Abstract
Enterocytozoon bieneusi is the most common microsporidia in humans worldwide, in addition to infecting a wide range of animals. However, there is limited information about this pathogen in children in Egypt. Here, we carried out a molecular epidemiological study of E. bieneusi in child care centers in three provinces in Egypt. Altogether, 585 fresh fecal samples were collected from children attending 18 child care centers in El-Dakahlia, El-Gharbia, and Damietta provinces in Northeast Egypt during March 2015 to April 2016. PCR and sequence analyses of the ribosomal internal transcribed spacer (ITS) were used to detect and genotype E. bieneusi. Twenty-seven fecal samples (4.6%, 27/585) were positive for E. bieneusi. Five genotypes were identified, including type IV (n = 13), Peru8 (n = 9), Peru6 (n = 2), Peru11 (n = 2), and D (n = 1). Phylogenetic analysis indicated that the five genotypes of E. bieneusi detected in this study were clustered into zoonotic group 1. These data provide important information on the prevalence and genetic diversity of E. bieneusi in children in this country. Further epidemiological studies should be conducted to elucidate the role of zoonotic transmission in human E. bieneusi infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microsporidia encompass a special group of obligate intracellular protists that are genetically related to fungi and commonly found in the environment with a broad range of hosts, including invertebrates and vertebrates (Vávra and Lukeš 2013; Stentiford et al. 2016). Among the nearly 1700 known microsporidia species in over 200 genera, 17 have been reported to cause human infections (Han et al. 2021). Of these, Enterocytozoon bieneusi most commonly infects humans worldwide, with most infections limited to the gastrointestinal tract (Li et al. 2022). In immunocompetent individuals, infections are often asymptomatic or cause self-limiting diarrhea (Santin 2015). Symptomatic infection occurs most commonly in patients with AIDS and other immunocompromised individuals such as organ transplant recipients and cancer patients, in whom it presents usually with diarrhea, which can lead to weight loss and wasting syndrome (Matos et al. 2012; Weiss and Becnel 2014; Li et al. 2019b). E. bieneusi infections in children with and without diarrhea have been reported in many countries around the world (Wang et al. 2017; Ding et al. 2018; Huibers et al. 2018; Rogawski et al. 2018; Shen et al. 2020; Muadica et al. 2020; Qi et al. 2020).
The transmission of E. bieneusi is thought to occur through the fecal–oral route by ingestion of food and water contaminated with spores or direct contact with infected humans and animals. Thus, the pathogen can be either anthroponotic or zoonotic in origin (Li and Xiao 2021). Genotyping tools have been used to determine the contribution of anthroponotic and zoonotic transmission in human microsporidiosis caused by E. bieneusi in various geographic and socioeconomic settings. This is mostly done through sequence analysis of the polymorphic ribosomal internal transcribed spacer (ITS) (Li et al. 2019a). Over 500 genotypes have been identified based on nucleotide sequence differences at this genetic locus. Phylogenetically, genotypes belong to 11 distinct groups with different host ranges (Li et al. 2019b; Zhang et al. 2021). Humans are infected with over 100 genotypes, most of which belong to group 1, and half of which have been found in animals (Li et al. 2019b). Some genotypes in groups 1 and 2 cause infections in a broad range of mammalian species, including humans, and thus are suspected to pose a major zoonotic risk. In contrast, genotypes in groups 3–11 are mostly found in specific groups of hosts, with reduced zoonotic potential (Li et al. 2019a).
Despite an increase in epidemiological studies in the last decade on the genetic diversity of E. bieneusi in humans around the world (Muadica et al. 2020; Li et al. 2022), data on the molecular epidemiology of E. bieneusi in humans in most African countries, including Egypt, remains limited. A recent microscopy-based study in Egypt showed a high prevalence (29%) of microsporidia, including E. bieneusi, in children with leukemia (Shehab et al. 2021). Unfortunately, no genotyping of E. bieneusi was conducted. Therefore, the present study aimed to examine the prevalence and genotypic identity of E. bieneusi in healthy children aged ≤ 3–8 years in several areas in Egypt.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of the Faculty of Veterinary Medicine, Mansoura University, Egypt. All parents or guardians of the children were informed of the objective of the survey and were asked to voluntarily participate in the study. Informed consent was obtained from the legal guardians of the participants before the sample and data collection.
Sample collections
This study was carried out in 18 child care centers in three provinces (El-Dakahlia, El-Gharbia, and Damietta) in Northeast Egypt during March 2015 to April 2016, as previously described (Naguib et al. 2018). Altogether, 585 children aged ≤ 3–8 years participated in the study. A stool sample was collected from each child in a sterile plastic box and transported to the laboratory in coolers. Information on age, gender, diarrhea and health status, animal contact history, and residence of the study children was collected from parents or guardians using a structured questionnaire. The samples were stored in 70% ethanol at 4 °C prior to molecular analyses.
DNA extraction and PCR amplification
The stool samples were washed twice with distilled water by centrifugation to remove ethanol. Genomic DNA was extracted from 0.2 g of fecal material using the FastDNA SPIN Kit for Soil (MP Biomedicals, Irvine, CA) and manufacturer-recommended procedures. The DNA was eluted with 100 µl of reagent-grade water and stored at − 20 °C prior to molecular analysis. E. bieneusi in the samples was detected by nested PCR analysis of a ∼392-bp fragment containing the entire ITS and parts of the small and large subunits of the rRNA gene as described (Sulaiman et al. 2003). Each PCR analysis was conducted in duplicate, using DNA of the PtEbIX genotype of E. bieneusi from a dog as the positive control and reagent-grade water as the negative control.
Sequence analyses
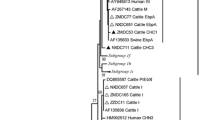
All positive secondary PCR products generated in the study were purified using Montage Filter PCR (Millipore, Bedford, MA) and bidirectionally sequenced using the secondary PCR primers. The DNA sequencing was conducted on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA) using the ABI BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems). The nucleotide sequences obtained were assembled and edited using the ChromasPro software version 1.5 (www.technelysium.com.au/ChromasPro.html). They were aligned with reference sequences downloaded from GenBank using ClustalX software (http://www.clustal.org/) to determine the genotype identity of E. bieneusi. The maximum likelihood algorithm implemented in MEGA 7.0.26 (www.megasoftware.net) was used to evaluate the phylogenetic relationship between the E. bieneusi genotypes identified in this study and known genotypes based on substitution rates calculated with the general time-reversible model. Bootstrap analysis with 1000 replicates was carried out to assess the reliability of cluster formation in the phylogenetic tree.
Statistical analysis
The Chi-square test was used to compare E. bieneusi occurrence between children of different localities, age groups, gender, residency, and with and without gastrointestinal complaints (diarrhea and abdominal pain) or animal contact history. The statistical analysis was conducted using the SPSS version 20.0 (IBM, Armonk, NY, USA). P-values of < 0.05 were considered significant.
Nucleotide sequence accession numbers
Representative nucleotide sequences generated in this study were deposited in GenBank under accession numbers MZ576250, MZ576253-MZ576260, ON420776, and ON420777.
Results
Prevalence of E. bieneusi
All 585 fecal samples were examined for E. bieneusi by PCR amplification of the ITS region of the rRNA gene. Among them, 27 (4.6%) were PCR-positive for E. bieneusi, which was confirmed by sequence analysis of the PCR products. E. bieneusi was detected in individuals from all three provinces. The infection rate in Damietta (5.6%) was slightly higher than in El-Dakahlia (4.8%) and El-Gharbia (3.7%) provinces (χ2 = 0.672, P = 0.715 for the overall comparison) (Table 1).
Risk factors for E. bieneusi occurrence
E. bieneusi was identified in all ages of children except 7-year-olds (Table 2). The highest infection rate was recorded in children aged 4 years (6.4%), followed by children aged 8 (5.9%), 5 (5.3%), ≤ 3 (4.1%), and 6 (2.9%) years (χ2 = 3.469, P = 0.628 for the overall comparison). The infection rates were 3.8 and 5.4% in girls and boys, respectively (χ2 = 0.849, P = 0.357). The infection rate was slightly higher in children with diarrhea (7.9%) than in those without diarrhea (4%) (χ2 = 2.518, P = 0.113). In addition, children without abdominal pain had a slightly higher infection rate (5.6%) than children with abdominal pain (4%) (χ2 = 0.783, P = 0.376). The infection rate of E. bieneusi was 4.7% in children with animal contact and 4.6% in children without animal contact. Children who live in rural areas had significantly higher infection rates (6%) than those living in urban areas (2.8%) (χ2 = 29.648, P = 0.029).
Genotypes of E. bieneusi
The comparative analysis of nucleotide sequences from the 27 PCR-positive samples led to the identification of five genotypes of E. bieneusi. Among them, type IV and Peru8 were the dominant genotypes, found in 13/27 (48%) and 9/27 (33%) E. bieneusi-positive children, respectively. Other genotypes included Peru6, Peru11, and D, which were detected in 2/27 (7%), 2/27 (7%), and 1/27 (4%) E. bieneusi-positive children, respectively. There were no mixed infections with multiple E. bieneusi genotypes in the study children.
The nucleotide sequences obtained from 13 type IV-positive samples were identical to a GenBank sequence AF242478. Among the sequences from nine Peru8-positive samples, eight were identical to AY371283, and the remaining one with some sequence ambiguity (ON420777) had one SNP. The sequences from two Peru6-positive samples were identical to AY371281. Similarly, the sequences obtained from two Peru11-positive samples were identical to a GenBank sequence AY371286, and the sequence obtained from the genotype D-positive sample (ON420776) was identical to DQ683751. In phylogenetic analysis of these and reference sequences, all study sequences were placed in group 1.
Discussion
To the best of our knowledge, this is the first molecular epidemiological work characterizing the genetic diversity of E. bieneusi in sampled children (≤ 3–8 years old) in three provinces in Northeast Egypt. The overall infection rate of E. bieneusi in children was 4.6% (27/585), with similar infection rates in the three provinces surveyed (3.7–5.6%). This is lower than the 22.0% infection rate of E. bieneusi by PCR seen in leukemic children receiving chemotherapy (Shehab et al. 2021). Our finding is similar to the 5.9% (36/609) infection rate in healthy children in Xinjiang, China obtained using PCR (Qi et al. 2020), 6.4% (19/299) in healthy children in Bangladesh using PCR (Karim et al. 2021), and 6.8% (3/44) in children who had undergone a liver transplant in Iran using trichrome staining and PCR (Agholi et al. 2013). Two studies in Uganda and Malawi, however, reported a much higher infection rate of E. bieneusi in children (30.4% or 68/224 and 37.1% or 13/35, respectively) (Mor et al. 2009; Huibers et al. 2018). The differences in infection rates of E. bieneusi among countries might reflect variations in the sample size or its representativeness, geographical locations, dietary habits, immune and health status, access to hygiene resources, socioeconomic development, diagnostics methods, and exposures to animals (Matos et al. 2012).
A significantly higher infection rate of E. bieneusi (6.0% versus 2.8%, P = 0.029) was observed among children from rural areas than those from urban areas. This supports similar observations in studies conducted on children in Mozambique and HIV-positive patients in China, which reported higher E. bieneusi infection rates in those living in rural areas (Liu et al. 2017; Muadica et al. 2020). These differences could be attributed to the more frequent contact with animals by rural children, which facilitates the occurrence of zoonotic transmission of E. bieneusi. However, they could also reflect the area’s low hygiene resource access.
The genotyping results of study isolates support the role of zoonotic transmission in E. bieneusi infection in study children. The five E. bieneusi genotypes identified in the study, including type IV, Peru8, Peru6, Peru11, and D, all belong to group 1, which is known to have low host specificity (Santín and Fayer 2009; Li et al. 2019a, b). The most common genotype in this study, type IV, has been found frequently in diverse animal species such as nonhuman primates, dogs, cats, cattle, rabbits, various rodents, deer, foxes, birds, or snakes (Li et al. 2019b). Similarly, the second most prevalent genotype in our study, Peru8, was previously detected in nonhuman primates, dogs, horses, farmed raccoon dogs, foxes, or chickens (Li et al. 2019a). The other less frequent genotypes, including Peru6, Peru11, and D, are also well-known zoonotic E. bieneusi genotypes (Li et al. 2019b). As these genotypes are also common ones in humans, further studies with both sampling of animals and more advanced typing of the pathogen are needed to elucidate the exact role of zoonotic transmission in microsporidiosis in children in Egypt. Since this study was limited to the small number of children sampled in three provinces in Egypt, these findings cannot be generalized to the overall child population of Egypt. Additional studies in other areas with better epidemiological design should be conducted to further assess the prevalence and genotype distribution of E. bieneusi and the associated risk factors.
Conclusions
E. bieneusi infections appear to be reasonably common in children aged ≤ 3–8 years in three provinces in Northeast in Egypt. All genotypes of E. bieneusi detected in this study belong to the zoonotic group 1. This and the higher occurrence of infection in rural children suggest contact with animals or contamination of pathogen from animals could be a source of infection for children. Further epidemiological studies in other provinces and with children in other setting should be conducted to clarify the prevalence and infection sources of E. bieneusi in humans in Egypt.
References
Agholi M, Hatam GR, Motazedian MH (2013) Microsporidia and coccidia as causes of persistence diarrhea among liver transplant children: incidence rate and species/genotypes. Pediatr Infect Dis J 32:185–187. https://doi.org/10.1097/INF.0b013e318273d95f
Ding S, Huang W, Qin Q et al (2018) Genotype identification and phylogenetic analysis of Enterocytozoon bieneusi isolates from stool samples of diarrheic children. J Parasitol 104:297–301. https://doi.org/10.1645/17-108
Han B, Pan G, Weiss LM (2021) Microsporidiosis in humans. Clin Microbiol Rev 34:e0001020. https://doi.org/10.1128/CMR.00010-20
Huibers MHW, Moons P, Maseko N et al (2018) Multiplex real-time PCR detection of intestinal protozoa in HIV-infected children in Malawi: Enterocytozoon Bieneusi is common and associated with gastrointestinal complaints and may delay BMI (nutritional status) recovery. Pediatr Infect Dis J 37:910–915. https://doi.org/10.1097/INF.0000000000001924
Karim MR, Rume FI, Li D, et al (2021) First molecular characterization of Enterocytozoon bieneusi in children and calves in Bangladesh. Transbound Emerg Dis https://doi.org/10.1111/tbed.14187
Li W, Feng Y, Santin M (2019) Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol 35:436–451. https://doi.org/10.1016/j.pt.2019.04.004
Li W, Feng Y, Xiao L (2022) Enterocytozoon bieneusi. Trends Parasitol 38:95–96. https://doi.org/10.1016/j.pt.2021.08.003
Li W, Feng Y, Zhang L, Xiao L (2019) Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect Genet Evol 75:104033. https://doi.org/10.1016/j.meegid.2019.104033
Li W, Xiao L (2021) Ecological and public health significance of Enterocytozoon bieneusi. One Health (amsterdam, Netherlands) 12:100209. https://doi.org/10.1016/j.onehlt.2020.100209
Liu H, Jiang Z, Yuan Z, Yin J, Wang Z, Yu B, Zhou D, Shen Y, Cao J (2017) Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang autonomous region. China BMC Infect Dis 17:684. https://doi.org/10.1186/s12879-017-2787-9
Matos O, Lobo ML, Xiao L (2012) Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res 2012:981424. https://doi.org/10.1155/2012/981424
Mor SM, Tumwine JK, Naumova EN et al (2009) Microsporidiosis and malnutrition in children with persistent diarrhea, Uganda. Emerg Infect Dis 15:49–52. https://doi.org/10.3201/eid1501.071536
Muadica AS, Messa AEJ, Dashti A et al (2020) First identification of genotypes of Enterocytozoon bieneusi (Microsporidia) among symptomatic and asymptomatic children in Mozambique. PLoS Negl Trop Dis 14:e0008419. https://doi.org/10.1371/journal.pntd.0008419
Naguib D, El-Gohary AH, Roellig D, et al (2018) Molecular characterization of Cryptosporidium spp. and Giardia duodenalis in children in Egypt. Parasit Vectors 11:403. https://doi.org/10.1186/s13071-018-2981-7
Qi M, Yu F, Zhao A et al (2020) Unusual dominant genotype NIA1 of Enterocytozoon bieneusi in children in Southern Xinjiang. China Plos Negl Trop Dis 14:e0008293. https://doi.org/10.1371/journal.pntd.0008293
Rogawski ET, Liu J, Platts-Mills JA et al (2018) Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health 6:e1319–e1328. https://doi.org/10.1016/S2214-109X(18)30351-6
Santin M (2015) Enterocytozoon bieneusi. In: Xiao L, Ryan U, Feng Y, editors. Biology of foodborne parasites. Boca Raton: CRC Press
Santín M, Fayer R (2009) Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol 56:34–38. https://doi.org/10.1111/j.1550-7408.2008.00380.x
Shehab AY, Moneer EA, Allam AF et al (2021) Intestinal microsporidia infection in leukemic children: microscopic and molecular detection. Acta Parasitol 66:346–353. https://doi.org/10.1007/s11686-020-00283-2
Shen Y, Gong B, Liu X et al (2020) First identification and genotyping of Enterocytozoon bieneusi in humans in Myanmar. BMC Microbiol 20:10. https://doi.org/10.1186/s12866-019-1694-1
Stentiford GD, Becnel JJ, Weiss LM et al (2016) Microsporidia - emergent pathogens in the global food chain. Trends Parasitol 32:336–348. https://doi.org/10.1016/j.pt.2015.12.004
Sulaiman IM, Fayer R, Lal AA et al (2003) Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol 69:4495–4501. https://doi.org/10.1128/AEM.69.8.4495-4501.2003
Vávra J, Lukeš J (2013) Microsporidia and “the art of living together.” Adv Parasit 82:253–319. https://doi.org/10.1016/B978-0-12-407706-5.00004-6
Wang T, Fan Y, Koehler A, v, et al (2017) First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol 51:127–131. https://doi.org/10.1016/j.meegid.2017.03.006
Weiss LM, Becnel -J J (2014) Microsporidia: pathogens of opportunity: fisrt. Wiley Blackwell
Zhang Y, Koehler A, v, Wang T, Gasser RB (2021) Enterocytozoon bieneusi of animals-with an “Australian twist.” Adv Parasit 111:1–73. https://doi.org/10.1016/bs.apar.2020.10.001
Acknowledgements
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
Funding
This study was supported in part by the National Natural Science Foundation of China (31820103014), 111 Project (D20008), and the Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Contributions
Doaa Naguib and Lihua Xiao conceived the idea of the study and designed the study protocol. Nagah Arafat collected data and samples. Doaa Naguib performed laboratory investigations. Doaa Naguib and Lihua Xiao analyzed data and drafted the manuscript. Dawn M. Roellig and Lihua Xiao supervised the work. All authors revised the final draft of the submitted manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of the Faculty of Veterinary Medicine, Mansoura University, Egypt. All parents or guardians of the children were informed of the objective of the survey and were asked to voluntarily participate in the study. Informed consent was obtained from the legal guardians of the participants before the sample and data collection.
Consent to participate
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Una Ryan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Naguib, D., Roellig, D.M., Arafat, N. et al. Prevalence and genetic characterization of Enterocytozoon bieneusi in children in Northeast Egypt. Parasitol Res 121, 2087–2092 (2022). https://doi.org/10.1007/s00436-022-07546-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07546-z