Abstract
Background
Microsporidia infection was originally described as an immunocompromised associated pathogen. Limitations to correct microscopic diagnosis of microsporidia include size of the organism presenting a challenge even to a highly competent laboratory expert.
Objective
The present study aimed to detect microsporidia infection among leukemic children. The performance of modified trichrome stain and PCR in the diagnosis of microsporidia was evaluated with further speciation.
Methods
Stool samples of 100 leukemic children on chemotherapy were examined microscopically for microsporidia. DNA was extracted from all samples. Amplification was performed by conventional and nested PCR. Sequencing of amplified products was performed on unspeciated samples.
Results
Microsporidia were detected in 23% of the children by MTS and 29% by PCR. The 29 positive samples were subjected to PCR for speciation. Enterocytozoon bieneusi was found to predominate in 20 cases, Encephalitozoon intestinalis in three cases, two cases had co-infection, and the remaining four samples were not amplified with either E. bieneusi or E. intestinalis specific primers. By DNA sequencing of the unspeciated samples, three samples shared high homology with Encephalitozoon hellem and one sample with Encephalitozoon cuniculi. Referring to PCR as a gold standard, MTS exhibited 72.4% sensitivity and 97.2% specificity with 90% accuracy. Among a number of studied variables, diarrhea and colic were significantly associated with microsporidia infection when diagnosed by either technique.
Conclusion
The use of sensitive and discriminative molecular tools will contribute to determining the true prevalence of microsporidiosis and possibly their potential transmission source depending on species identification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microsporidia are obligate intracellular opportunistic pathogens, ubiquitous in nature and are globally found. Over 187 genera and 1500 species of microsporidia are classified in the phylum Microsporidia, primarily reported in fish, insects and mammals, of which only 16 species are known to infect humans both immunocompromised and immunocompetent [1]. Enterocytozoon bieneusi and Encephalitozoon intestinalis are the two most common species causing infection in humans. Microsporidial infections have been described to occur in severely immunocompromised individuals, mainly HIV patients, organ transplantation recipients or oncology patients [2,3,4].
Microsporidia clinical illness differs according to the pathogen species and the immunological condition of the host [5]. The spectrum of the disease caused by different species of microsporidia is broad and embraces various forms of inflammations as hepatitis, peritonitis, keratoconjunctivits, sinusitis, bronchitis, pneumonia, cystitis, nephritis, myositis, encephalitis, and further cerebral infections [6].
Diagnosis of microsporidiosis is generally based on microscopic examination of stools with detection of spores. Restrictions to correct diagnosis include size of the organism, its resemblance to bacteria and fungi found in stools and sometimes the large number of samples to be examined, presenting a challenge even to a highly trained laboratory technician. In addition, microscopy offers no information on the infecting species. With the advent of molecular tools, detection of microsporidia with further speciation and sequencing was made possible [7].
Human microsporidiosis occurs worldwide, but data on the prevalence and geographic distribution of microsporidial infections are still incomplete and very diverse due to the use of different diagnostic methods, lack of specialized personnel and the focus on more common parasites. In Egypt, merely few studies concerning the prevalence of microsporidia in immunocompromised patients are found [8, 9].
The aim of the present work was to detect microsporidia infection in leukemic children. The diagnostic performance of MTS and conventional PCR was evaluated. Species identification was then performed using specific primers and sequencing techniques.
Materials and Methods
Ethical Considerations
For conduction of the present work, ethical clearance was permitted by the Ethical Committee of the Medical Research Institute, Alexandria University (IORG 0008812). Informed consents from the parents/guardians as well as from the hospital authority were obtained.
One hundred confirmed leukemic children admitted to El-Shatby hospital, Alexandria University participated in the present study. Participating children were below 12 years of age of which 46% were less than the age of six, 55 were males and 45 females, the majority of cases (72%) came from rural areas. Subjects with a history of intake of anti-parasitic drugs in the preceding 2 weeks were excluded. Children were examined clinically, fresh stool samples were collected and each sample was subjected to the following:
Microscopic Examination
Formol ethyl acetate concentration of fecal samples was done followed by permanent staining of fecal smears using MTS (Kokoskan hot method) [10].
Molecular Detection
DNA was extracted from stool samples using Zymo Research Fecal DNA Mini Kit, USA according to manufacturer’s protocol and stored at − 20 °C.
Conventional PCR
It was done using primers targeting the entire conserved region of SSU rRNA gene of the four common intestinal microsporidia in humans: E. bieneusi, E. intestinalis, E. cuniculi and E. hellem. The amplification protocol included an initial denaturation at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, elongation at 72 °C for 1 min and 5 min of extension at 72 °C after 30 cycles [11]. Amplified DNA was detected by electrophoresis apparatus using 1.5% agarose gel stained with ethidium bromide.
Nested PCR
Samples showing a specific band of 1200 bp were subjected to PCR for speciation using species-specific primers. Species determination for E. bieneusi and E. intestinalis were done using species-specific primers (Table 1) [12]. Cycling conditions included an initial denaturation of DNA at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 50.2 °C for 1 min (E. bieneusi)/58 °C for 1 min (E. intestinalis), elongation at 72 °C for 1 min and 5 min of extension at 72 °C after 35 cycles.
Species Identification
Species identification of samples negative for both E. bieneusi and E. intestinalis was performed by further DNA sequencing technique for amplified PCR products. PCR products were sequenced using C1 forward primer (24.4 µM, 56 Tm) after purification with Gene JETTM purification kit to identify the species of samples positive by general primer, negative with specific primers. Sequencing reactions of purified PCR products were performed using ABI 3730XL DNA sequence [Applied Biological Materials (abm) Richmond, BC Canada].
Statistical Analysis
Data were analyzed using IBM SPSS for Windows, Version 22.0. The statistical program was utilized for both data presentation and statistical analysis of the results. For descriptive analysis, the prevalence by different methods was articulated in percentages. Confidence interval (CI) of 95% was applied as a measure of central tendency and dispersion respectively for normally distributed quantitative data. X2 test was used for quantitative analysis. Agreement between two qualitative variables was done by Cohen’s Kappa agreement test [13]. By establishing PCR as a gold standard, sensitivity, specificity, predictive values and accuracy were calculated at 95% CI using standard formulae.
Results
Microscopic Detection of Microsporidia
Among the 100 children included in the study, 23% were found positive for microsporidia by MTS. Pinkish-red oval or rounded small refractile bodies with polar bodies at one end and a characteristic posterior vacuole at the other end were identified as microsporidial spores (Fig. 1).
Molecular Detection of Microsporidial Infection
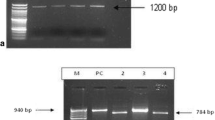
Amplification showed that 29 cases (29%) were positive for microsporidia utilizing pan-specific primers, while 71 samples gave negative results. Agarose gel electrophoresis revealed the amplification of a DNA fragment with an average size of 1200 bp (Fig. 2).
Sociodemographic and Clinical Characteristics and Their Relation to Microsporidiosis
Table 2 shows the distribution of microsporidia positive cases diagnosed by either MTS or PCR according to different demographic aspects, behavioral dynamics and clinical manifestations. Age, gender, residence, source of water, animal contact and fever showed no statistical significant relation to microsporidia infection. A statistically significant number of microsporidia positive children had diarrhea for more than 6 days compared to those with diarrhea for less than 6 days regardless of the technique used for diagnosis. A significant association between colic and microsporidia was found.
Agreement Between MTS and PCR in Diagnosis of Microsporidiosis
Agreement between MTS and PCR in the diagnosis of microsporidiosis was studied by comparing the results of both techniques, the following was revealed: 21 out of the 100 examined cases were positive by both techniques and were considered as true positives (concordant positive results). Sixty nine cases were true negatives (concordant negative results). By analysis of the discordant results, eight cases positive by PCR assay were missed by MTS, on the other hand two cases diagnosed positive by MTS were negative by PCR. Statistical analysis showed a Kappa index of 0.74 indicating a good agreement between MTS and PCR in diagnosing microsporidal infections (Table 3).
Measures of MTS Diagnostic Accuracy
Asserting PCR as a reference method, MTS showed 21 (72.4%) true positives and 69 (97.2%) true negative cases. There were two (2.8%) false positives and eight (27.6%) false negative cases. The positive predictive value (PPV) and negative predictive value (NPV) for MTS in comparison to PCR were 91.3% and 89.6% respectively with 90% accuracy (Table 3).
Microsporidia Speciation Using Specific Primers
Twenty two of the 29 amplified positive samples were diagnosed as E. bieneusi infection with E. bieneusi specific primer. Agarose gel electrophoresis revealed specific bands at 353 bp (Fig. 3). On retesting the 29 microsporidia positive samples using E. intestinalis specific primer, only five of the total amplified samples were diagnosed as E. intestinalis. Amplification of templates resulted in a 375 bp specific DNA fragment (Fig. 4). Two children were found to be infected by both species. The two samples gave positive results using the two specific primers. The remaining four cases were not amplified with either E. bieneusi or E. intestinalis specific primers, lasting unspeciated (Table 4). They possibly could be due to either E. cuniculi or E. hellem. DNA sequencing for species differentiation for E. cuniculi and E. hellem was done for these four unspeciated samples.
DNA Sequencing
Sequencing results of the1200 bp amplicon of the four unspeciated samples were submitted in BLASTN. Three of the sequenced samples had great identity with E. hellem. One sample exhibited 100% homology to sequences in GenBank with accession number (CP002715), while two samples displayed 80% identity with accession numbers (AF118143.1and MG584869.1). The last sample was diagnosed as E. cuniculi with 90% identity (Table 5).
Discussion
Human microsporidiosis represents an important disease, affecting mainly but not utterly HIV-positive and immunosuppressed patients [14, 15]. Data is mutable not only because of the troubles in identification of the organism but also because parasite pursuit tends to focus on more common ones. The goal of conducting the present study was to put emphasis on the importance of microsporidiosis as an emerging infection in leukemic children on chemotherapy that adds to their clinical complications, and to evaluate MTS and PCR as methods of detection of microsporidia.
In the present study, the prevalence of microsporidia was 23% and 29% as diagnosed by MTS and PCR respectively. Results of the sociodemographic data, concerning children aged from 6 to 12, males, residing in rural areas, drinking filtered water and having contact with animals revealed higher microsporidia infection rates than all leukemic children, yet, the difference was not significant. On the other hand, a clear significant association between the presence of microsporidia, diarrhea and colic was detected. The association between microsporidia infection and diarrhea was recognized by many authors among children with malignancy and HIV-infected patients [16,17,18]. Discrepancies in various studies concerning epidemiological features can be linked to the study settings, subjects under study and the used diagnostic methods [9, 16, 19].
As for laboratory diagnosis of microsporidiosis, it is mainly based on the detection of spores in stool specimens by light microscopy using special stains. The percentage of microsporidia infection in the study at hand was 23% using the MTS. The choice of this technique depended on its simplicity and its capacity to detect microsporidial spores in stools. Stool smears must be very thin to observe the internal structure of the microsporidia. Yet, they may be easily missed if the pathogen burden is low. In addition, small yeasts and some bacteria in stool may also stain pink, which may complicate the diagnosis [10]. This may thus lead to false negative as well as false positive results.
Few studies investigating the prevalence of microsporidiosis among patients with various forms of immunosuppression were carried out. In Mexico, García et al.[19] reported that 20% of leukemic children were diagnosed with microsporidia using coproparasitoscopic methods including trichrome stain. In Iran, Microsporidia spp. were detected in 11.6% of HIV patients, and in 7% of cancer patients using trichrome staining [20]. Spores were reported in 6% of the stool samples of chronic renal failure patients examined using special stains [21]. The variability in the positive rates of microsporidia cases obtained from the previous studies may be an evidence that its rate fluctuates among different geographical areas with different environmental and seasonal factors and different types of immunosuppression.
In the present study, to confirm diagnosis following microscopy, all stool samples were subjected to conventional PCR amplification using a general primer pair. Out of the 100 stool samples examined, 29% were positive for microsporidial spores. This relatively high percentage points to the importance of diagnosing microsporidia in children undergoing chemotherapy as this parasite can decrease the quality of life of those children and might also cause serious complications.
The present results corroborate several previously published reports. In Tunisia, microsporidia was found in 10.5% of the examined immunocompromised patients diagnosed by PCR, while 5.8% were positive by microscopy [22]. On the same line, in India, higher prevalence of microsporidia was detected by PCR (14.4%) compared to microscopy (5.6%) [23]. Conversely, El-Mahallawy et al. [9] found more positive Microsporidia cases by staining methods versus PCR.
Comparing the results of both techniques, the current study showed that MTS and PCR shared the detection of 22 affirmed positive cases as well as 69 negative ones elucidating a good agreement between the two techniques in diagnosing microsporidiosis. However, MTS revealed two positive samples not detected by PCR, while eight cases diagnosed positive by PCR were missed by MTS. A possible explanation for the few cases missed by microscopy is the small size of the microsporidial spores and the close similarity to other fungal spores and some bacteria that might contribute to the uncertainty of microscopic results. Also this difference might probably be the result of the limited sensitivity of light microscopy estimated between 104 and 106 spores per gram stool, whereas PCR can detect pathogen loads of 102 spores per gm stool [24]. On the other hand, the two samples diagnosed positive only by light microscopy cannot be overlooked. Variation in size and staining characteristics of microsporidia, makes a definitive identification exceptionally difficult, raising the possibility of false positive results. Yet, a false negative PCR result concerning these two cases is also possible and prone to occur due to storage conditions of the samples prior to performing the PCR analysis which might have resulted in degradation of the spores and consequently of microsporidial DNA. The presence of inhibitors during DNA extraction may play a role as well.
Kaushik et al. [25] stated that PCR showed almost perfect agreement with MTS, suggesting screening of stool samples with MTS followed by PCR confirmation for maximum detection. Furthermore, El-Mahallawy et al. [9] reported that 15 out of 271 diarrheic samples from pediatric patients with cancer had microsporidiosis. Infection was confirmed in 13 out of 15 cases positive by both PCR and acid-fast trichrome staining, two samples were positive only by stain.
Many authors reported that PCR had 100% sensitivity and specificity compared to microscopic techniques in detecting microsporidial infections [25,26,27]. Thus, in the current study, the accuracy of MTS as a diagnostic tool in detecting microsporidial spores was calculated taking in consideration PCR as the reference standard method of assumed 100% sensitivity and specificity. Values for MTS were 72.4% and 97.2% for sensitivity and specificity respectively with 90% accuracy. Many previously published studies have reported variable sensitivities of MTS ranging from 54 to 100% bearing in mind that sensitivity depended principally on technical expertise. In addition, a long time is needed for the initial standardization of the staining technique due to thick spore walls. As for specificity, comparable values were reported earlier and ranged from 82.8 to 100% [26,27,28].
Microsporidia characterization and speciation are necessary for the appropriate therapeutic management of patients. Microsporidial species appear to differ in their susceptibilities to antimicrobial agents. Albendazole was found effective against Encephalitozoon spp. [29, 30], while fumagillin has been shown to be effective against E. bieneusi [31, 32]. For a long time transmission electron microscopy (TEM) was conventionally the gold standard to diagnose microsporidiosis on the basis of morphological characteristics but it failed to differentiate between the morphologically identical species, in addition it is difficult to perform TEM on routine basis [7, 33]. PCR was thus suggested as a reference tool for identification of microsporidia to the species level [23]. It may therefore be indispensable to perform a species-specific PCR before treatment regimens are decided upon, especially for E. bieneusi infections; the only species to show resistance to albendazole [34].
The molecular characterization of different microsporidia has widely relied on sequencing and analyzing SSU rRNA gene, which also can be used as a molecular marker for estimating relationships among microsporidia because of its relatively highly variable regions pointing to the critical role played by SSU rRNA gene in phylogenetic research [35, 36]. In the present study, speciation performed on the 29 positive samples by conventional analysis corresponded to: 20 E. bieneusi, three E. intestinalis cases, two E. bieneusi/E. intestinalis co-infection and 4 cases remained unspeciated.
A study conducted in Iran among cancer patients revealed that E. bieneusi was the most frequent intestinal microsporidial species amounting to 61% [20]. Furthermore, Espern et al. [37] and Akinbo et al. [38] reported the dominance of E. bieneusi among all other microsporidial species. On the contrary, among 93 cancer patients, 69.9% of patients were positive for Microsporidia comprising 46.2% E. intestinalis, 9.7% E. bieneusi and 14% mixed infections [39].
Based on the DNA sequence data, it was extremely remarkable to note the presence of E. hellem and E. cuniculi among the studied participants. To our knowledge, this is the first documented cases of these species in man in Egypt. Similar sequencing results were reported by Polly et al. [40] and Kazemi et al. [20].
Regarding the knowledge that birds are natural hosts of E. hellem lead to the hypothesis that human infections with E. hellem originate from exposure to birds [41]. E. cuniculi was the first microsporidium to be recognized as a parasite of mammals. Rabbits are the main host for E. cuniculi [42]. The two species of Encephalitozoon; E. cuniculi and E. hellem, which are morphologically similar by light and electron microscopy, are rarely detected in feces and usually cause systemic diseases; these species are primarily shed in urine rather than feces.
In conclusion, staining of stool specimens could be suggested as an initial routine test for leukemic children with the advantage of being low-cost and simpler technique than PCR. The use of sensitive and discriminative molecular tools will contribute to determining the true prevalence of microsporidiosis and the possible potential transmission source.
References
Simpson AGB, Slamovits CH, Archibald JM (2017) Protist diversity and eukaryote phylogeny. Handbook of the Protists, vol 2. Springer International Publishing, Berlin, pp 1–20. https://doi.org/10.1007/978-3-319-28149-0_45
Carlson J, Helton C, Munn R, Wasson K, Perez R, Gallay B, Finkbeiner WE (2004) Disseminated microsporidiosis in a pancreas/kidney transplant recipient. Arch Pathol Lab Med 128(3):41–42. https://doi.org/10.1043/1543-2165(2004)128<e41:DMIAKT>2.0.CO.2
Viriyavejakul P, Nintasen R, Punsawad C, Chaisri U, Punpoowong B, Riganti M (2009) High prevalence of Microsporidium infection in HIV-infected patients. Southeast Asian J Trop Med Public Health 40(2):223–228
Dikman AE, Schonfeld E, Srisarajivakul NC, Poles MA (2015) Human immunodeficiency virus-associated diarrhea: still an issue in the era of antiretroviral therapy. Digest Dis Sci 60(8):2236–2245. https://doi.org/10.1007/s10620-015-3615-y
Didier ES, Weiss LM (2006) Microsporidiosis: current status. Curr Opin Infect Dis 19(5):458–492. https://doi.org/10.1097/01.qco.0000244055.46382.23
Orenstein JM, Gaetz HP, Yachnis AT, Frankel SS, Mertens RB, Didier ES (1997) Disseminated microsporidiosis in AIDS: are any organs spared? AIDS 11(3):385–386
Garcia LS (2002) Laboratory identification of the microsporidia. J Clin Microbiol 40(6):1892–1901. https://doi.org/10.1128/jcm.40.6.1892-1901.2002
Ali M, Mahmoud LA, Abaza B, Ramadan M (2000) Intestinal spore-forming protozoa among patients suffering from chronic renal failure. J Egypt Soc Parasitol 30(1):93–100
El-Mahallawy H, Zaki MM, El-Arousy M, Shalabi L, Mansour T (2011) Diagnosis of intestinal microsporidiosis in pediatric oncology patients in Egypt using modified acid fast trichrome staining versus PCR. Acta Parasitol 56(4):348–352. https://doi.org/10.2478/s11686-011-0072-4
Kokoskin E, Gyorkos TW, Camus A, Cedilotte L, Purtill T, Ward B (1994) Modified technique for efficient detection of microsporidia. J Clin Microbiol 32(4):1074–1075. https://doi.org/10.1128/JCM.32.4.1074-1075.1994
Raynaud L, Delbac F, Broussolle V, Rabodonirina M, Girault V, Wallon M et al (1998) Identification of Encephalitozoon intestinalis in travelers with chronic diarrhea by specific PCR amplification. J Clin Microbiol 36(1):37–40. https://doi.org/10.1128/JCM.36.1.37-40.1998
Coyle C, Wittner M, Kotler D, Noyer C, Orenstein J, Tanowitz H et al (1996) Prevalence of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon (Septata) intestinalis among patients with AIDS-related diarrhea: determination by polymerase chain reaction to the microsporidial small-subunit rRNA gene. Clin Infect Dis 23(5):1002–1006. https://doi.org/10.1093/clinids/23.5.1002
Viera AJ, Garrett JM (2005) Understanding inter observer agreement: the kappa statistic. Fam Med 37(5):360–363
Didier ES (1998) Microsporidiosis. Clin Infect Dis 27(1):1–7. https://doi.org/10.1086/514607
Aini UN, Al-Mekhlafi MH, Anisah N, Fatmah MS, Azlin M, Rozlida AR et al (2008) A preliminary study on the prevalence of intestinal microsporidiosis in patients with and without gastrointestinal symptoms in Malaysia. Trans R Soc Trop Med Hyg 102(12):1274–1278. https://doi.org/10.1016/j.trstmh.2008.05.019
Nadham KM, Maysloon AAS (2012) Microsporidiosis among children with malignant diseases in Basrah, Iraq. Pak J Med Sci 28(4):621–624
Centers of Diseases Control and prevention (2009) Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents. MMWR 58(10):3–59
Muller A, Bialek R, Kamper A, Fatkenheuer G, Salzberger B, Franzen C (2001) Detection of microsporidia in travelers with diarrhea. J Clin Microbiol 39:1630–1632. https://doi.org/10.1128/JCM.39.4.1630-1632.2001
García L, Cano-Estrada A, Cortés-Campos A, Medina-Sansón A, Jiménez-Cardoso E (2013) Identification of Microsporidium spp in patients with acute lymphoblastic leukemia. Boletin Medico del Hospital Infantil de Mexico 70(1):24–28
Kazemi E, Tavalla M, Maraghi S, Jafaryad M, Latifi M (2017) Frequency of microsporidial infection in immunocompromised patients with staining and molecular methods based on internal transcribed spacer region gene in two cities of Southwest Iran during 2013–2014. AJPRHC 9(1):7–16. https://doi.org/10.18311/ajprhc/2017/6014
Fadl HO, El-gabrty SAE, Hanafy NA, Khairy H, Ali N (2015) Detection of microsporidiosis in cases with chronic renal failure. J Life Appl Sci 2(1):43–51
Chabchoub N, Abdelmalek R, Mellouli F, Kanoun F, Thellier M, Bouratbine A et al (2009) Genetic identification of intestinal microsporidia species in immunocompromised patients in Tunisia. Am J Trop Med Hyg 80(1):24–27. https://doi.org/10.4269/ajtmh.2009.80.24
Saigal K, Khurana S, Sharma A, Sehgal R, Malla N (2013) Comparison of staining techniques and multiplex nested PCR for diagnosis of intestinal microsporidiosis. Diagn Microbiol Infect Dis 77(3):248–249. https://doi.org/10.1016/j.diagmicrobio.2013.07.004
Müller AK, Stellermann P, Hartmann M, Schrappe G, Fätkenheuer B, Salzberger V (1999) A powerful DNA extraction method and PCR for detection of microsporidia in clinical stool specimens. Clin Diagn Lab Immunol 6:243–246
Kaushik S, Saha R, Das S, Ramachandran VG, Goel A (2018) Pragmatic combination of available diagnostic tools for optimal detection of intestinal microsporidia. Adv Exp Med Biol 1057:85–94. https://doi.org/10.1007/5584_2017_97
Rinder H, Janitschke K, Aspöck H, Da Silva AJ, Deplazes P, Fedorko D (1998) Blinded, externally controlled multicenter evaluation of light microscopy and PCR for detection of Microsporidia in stool specimens. The diagnostic multicenter study group on microsporidia. J Clin Microbiol 36(6):1814–1818. https://doi.org/10.1128/JCM.36.6.1814-1818.1998
Subrungruang I, Mungthin M, Chavalitshewinkoon-Petmitr P, Rangsin RS, Naaglor T, Leelayoova S (2004) Evaluation of DNA extraction and PCR methods for detection of Enterocytozoon bienuesi in stool specimens. J Clin Microbiol 42(8):3490–3494. https://doi.org/10.1128/JCM.42.8.3490-3494.2004
Mohammed H, Endeshaw T, Kebede A, Defera M (2009) Comparison of chromotrope 2R and Uvitex 2B for the detection of intestinal microsporidial spores in stool specimens of HIV patients attending Nekempte Hospital, West Ethiopia. Ethiop Med J 47(3):233–237
Conteas CN, Berlin OG, Ash LR, Pruthi JS (2000) Therapy for human gastrointestinal microsporidiosis. Am J Trop Med Hyg 63(4–6):121–127. https://doi.org/10.4269/ajtmh.2000.63.121
Kotkova M, Sak B, Kvetonova D, Kvac M (2013) Latent microsporidiosis caused by Encephalitozoon cuniculi in immunocompetent hosts: a murine model demonstrating the ineffectiveness of the immune system and treatment with albendazole. PLoS One 8(4):e60941. https://doi.org/10.1371/journal.pone.0060941
Molina JM, Goguel J, Sarfati C, Michiels JF, Desportes-Livage I, Balkan S et al (2000) Trial of oral fumagillin for the treatment of intestinal microsporidiosis in patients with HIV infection. ANRS 054 Study Group. Agence Nationale de Recherche sur le SIDA. AIDS 14(10):1341–1348. https://doi.org/10.1097/00002030-200007070-00006
Champion L, Durrbach A, Lang P, Delahousse M, Chauvet C, Sarfati C (2010) Fumagillin for treatment of intestinal microsporidiosis in renal transplant recipients. Am J Transplant 10(8):1925–1930. https://doi.org/10.1111/j.1600-6143.2010.03166.x
Larsson JI (2005) Fixation of microsporidian spores for electron microscopy. J Invertebr Pathol 90(1):47–50. https://doi.org/10.1016/j.jip.2005.06.016
De Groote MA, Visvesvara G, Wilson ML, Pieniazek NJ, Slemenda SB, daSilva AJ et al (1995) Polymerase chain reaction and culture confirmation of disseminated Encephalitozoon cuniculi in a patient with AIDS: successful therapy with albendazole. J Infect Dis 171(5):1375–1378. https://doi.org/10.1093/infdis/171.5.1375
Tsai ST, Huang WF, Wang CH (2005) Complete sequence and gene organization of the Nosema spodopterae rRNA gene. J Eukaryot Microbiol 52(1):52–54. https://doi.org/10.1111/j.1550-7408.2005.3291rr.x
Tay W, Mahony EM, Paxton RJ (2005) Complete rRNA gene sequences reveal that the microsporidium Nosema bombi infects diverse Bumblebee (Bombus spp.) hosts and contains multiple polymorphic sites. J Eukaryot Microbiol 52(6):505–513. https://doi.org/10.1111/j.1550-7408.2005.00057.x
Espern A, Morio F, Miegeville M, Illa H, Abdoulaye M, Meyssonnier V et al (2007) Molecular study of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon intestinalis among human immunodeficiency virus-infected patients from two geographical areas: Niamey, Niger, and Hanoi, Vietnam. J Clin Microbiol 45(9):2999–3002. https://doi.org/10.1128/JCM.00684-07
Akinbo FO, Okaka CE, Omoregie R (2010) Prevalence of intestinal parasitic infections among HIV patients in Benin City, Nigeria. Libyan J Med 5(1):1–6. https://doi.org/10.3402/ljm.v5i0.5506
Hamamcı B, Çetinkaya Ü, Berk V, Kaynar L, Kuk S (2015) Prevalence of Encephalitozoon intestinalisand Enterocytozoon bieneusi in cancer patients under chemotherapy. Mikrobiyol Bul 49(1):105–113. https://doi.org/10.5578/mb.8787
Polley SD, Boadi S, Watson J, Curry A, Chiodini PL (2011) Detection and species identification of microsporidial infections using SYBR Green real-time PCR. J Med Microbiol 60(4):459–466. https://doi.org/10.1099/jmm.0.026781-0
Childs-Sanford SE, Garner MM, Raymond JT, Didier ES, Kollias GV (2006) Disseminated microsporidiosis due to Encephalitozoon hellem in an Egyptian fruit bat (Rousettus aegyptiacus). J Comp Pathol 134(4):370–373. https://doi.org/10.1016/j.jcpa.2006.01.004
Künzel F, Joachim A (2010) Encephalitozoonosis in rabbits. Parasitol Res 106(2):299–309. https://doi.org/10.1007/s00436-009-1679-3
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shehab, A.Y., Moneer, E.A., Allam, A.F. et al. Intestinal Microsporidia Infection in Leukemic Children: Microscopic and Molecular Detection. Acta Parasit. 66, 346–353 (2021). https://doi.org/10.1007/s11686-020-00283-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-020-00283-2