gene (Alves et al. 2003). The primer sequences used were chosen according to previous studies (Xiao et al. 2001; Li et al. 2014; Fan et al. 2021). The products of the secondary PCR were detected using 1% agarose gel electrophoresis containing DNAGREEN (Tiandz, Inc., Beijing, China). An Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) was used to amplify the Cryptosporidium SSU rRNA gene. This was achieved in 25 μl volumes, including 1 μl template DNA or primary PCR product, 2.5 μl 10× KOD-Plus PCR buffer, 2.5 μl dNTPs (2 nM), 1.5 μl MgSO4 (25 nM), 0.5 μl of each primer (25 nM), 16 μl double distilled water, and 0.5 μl KOD-Plus amplification enzyme (1 unit/μl) (ToYoBo Co., Ltd., Osaka, Japan). A total of 35 cycles were carried out; each of these consisted of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 1 min (90 s for C. ubiquitum and C. xiaoi gp60 gene). There was also an initial hot start at 94 °C for 5 min and a final extension at 72 °C for 7 min (10 min for C. ubiquitum and C. xiaoi gp60 gene). The secondary cycling conditions were identical to those used in the primary PCR. Both the positive and negative controls were included in each PCR amplification, positive controls were the genomic DNA of sheep fecal samples, these samples were PCR-positive for Cryptosporidium spp., negative controls were 24 μl volumes without 1 μl template DNA.
Sequence analysis
The secondary PCR products of the SSU rRNA gene were sequenced bidirectionally by SinoGenoMax Biotechnology CO., Ltd (Beijing, China). Sequence accuracy was confirmed by two-directional sequencing and by sequencing a new PCR product if necessary.
To infer the phylogenetic relationships of the detected samples, neighbor-joining (NJ) trees were constructed with the MEGA 7.0 software (http://www.megasoftware.net) based on evolutionary distances calculated with the Kimura 2-parameter model. The reliability of these trees was assessed with a bootstrap analysis of 1000 replicates.
Statistical analysis
The prevalence of parasitic infections, with a 95% confidence interval (CI), was calculated. The chi-square test was used to compare differences in infection rates between different age groups and clinical symptoms. A two-tailed p-value < 0.05 was considered statistically significant.
Results
Prevalence of Cryptosporidium species
A total of 1032 fecal samples (491samples of sheep, 541samples of goats) were collected from 16 sheep and goat farms. Of the total, 23 specimens were PCR-positive for Cryptosporidium spp., and the overall infection rate was 2.23%. The highest infection rate (20.00%, 4/20) was detected on site 1, followed by site 8 (7.84%, 4/51), site 13 (7.14%, 3/42) and site 5 (7.00%, 4/57), and the infection rates in site 7, site 3, site 2 and site 11 were 4.76% (n = 1), 3.30% (n = 1), 2.92% (n = 4), 1.11% (n = 2) respectively. There was no Cryptosporidium detected in the other 8 farms (Table 1).
Distribution and subtype of Cryptosporidium
Twenty-three isolates were identified as three Cryptosporidium species: C. andersoni, C. xiaoi and C. ubiquitum through DNA sequence analysis of the SSU rRNA gene in Cryptosporidium spp., and C. xiaoi was the predominant species. It accounted for 43.47% of all Cryptosporidium positive specimens, and it was detected in four farms. C. ubiquitum and C. andersoni were detected in three farms, respectively. In the analysis of subtypes, the C. ubiquitum isolates were all identified as subtype family XIIa, and nine C. xiaoi isolates were identified as subtypes XXIIIc, XXIIIf and XXIIIg (Table 1).
Sequence analysis
From the Cryptosporidium SSU rRNAgene, eight isolates shared 100% homology to Indian cattle C. ubiquitum isolate, Chinese sheep C. ubiquitum isolate and Pacific Northwest mountain beaver isolate (GenBank Accession NO.MT044147, MH059802, MT524974). Ten isolates shared 100% homology to Chinese sheep and goat isolates (GenBank Accession NO.MH049731, MG 602,953), and five isolates shared 100% homology to Bangladeshi calf C. andersoni isolate (GenBank Accession NO.MK982465).
Nine C. xiaoi isolates and five C. ubiquitum isolates were successfully identified at the gp60 locus: three isolates from site 1, one isolate from site 3, one isolate from site 2, one isolate from site 3, one isolate from site 7, and four isolates from site 8. Five isolates belonged to subtype XXIIIg, three isolates belonged to subtype XXIIIf, and one isolate belonged to subtype XXIIIc. Three C. xiaoi XXIIIg subtype isolates were found in this study, and they have 10 single nucleotide polymorphisms (SNP) in comparison with Chinese goat XXIIIg subtype isolate (GenBank Accession NO. MW815228), and two C. xiaoi XXIIIg subtype isolates found in this study have only one nucleotide difference in comparison with Chinese goat XXIIIg subtype isolates (GenBank Accession NO.MW815228). The C. xiaoi XXIIIc and XXIIIf subtypes isolates found in this study shared 100% homology to Chinese goat XXIIIc and XXIIIf subtypes isolates, respectively (GenBank Accession NOs.MW815204 and MW815225).
All of the eight C. ubiquitum sequences belonged to subtype XIIa, which shared 100% homology to Chinese goat, sheep and Czechic Mustela vison (GenBank Accession NOs.KM199742, MH049733 and KY596689), and one nucleotide difference in comparison with the polish Chinchilla lanigera XIIa subtype isolate, Swedish Homo sapiens XIIa-1 subtype isolate, and Czechic Republic Struthio camelus (GenBank Accession NOs.KY596686, KU852740 and MN973963).
Correlation analysis
Four Cryptosporidium spp. positive samples were detected in 491 sheep, and 19 Cryptosporidium spp. positive samples were detected in 541 goats. Cryptosporidium ubiquitum and C. xiaoi were detected as positive in both sheep and goats, while C. andersoni was only found in goats (Table 1).
The Cryptosporidium spp. infection rate of goats (3.51%, 19/541) was higher than the infection rate in sheep (0.81%, 4/491), and the infection rates in goats and sheep were significantly different (χ2 = 8.594, p = 0.003) (Table 2).
A total of 873 fecal samples were collected from pastured sheep and goats, and 19 Cryptosporidium spp. positive samples were detected. A total of 159 fecal samples were collected from captive sheep and goats, and four Cryptosporidium spp. positive samples were detected (Table 2).
The Cryptosporidium spp. infection rate of pastured animals (2.18%, 19/873) was lower than the infection rate of captive animals (2.52%, 4/159), but the Cryptosporidium spp. infection rates in pasture and captive animals were not significantly different (χ2 = 0.071, p = 0.790) (Table 2). Considering the ages of sheep and goats, 969 samples were collected from adult sheep and goats, and 23 Cryptosporidium spp. positive samples were found. Among the 63 samples collected from lambs, Cryptosporidium spp. positive samples were not detected (Table 1). The Cryptosporidium spp. infection rate of adult sheep and goats (2.37%, 23/969) was higher than the infection rate of lambs (0, 0/63). The Cryptosporidium spp. infection rates of two age groups were not significantly different (χ2 = 1.529, p = 0.216) (Table 2).
A total of 63 samples were collected in summer, 289 samples were collected in autumn, and 650 samples were collected in winter. The Cryptosporidium spp. infection rate in summer (4.63%, 5/93) was higher than the rates in autumn (3.80%, 13/289) and winter (0.77%, 5/650). The Cryptosporidium spp. infection rates in different seasons were significantly different (χ2 = 14.796, p = 0.001) (Table 1 and 3).
Discussion
In this study, the overall infection rate of Cryptosporidium spp. was 2.23%, which was lower than the infection rate previously reported for Cryptosporidium spp. in sheep and goats, such as Spain (Díaz et al. 2015) (37.72%,109/289), and Turkey (Kabir et al. 2020) (25.6%,106/415), and higher than the infection rate in Italy (Dessì et al. 2020) (1.64%,15/915) and Brazil (Fiuza et al. 2011) (1.60%,2/125). In China, some higher infection rates of Cryptosporidium spp. in sheep and goats have been reported in previous studies of Shandong (Zhu et al. 2018) (6.76%, 15/222), Ningxia (Yang 2018) (28.33%, 136/480), and Sichuan (Zhong et al. 2018) (4.7%, 16/342).
Various Cryptosporidium species were found in sheep and goats in previous studies, including C. xiaoi, C. ubiquitum, C. andersoni, C. parvum, C. bovis, and C. hominis, and the distribution of Cryptosporidium spp. in sheep and goats was related to geographic locations. In sheep, from China, the dominant Cryptosporidium species were previously reported to be C. ubiquitum and C. xiaoi in Henan (Wang et al. 2010), Sichuan (Zhong et al. 2018), Anhui (Li et al. 2019), Inner Mongolia (Ye et al. 2013), and Ningxia (Yang 2018). The dominant Cryptosporidium species was C. parvum in Italy (Dessì et al. 2020), Spain (Díaz et al. 2015), the UK (Pritchard et al. 2007), and most other countries. However, in goats, from China, the dominant Cryptosporidium species were previously reported to be C. ubiquitum and C. xiaoi (Wang et al. 2010; Yang 2018). In contrast, the dominant species in most other countries were C. parvum. In this study, C. ubiquitum was the dominant Cryptosporidium species in sheep, and C. xiaoi was the dominant species in goats. The results were similar to the results of previous studies in most regions of China, and different from the reports in most of other countries. A previous study (Ye et al. 2013), presented results that were partly identical to the results in this study, and no C. parvum was detected in this study. The difference in distribution of Cryptosporidium species between China and other countries may be attributed to a variety of factors: although Hulunbeier and Alxa were in the same autonomous region, they are exactly distant, and the time of sample collection were not identical in the two studies, or different feeding models and so on.
Cryptosporidium parvum has been detected in sheep and goats of lots of other countries and in some regions of China, such as Spain (Díaz et al. 2015), the UK (Pritchard et al. 2007), and India (Maurya et al. 2013), and Anhui (Li et al. 2019), Ningxia (Yang 2018) of China (Table 3 and Table 4). In addition, C. andersoni, C. scrofarum, C. hominis and C. rat genotype II were found in China and other countries, but their dominant hosts are not sheep and goats. Therefore, the infections of these Cryptosporidium species in sheep and goats may result from sharing feeding grounds with other animals or farmers (Koinari et al. 2014).
In this study, Cryptosporidium spp. in sheep and goats showed seasonal variation, C. xiaoi was only detected in summer and C. ubiquitum was only detected in winter. Cryptosporidium andersoni, C. xiaoi and C. ubiquitum were detected in autumn. The infection rate of Cryptosporidium spp. in summer was the highest and that in winter was the lowest among the three seasons, which was similar to the results reported in Henan (Wang et al. 2010), Shandong (Zhu et al. 2018), Jilin and Liaoning (Mi et al. 2014). In this study, the infection rate of Cryptosporidium spp. in summer was 6.4%, and there was no infection in winter. In India, the infection rate reached to the highest in post-monsoon (Maurya et al. 2013). However, in Anhui (Li et al. 2019), Chongqing (Wang et al. 2014), Henan (Wang et al. 2010), and Inner Mongolia (Ye et al. 2013), the infection rate of Cryptosporidium spp. in autumn was higher than in other seasons, and there was no infection in summer. The variation illustrated in the results suggested that the infection of Cryptosporidium spp. was not directly related to different seasons.
In this study, the Cryptosporidium spp. infection rate of captive sheep and goats (2.52%) was slightly higher than that of pastoral sheep and goats (2.18%), and there was no significant difference between the two feeding models. The results were similar to those of the other nine regions (Table 4), which were probably correlated to the sanitary conditions and breeding density of sheep and goats in sheepfolds. In addition, the Cryptosporidium spp. infection rate of pastoral sheep and goats was higher in autumn and winter, because of the lack of forage grass (Gao et al. 2021).
In addition, different age patterns were related to the infection rate of Cryptosporidium spp. In this study, no Cryptosporidium spp. was detected in lambs and kid goats, and all 23 Cryptosporidium spp. positive samples were collected from adult sheep and goats. The infection rate of Cryptosporidium spp. (2.37%) in adult sheep and goats was lower than the infection rate of most provinces in China, and some other countries (Table 3 and Table 4). In previous studies, the infection rate of Cryptosporidium spp. declined with the increased age of sheep and goats. This finding was not consistent with the result of this study, probably related to the different breeding density, environments and seasons, or because the number of samples of lambs and kid goats was fewer than samples of adult goats and sheep (Table 2).
Cryptosporidium ubiquitum was named as cervine genotype several years ago, because of its great distribution range and various host species. C. ubiquitum has been detected in China, the USA, the UK, Canada, Spain and many other countries (Blanco et al. 2016). In previous studies, the nucleotide sequence of gp60 gene of C. ubiquitum formed six subtype families: XIIa to XIIf (Li et al. 2014). In this study, the eight C. ubiquitum isolates all belonged to subtype XIIa, which has been detected in ruminants, humans and other animals. A total of 59 C. ubiquitum infection cases in humans were all caused by subtypes XIIa to XIId, and mainly by XIIa. In the USA, the UK, Canada, Australia and Wales, C. ubiquitum subtypes XIIb and XIId were detected in water (Li et al. 2014). However, this may not entirely indicate the reason of C. ubiquitum infections in humans, which were caused by C. ubiquitum susceptible animals or contaminated water, because C. ubiquitum subtypes XIIa to XIId can also be found in other animals.
Cryptosporidium xiaoi was commonly found in sheep and goats, in previous studies, C. xiaoi was found in Papua New Guinea (Koinari et al. 2014), Kuwait (Majeed et al. 2018), and Spain (Díaz et al. 2015). In China, C. xiaoi was the dominant Cryptosporidium species in sheep of Guangdong, Hubei, Shandong and Shanghai (Mi et al. 2014). It was the dominant Cryptosporidium species of goats in Inner Mongolia (Ye et al. 2013), Anhui (Li et al. 2019), Ningxia (Yang 2018) and Qinghai (Li et al. 2016). The distribution of C. xiaoi subtypes was correlated with different hosts. In previous studies. C. xiaoi subtype families, XXIIIa, XXIIIc, XXIIIg and XXIIIj, were only detected in goats; the other eight subtype families were found in both goats and sheep (Fan et al. 2021). In this study, nine C. xiaoi isolates were successfully subtyped, and all belonged to XXIIIf (n = 3), XXIIIg (n = 5) and XXIIIc (n = 1) subtypes. The three subtypes were found in sheep and goats, which differed from the results of previous studies, subtype family XXIIIg was only found in sheep. In this study, geographic locations were not a significant factor to the distribution of C. xiaoi subtypes. In addition, C. xiaoi was found in two AIDS patients in Ethiopia, suggesting that C. xiaoi presented a zoonotic risk (Adamu et al. 2014).
Cryptosporidium andersoni has a wide host range, cattle and Bactrian camels being major hosts, sheep and goats being minor hosts (Xiao et al. 2004). Cryptosporidium andersoni has been found in sheep from Henan Province of China (Wang et al. 2010), and Papua New Guinea (Koinari et al. 2014). It was found in goats in Henan and Sichuan (Zhong et al. 2018) Provinces of China. Cryptosporidium andersoni has been identified in seven sporadic cases (Xiao et al. 2004), with low public health risk, and its clinical symptoms were not obvious.
In conclusion, we detected C. ubiquitum, C. andersoni, and C. xiaoi in sheep and goats from Inner Mongolia, China. Cryptosporidium ubiquitum and C. xiaoi were the dominant Cryptosporidium spp., and C. xiaoi XXIIIc subtype was first detected in sheep. Moreover, C. ubiquitum belonged to the zoonotic XIIa subtype, which has the potential threat for human health in Inner Mongolia. More studies are required to understand the differences in the transmission and the human public health significance of cryptosporidiosis in sheep and goats.
Data availability
All of the data generated and analyzed during this study are included in this published manuscript. The nucleotide sequences of C. xiaoi for the gp60 gene obtained in this study have been deposited in GenBank, GenBank accession numbers: ON809515—ON809517.
References
Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L (2014) Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis 8:e2831. https://doi.org/10.1371/journal.pntd.0002831
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41:2744–2747. https://doi.org/10.1128/jcm.41.6.2744-2747.2003
Blanco MA, de Lucio A, Fuentes I, Carmena D (2016) Cryptosporidium ubiquitum in Venezuela: first report in a paediatric patient with acute diarrhoea. Enferm Infecc Microbiol Clin 34:142–143. https://doi.org/10.1016/j.eimc.2015.05.010
Dessì G, Tamponi C, Varcasia A, Sanna G, Pipia AP, Carta S, Salis F, Díaz P, Scala A (2020) Cryptosporidium infections in sheep farms from Italy. Parasitol Res 119:4211–4218. https://doi.org/10.1007/s00436-020-06947-2
Díaz P, Quílez J, Prieto A, Navarro E, Pérez-Creo A, Fernández G, Panadero R, López C, Díez-Baños P, Morrondo P (2015) Cryptosporidium species and subtype analysis in diarrhoeic pre-weaned lambs and goat kids from north-western Spain. Parasitol Res 114:4099–4105. https://doi.org/10.1007/s00436-015-4639-0
Fan Y, Huang X, Guo S, Yang F, Yang X, Guo Y, Feng Y, Xiao L, Li N (2021) Subtyping Cryptosporidium xiaoi, a common pathogen in sheep and goats. Pathogens 10:800. https://doi.org/10.3390/pathogens10070800
Fiuza VR, Cosendey RI, Frazão-Teixeira E, Santín M, Fayer R, de Oliveira FC (2011) Molecular characterization of Cryptosporidium in Brazilian sheep. Vet Parasitol 175:360–362. https://doi.org/10.1016/j.vetpar.2010.10.036
Gao H, Wang J, Kang X, Wang P, Wu X, Li Y (2021) Prevalence and risk factors of Cryptosporidium in sheep and goat of China. Chinese Journal of Veterinary Science 41:401–06. https://doi.org/10.16303/j.cnki.1005-4545.2021.02.33
Holubová N, Zikmundová V, Limpouchová Z, Sak B, Konečný R, Hlásková L, Rajský D, Kopacz Z, McEvoy J, Kváč M (2019) Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur J Protistol 69:70–87. https://doi.org/10.1016/j.ejop.2019.03.001
Jezkova J, Horcickova M, Hlaskova L, Sak B, Kvetonova D, Novak J, Hofmannova L, McEvoy J, Kvac M (2016) Cryptosporidium testudinis sp. n., Cryptosporidium ducismarci Traversa, 2010 and Cryptosporidium tortoise genotype III (Apicomplexa: Cryptosporidiidae) in tortoises, Folia Parasitol (Praha), 63.https://doi.org/10.14411/fp.2016.035
Kabir MHB, Ceylan O, Ceylan C, Shehata AA, Bando H, Essa MI, Xuan X, Sevinc F, Kato K (2020) Molecular detection of genotypes and subtypes of Cryptosporidium infection in diarrheic calves, lambs, and goat kids from Turkey. Parasitol Int 79:102163. https://doi.org/10.1016/j.parint.2020.102163
Koinari M, Lymbery AJ, Ryan UM (2014) Cryptosporidium species in sheep and goats from Papua New Guinea. Exp Parasitol 141:134–137. https://doi.org/10.1016/j.exppara.2014.03.021
Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y (2014) Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis 20:217–224. https://doi.org/10.3201/eid2002.121797
Li P, Cai J, Cai M, Wu W, Li C, Lei M, Xu H, Feng L, Ma J, Feng Y, Xiao L (2016) Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai. China, Vet Parasitol 215:58–62. https://doi.org/10.1016/j.vetpar.2015.11.009
Li W, Wang K, Tang L, Chen M, Li H, Kan Z, Gu Y (2019) Molecular characterization of Cryptosporidium species in sheep and goats in Anhui Province and neighboring provinces. Chinese J Schistosomiasis Control 31:474–78+85. https://doi.org/10.16250/j.32.1374.2018043
Majeed QAH, El-Azazy OME, Abdou NMI, Al-Aal ZA, El-Kabbany AI, Tahrani LMA, AlAzemi MS, Wang Y, Feng Y, Xiao L (2018) Epidemiological observations on cryptosporidiosis and molecular characterization of Cryptosporidium spp. in sheep and goats in Kuwait. Parasitol Res 117:1631–1636. https://doi.org/10.1007/s00436-018-5847-1
Maurya PS, Rakesh RL, Pradeep B, Kumar S, Kundu K, Garg R, Ram H, Kumar A, Banerjee PS (2013) Prevalence and risk factors associated with Cryptosporidium spp. infection in young domestic livestock in India. Trop Anim Health Prod 45:941–946. https://doi.org/10.1007/s11250-012-0311-1
Mi R, Wang X, Huang Y, Zhou P, Liu Y, Chen Y, Chen J, Zhu W, Chen Z (2014) Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PLoS ONE 9:e111164. https://doi.org/10.1371/journal.pone.0111164
Mi R, Wang X, Huang Y, Mu G, Zhang Y, Jia H, Zhang X, Yang H, Wang X, Han X, Chen Z (2018) Sheep as a potential source of zoonotic cryptosporidiosis in China. Appl Environ Microbiol 84:e00868-18. https://doi.org/10.1128/aem.00868-18
Morgan-Ryan UM, Fall A, Ward LA, Hijjawi N, Sulaiman I, Fayer R, Thompson RC, Olson M, Lal A, Xiao L (2002) Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J Eukaryot Microbiol 49:433–440. https://doi.org/10.1111/j.1550-7408.2002.tb00224.x
Pritchard GC, Marshall JA, Giles M, Chalmers RM, Marshall RN (2007) Cryptosporidium parvum infection in orphan lambs on a farm open to the public. Vet Rec 161:11–14. https://doi.org/10.1136/vr.161.1.11
Ryan UM, Feng Y, Fayer R, Xiao L (2021) Taxonomy and molecular epidemiology of Cryptosporidium and Giardia - a 50 year perspective (1971–2021). Int J Parasitol 51:1099–1119. https://doi.org/10.1016/j.ijpara.2021.08.007
Santín M (2013) Clinical and subclinical infections with Cryptosporidium in animals. N Z Vet J 61:1–10. https://doi.org/10.1080/00480169.2012.731681
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM (2011) Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis 17:7–15. https://doi.org/10.3201/eid1701.p11101
Wang Y, Feng Y, Cui B, Jian F, Ning C, Wang R, Zhang L, Xiao L (2010) Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol Res 106:341–347. https://doi.org/10.1007/s00436-009-1664-x
Wang R, Li G, Cui B, Huang J, Cui Z, Zhang S, Dong H, Yue D, Zhang L, Ning C, Wang M (2014) Prevalence, molecular characterization and zoonotic potential of Cryptosporidium spp. in goats in Henan and Chongqing. China, Exp Parasitol 142:11–16. https://doi.org/10.1016/j.exppara.2014.04.001
Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A (2001) Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol 67:1097–1101. https://doi.org/10.1128/aem.67.3.1097-1101.2001
Xiao L, Fayer R, Ryan U, Upton SJ (2004) Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev 17:72–97. https://doi.org/10.1128/cmr.17.1.72-97.2004
Yang, J (2018) 'Investigation on Cryptosporidium infections of cattle, goats and sheep in part areas of Ningxia', Northwest A&F University
Ye J, Xiao L, Wang Y, Wang L, Amer S, Roellig DM, Guo Y, Feng Y (2013) Periparturient transmission of Cryptosporidium xiaoi from ewes to lambs. Vet Parasitol 197:627–633. https://doi.org/10.1016/j.vetpar.2013.07.021
Zahedi A, Bolland SJ, Oskam CL, Ryan U (2021) Cryptosporidium abrahamseni n sp (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp Parasitol 223:108089. https://doi.org/10.1016/j.exppara.2021.108089
Zhang Z, Chen D, Zou Y, Hou J, Sun L, Li Z, Yang J, Zou F, Zhu X (2020) First report of Cryptosporidium spp. infection and risk factors in black-boned goats and black-boned sheep in China. Parasitol Res 119:2813–2819. https://doi.org/10.1007/s00436-020-06781-6
Zhong Z, Tu R, Ou H, Yan G, Dan J, Xiao Q, Wang Y, Cao S, Shen L, Deng J, Zuo Z, Ma X, Zhou Z, Liu H, Yu S, Ren Z, Hu Y, Peng G (2018) Occurrence and genetic characterization of Giardia duodenalis and Cryptosporidium spp from adult goats in Sichuan Province. China. PLoS One 13:e0199325. https://doi.org/10.1371/journal.pone.0199325
Zhu W, Mi R, Wang J, Wang C, Huang Y, Zhang H, Jia H, Zhang X, Han X, Chen Z (2018) Detection and actin sequence analysis of Cryptosporidium infection in sheep and goats in Tengzhou Shandong Province. Chinese J Animal Infect Dis 26:69–75. https://doi.org/10.1128/AEM.00868-18
Acknowledgements
We thank Let Pub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (U1904203), and the Leading talents of the Thousand Talents Program of Central China (19CZ0122). The sponsors played no role in study design, in the collection, analysis, or interpretation of the data, in writing the report, or in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Longxian Zhang contributed to the conception and design of the experiments. Jiashu Lang, Han Han and Heping Dong performed the experiments. Ziyang Qin and Huikai Qin helped in interpretation of data. Yin Fu and Junchen Zhang collected fecal samples. Junqiang Li, Xiaoying Li, Guanghui Zhao and Jinfeng Zhao interpreted the results and drafted the manuscript. All of the authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. The research protocol was reviewed and approved by the Research Ethics Committee of Henan Agricultural University. Permission was obtained from farm owners before the collection of animal fecal samples.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Yaoyu Feng
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key findings
• The infection rate of Cryptosporidium spp. was 2.23% in sheep and goats.
• Cryptosporidium ubiquitum, Cryptosporidium andersoni and Cryptosporidium xiaoi were identified in sheep and goats.
• Cryptosporidium ubiquitum subtype XIIa, and C. xiaoi subtypes XXIIIc, XXIIIf and XXIIIg were found in this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lang, J., Han, H., Dong, H. et al. Molecular characterization and prevalence of Cryptosporidium spp. in sheep and goats in western Inner Mongolia, China. Parasitol Res 122, 537–545 (2023). https://doi.org/10.1007/s00436-022-07756-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07756-5