Abstract
Species of Cosmocerca Diesing, 1861 (Ascaridomorpha: Cosmocercoidea), are common nematode parasites of amphibians. In the present study, a new species of Cosmocerca, namely C. simile n. sp., was described using light and scanning electron microscopy, and sequencing different nuclear and mitochondrial genetic markers (i.e. small ribosomal DNA (18S), large ribosomal DNA (28S), internal transcribed spacer (ITS) and cytochrome c oxidase subunit 1 (cox1)). Cosmocerca simile n. sp. differs from its congeners based on body size, morphology and number of plectanes, relative length of spicules and gubernaculum and spicules to total body length and morphology and length of tail. Molecular analysis showed no nucleotide polymorphisms among different individuals of the new species regarding nuclear DNA. Very low intraspecific nucleotide variation (0.52–0.78%) was detected in cox1 mtDNA. In contrast, the level of interspecific nucleotide variation between C. simile n. sp. and its congeners were distinctly higher (2.74–18.1% in the partial ITS region and 10.2–13.5% in the partial cox1 region, respectively) than that of intraspecific variation. Phylogenetic analyses using maximum likelihood (ML) inference based on the partial ITS and cox1 sequence data both supported the new species to be a member of the genus Cosmocerca, and formed a sister relationship to C. japonica. The newly obtained genetic data are important for further studies of DNA-based taxonomy, population genetics and phylogenetics of the Cosmocercoidea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Species of Cosmocerca Diesing, 1861 (Ascaridomorpha: Cosmocercoidea), are common nematode parasites occurring in the digestive tract of various amphibians (Yamaguti 1938; Skrjabin et al. 1961; Baker and Green 1988; Moravec and Sey 1985; Moravec and Baruš 1990; Bursey et al. 2015; Sou and Nandi 2015). To date, approximately 30 species of Cosmocerca have been reported worldwide (Rizvi et al. 2011; Bursey et al. 2015; Sou et al. 2018a). Of them, only C. japonica Yamaguti 1938 and C. ornata (Dujardin, 1845) were recorded in China.
In recent years, molecular approaches have been widely used for exact identification of ascaridid nematodes and treatment of some unsolved taxonomical problems with morphological methods (i.e. diagnosis of eggs and larvae, discovery of cryptic species and delimitation of phenotypic variation) (Li et al. 2012, 2014, 2017; Sato et al. 2015; Zhang et al. 2018; Chen et al. 2018; Zhao et al. 2017, 2018). However, our present knowledge of molecular identification of the cosmocercoid nematodes remains very limited, due to the paucity of genetic data. Currently, only 20 species of Cosmocercoidea have been genetically characterized (i.e. 18S, 28S, ITS, 12S, cox1 and cox2) in the GenBank database. This situation has hindered the further studies of DNA-based taxonomy, population genetics and phylogenetics of the referred superfamily.
During a helminthological survey in Chinese amphibians, some nematodes were collected from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura). An integrative approach, including light and scanning electron microscopy, sequencing and analysis of the different nuclear and mitochondrial genetic markers (i.e. small ribosomal DNA (18S), large ribosomal DNA (28S), internal transcribed spacer (ITS) and cytochrome c oxidase subunit 1 (cox1)), was used to accurately characterize these parasites. Moreover, in order to determine the systematic position of these nematodes, the phylogenetic analyses were performed using maximum likelihood (ML) inference based on the partial ITS and cox1 sequence data, respectively.
Materials and methods
Light and scanning electron microscopy
A total of 542 individuals of the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura) collected in Zhejiang Province were dissected for parasites. Nematodes found in the intestine were fixed and stored in 80% ethanol prior to study. For light microscopy studies, nematodes were cleared in lactophenol. Drawings were made with the use of a Nikon microscope drawing attachment. For scanning electron microscopy (SEM), the anterior and posterior ends of nematodes were re-fixed in 4% formaldehyde solution, post-fixed in 1% OsO4, dehydrated via an ethanol series and acetone and then critical point dried. Samples were coated with gold and examined using a Hitachi S-4800 scanning electron microscope at an accelerating voltage of 20 kV. Measurements (range, followed by the mean in parentheses) are given in micrometres (μm) unless otherwise stated. Type specimens were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
Molecular procedures
One male and seven females of the new species, one female of C. ornata collected from Hylarana spinulosa (Smith) (Anura: Ranidae) in Dayaoshan, Guangxi Province, China, and one female of Cosmocerca sp. collected from Bufo melanostictus Schneider (Anura: Bufonidae) in Jinghong, Yunnan Province, China, were used for molecular analysis. Genomic DNA from the 10 samples was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer’s instructions. The partial 18S region was amplified by polymerase chain reaction (PCR) using the forward primer 18S-F (5′-CGCGAATRGCTCATTACAACAGC-3′) and the reverse primer 18S-R (5′-GGGCGGTATCTGATCGCC-3′) (Floyd et al. 2005). The partial 28S region was amplified by PCR using the forward primer 28S-F (5′-AGCGGAGGAAAAGAAACTAA-3′) and the reverse primer 28S-R (5′-ATCCGTGTTTCAAGACGGG-3′) (Nadler and Hudspeth 1998). The ITS-1 region of nuclear rDNA was amplified by PCR using the forward primer SS1 (5′-GTTTCCGTAGGTGAACCTGCG-3′) and the reverse primer SS2R (5′-AGTGCTCAATGTGTCTGCAA-3′). The ITS-2 region of nuclear rDNA was amplified by PCR using the forward primer NC13 (5′-ATCGATGAAGAACGCAGC-3′) and the reverse primer NC2 (reverse: 5′-TTAGTTTCTTTTCCTCCGCT-3′) (Zhu et al. 2000). The partial cox1 region was amplified by PCR using the forward primer COIF (5′-TTTTTTGGTCATCCTGAGGTTTAT-3′) and the reverse primer COIR (5′-ACATAATGAAAATGACTAACAAC-3′) (Lazarova et al. 2006). The cycling conditions were as described by Chen et al. (2018). PCR products were checked on GoldView-stained 1.5% agarose gels and purified with Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing was carried out using a Dye Deoxy Terminator Cycle Sequencing Kit (v.2, Applied Biosystems, CA, USA) and an automated sequencer (ABI-PRISM 377). Sequences were aligned using ClustalW2 and adjusted manually. The DNA sequences obtained herein were compared (using the algorithm BLASTn) with those available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov).
Phylogenetic analyses
Phylogenetic trees were constructed using maximum likelihood (ML) inference with MEGA 6 based on the partial ITS and cox1 sequence data, respectively. Falcaustra sinensis (Cosmocercoidea: Kathlaniidae) was treated as the outgroup. The ingroup includes the representatives of the Cosmocercidae with the ITS and cox1 sequence data available in the GenBank database (due to only partial ITS-1 sequence available, C. longicauda was not included in the phylogeny). We used a built-in function in the software MEGA 6 to select a best-fitting substitution model for the present sequences according to the Bayesian information criterion. The K2 (Kimura 2-parameter) + G model for the ITS sequence data and the HKY (Hasegawa-Kishino-Yano) + G + I model for the cox1 sequence data were identified as the optimal nucleotide substitution model, respectively. Reliabilities for ML trees were tested using 1000 bootstrap replications, and bootstrap values exceeding 50% were showed in the phylogenetic trees.
Results
Cosmocerca simile n. sp. (Figs. 1 and 2)
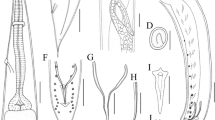
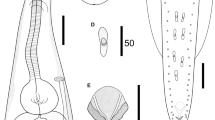
Small-sized, whitish nematodes. Body cylindrical, maximum width at about mid-body (Fig. 1A, E). Cuticle with fine transverse striations. Somatic papillae present (Fig. 2a, b). Narrow lateral alae present in both sexes (Fig. 2a, d, e). Oral aperture simple, somewhat triangular, surrounded by 3 small lips, each lip with inner flanges and several simple somatic papillae (Fig. 2b). Dorsal lip with one pair of large double cephalic papillae; subventral lips with single large double cephalic papilla and amphid (Fig. 2b). Oesophagus divided into anterior pharynx, cylindrical corpus and terminal posterior bulb with valves (isthmus indistinct) (Fig. 1A, B, E). Nerve ring located at about 1/2 of oesophageal length (Fig. 1A, B, E). Excretory pore at level of oesophageal bulb (Figs. 1A, B, E and 2a, f). Deirids not observed. Tail conical, with pointed tip in both sexes (Figs. 1A, D, E, I and 2d, e).
Cosmocerca simile n. sp. collected from Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura) in China (A–K). (A) body of female, lateral view; (B) anterior part of female, lateral view; (C) region of vulva, lateral view; (D) tail of female, lateral view; (E) body of male, lateral view; (F) spicules, ventral view; (G) gubernaculum, ventral view; (H) plectane, ventral view; (I) posterior end of male, ventral view; (J) egg; (K) larva; (L) spicules of C. ornata collected from Hylarana spinulosa (Smith) (Anura: Ranidae) in China, ventral view. Scale bars: A = 500 μm; B–E = 200 μm; F, G, J–L = 50 μm; H, I = 100 μm
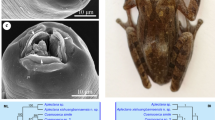
Scanning electron micrographs of Cosmocerca simile n. sp. collected from Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura) in China. a Anterior part of female (lateral ala and excretory pore arrowed, black arrows indicate somatic papillae), ventrolateral view. b Cephalic end of female (somatic papillae arrowed), apical view. c Magnified image of vulva, ventral view. d Tail of female (caudal alae arrowed), ventral view. e Posterior end of male (plectanes and caudal ala arrowed), ventral view. f Magnified image of excretory pore. g Larvae. h Magnified image of plectane. Abbreviations: a, lateral alae; ep, excretory pore; dl, dorsal lip; vl, ventrolateral lip
Male (based on 3 mature specimens)
Body 1.93–2.71 (2.42) mm long; maximum width 218–277 (248). Oesophagus 327–347 (333) in total length, representing 12.5–16.9 (14.1) % of body length; pharynx and corpus 257–287 (271) long; size of bulb 50–59 (56) × 59–79 (69). Nerve ring 129–168 (149) and excretory pore 267–347 (297) from anterior extremity, respectively. Lateral alae extending from some distance posterior to base of lips as far as level of third pair of pre-cloacal plectane (Fig. 2e). Posterior end of body distinctly ventrally curved (Figs. 1E and 2e). Spicules small and equal, well sclerotized, with distal end pointed, 79–99 (89) long, representing 3.28–4.10 (3.72) % of body length (Fig. 1E, F). Gubernaculum small and conical, well sclerotized, 50–69 (58) long (Fig. 1G). Five pairs of pre-cloacal plectanes present (Figs. 1E, I and 2e). Plectane consisting of central papilla with 5–6 cuticular tubercles on underlying sclerotized segments (Figs. 1H and 2h). Six pairs of postclocal, subventral, large, and simple papillae (distinguishable from somatic papillae) present. Tail 149–168 (158) long, representing 6.20–7.72 (6.53) % of body length, ending in a small spike (Figs. 1E, I and 2e).
Female (based on 10 mature specimens)
Body 4.65–6.21 (5.45) mm long; maximum width 446–614 (526). Oesophagus 495–545 (528) mm in total length, representing 7.97–11.3 (9.78) % of body length; pharynx and corpus 376–436 (408) long, size of bulb 109–139 (120) × 139–158 (145). Nerve ring 168–218 (193) and excretory pore 366–554 (457) from anterior extremity, respectively. Lateral alae extending from some distance posterior to base of lips and ending at base of tail tip (Fig. 2d). Vulva with transverse slit opening, with slightly salient lips, 2.12–3.06 (2.48) mm from anterior extremity, representing 41.4–49.3 (45.4) % of body length (Figs. 1C and 2c). Vagina muscular, each uterus full of larvated eggs (sometimes premature larvae also present in uterus); egg oval, thin-walled, with smooth surface, 59–119 (94) × 50–89 (66) (n = 20) (Figs. 1J, K and 2g). Tail 594–842 (780) long, representing 12.8–13.6 (14.3) % of body length, with long filamentous tip (Figs. 1A, D and 2d). Phasmids not observed.
Taxonomic summary
Type host. Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura).
Type locality. Yuyao, Zhejiang Province, China.
Site of infection. Intestine.
Level of infection: 27.7% (150 out of 542 B. gargarizans) were infected with intensity of 1–19 (mean 3.5) nematodes.
Type deposition. Holotype, male (HBNU–N-2019A021C-L); allotype, female (HBNU–N-2019A022C-L); paratypes: 2 males, 100 females (HBNU–N-2019A023C-L).
Etymology. The specific epithet is derived from the Latin word -similis and refers to the striking similarity with C. japonica Yamaguti, 1939 and C. novaeguineae Moravec & Sey, 1990.
Molecular characterization
18S region
The three 18S sequences of C. simile n. sp. were 1681 bp in length, representing only one genotype. Currently, there is no species of Cosmocerca with the 18S sequence in GenBank. Pairwise comparison of C. simile n. sp. with the other species of Cosmocercidae with the 18S sequenced available in GenBank, i.e. Cosmocercoides dukae (FJ516753), C. pulcher (LC018444, MH178322–MH178326), C. qingtianensis (MH032769–MH032771, MH178319–MH178321), C. tonkinensis (AB908160), C. wuyiensis (MK110872), Nemhelix bakeri (DQ118537), Neoxysomatium brevicaudatum (JF713457) and Raillietnema sp. (DQ503461), displayed 1.25–2.25% of nucleotide divergence. The 18S sequences of C. simile n. sp. (accession numbers MN839758–MN839760) were deposited in GenBank database (http://www.ncbi.nlm.nih.gov).
Partial ITS region
The eight ITS sequences of C. simile n. sp. were 961 bp in length, representing only one genotype. There are four species of Cosmocerca with the ITS sequenced available in GenBank, i.e. C. japonica Yamaguti, 1939 (LC052772–LC052782), C. ornata (Dujardin, 1845) (MT108302), Cosmocerca sp. (MT108303) and C. longicauda (Linstow, 1885) (only ITS-1 sequences available, MG594349–MG594351). Pairwise comparison of C. simile n. sp. with C. japonica, C. ornata and Cosmocerca sp. showed 2.74–3.37% (C. japonica) to 18.1% (C. ornata) of nucleotide divergence, respectively (Table 1). However, about 27.0% nucleotide divergence in the ITS-1 region was detected between C. simile and C. longicauda. Pairwise comparison of C. simile n. sp. with the other species of Cosmocercidae with the ITS sequenced available in GenBank, i.e. Cosmocercoides pulcher (MH178314–MH178318, LC018444), C. qingtianensis (MH178311–MH178313, MH032772–MH032774), C. tonkinensis (AB908160, AB908161) and C. wuyiensis (MK110871), displayed 27.6–28.2% of nucleotide divergence. The ITS sequences of C. simile n. sp. were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (accession numbers MN839761–MN839768).
Partial 28S region
The three 28S sequences of C. simile n. sp. were 738 bp in length, representing only one genotype. There is no species of Cosmocerca with the 28S sequenced in GenBank. Pairwise comparison of C. simile n. sp. with the other species of Cosmocercidae with the 28S sequenced available in GenBank, i.e. C. pulcher (LC018444) and C. tonkinensis (AB908160), displayed 10.4–10.7% of nucleotide divergence. The 28S sequences of C. simile n. sp. were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (accession numbers MN833301–MN833303).
Partial cox1 region
The three cox1 sequences of C. simile n. sp. were 384 bp in length, representing three different genotypes and showing 0.52–0.78% of nucleotide divergence. There are three species of Cosmocerca with cox1 sequenced available in GenBank, i.e. C. japonica (LC052756–LC052770), C. ornata (MT108304) and Cosmocerca sp. (MT108305). Pairwise comparison of C. simile n. sp. with the three species of Cosmocerca displayed 10.2–15.5% of nucleotide divergence, respectively (Table 1). Pairwise comparison of C. simile n. sp. with the other species of Cosmocercidae with the cox1 sequenced available in GenBank, including C. pulcher (MH178306–MH178310, LC052771) and C. qingtianensis (MH178303–MH178305, MH032775–MH032777), displayed 13.7–19.5% of nucleotide divergence. The cox1 sequences of C. simile n. sp. were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (accession numbers MN839755–MN839757).
Phylogenetic analyses
The phylogenetic trees were constructed using the partial ITS and cox1 sequence data, respectively, which showed similar topology. The representatives of the family Cosmocercidae were divided into two clades (genera), the genus Cosmocerca and the genus Cosmocercoides. Both of the phylogenetic trees displayed that Cosmocerca simile n. sp. clustered together with C. japonica, and C. ornata formed the most basal branch in Cosmocerca (Figs. 3 and 4).
Discussion
In the genus Cosmocerca, C. japonica Yamaguti, 1938, C. acanthurum Falcón-Ordaz et al., 2007, C. ornata (Dujardin, 1845), C. banyulensis Chabaud & Campana-Rouget, 1955, C. cruzi Rodrigues & Fabio, 1970, C. novaeguineae Moravec & Sey, 1990, C. paraguayensis Moravec & Kaiser, 1994, C. parva Travassos, 1925, C. podicipinus Baker & Vaucher, 1984, C. travassosi Rodrigues & Fabio, 1970; C. kalesari Rizvi, Bursey & Bhutia, 2011; C. microhylae Sou & Nandi, 2015 and C. bengalensis Sou, Sow & Nandi, 2018, have five pairs of plectanes, as same as in C. simile n. sp. (Chabaud and Campana-Rouget 1955; Rodrigues and Fabio 1970a, b; Baker and Vaucher 1984; Rizvi et al. 2011; Sou and Nandi 2015; Falcón-Ordaz et al. 2007; Sou et al. 2018b).
Among the above mentioned species, C. acanthurum reported from plethodontid salamanders in Mexico, could be easily distinguished from the new species by the unique spiny tail of females (vs female tail without spines in the new species) (Falcón-Ordaz et al. 2007). Cosmocerca simile n. sp. differs from C. parva, C. podicipinus, C. cruzi and C. travassosi by having different morphology of plectanes (i.e. plectanecentral papilla surrounded by 1–2 complete circles of tubercles vs plectane central papilla surrounded by tubercles forming about a quarter of a circle in the new species) (Rodrigues and Fabio 1970a, b; Baker and Vaucher 1984; González and Hamann 2008). The new species is also different from C. japonica and C. ornata by having well sclerotized spicules distinctly longer than gubernaculum (vs weakly sclerotized and thin spicules slightly shorter than gubernaculum in C. ornata and nearly completely atrophied spicules in C. japonica) (Fig. 1F, L) and much longer tail in females (0.59–0.84 mm, representing 12.8–13.6% of body length vs 0.19–0.45 mm, representing less than 10.0% of body length) (Yamaguti 1938; Kung and Wu 1945, Moravec and Sey 1985; Sou et al. 2018b). Cosmocerca simile n. sp. can be easily distinguished from C. bengalensis by having two separated spicules and small gubernaculum (vs spicules fused to a single spatulate body and gubernaculum absent according to Sou et al. (2018a)), shorter tail in males (0.15–0.17 mm, representing 6.20–7.72% of body length vs 0.29–0.36 mm, representing 13.4–15.4% of body length) and much longer tail in females (0.59–0.84 mm, representing 12.8–13.6% of body length vs 0.42–0.45 mm, representing 7.90–8.55% of body length) (Sou et al. 2018a). However, we considered that Sou et al. (2018a) mistook the gubernaculum as ‘fused’ spicules. The spicules of this species seems to be atrophied.
With lateral alae ending at base of caudal filament and relatively longer spicules (spicules representing 13.5–14.1% of body length) (Sou and Nandi 2015), C. microhylae also differs from the new species (vs lateral alae ending at level of third pair of pre-cloacal plectane and spicules representing 3.28–4.10% of body length). Cosmocerca banyulensis can be differentiated from the new species by having particular cuticular hood (vs absence of this structure in the new species), smaller body size and relatively longer tail in males (0.97 mm, tail length representing about 18.0% of body length vs 1.93–2.71 mm, tail length representing about 6.20–7.72% of body length) (Chabaud and Campana-Rouget 1955). Cosmocerca kalesari has weakly sclerotized spicules slightly shorter than gubernaculum and relatively shorter tail in females (tail length representing 8.04–10.6% of body length) (Rizvi et al. 2011), which is different from C. simile n. sp. (spicules well sclerotized, distinctly longer than gubernaculum; tail length representing 12.8–13.6% of body length).
Moravec and Sey (1990) originally described C. novaeguineae from Platymantis papuensis Meyer in Papua New Guinea. One of the present authors (Li. L) re-examined the type material of this species (Cat. No. N-387) deposited in Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic and concluded that the morphology and measurements of male of C. novaeguineae are very similar to that of the new species. However, C. simile n. sp. can be differentiated from C. novaeguineae by having well sclerotized spicules distinctly longer than gubernaculum (vs spicules atrophied, distinctly shorter than gubernaculum) and lateral alae extending to level of third pair of pre-cloacal plectanes in males (vs lateral alae extending to cloaca), and distinctly shorter tail in females (0.59–0.84 mm vs 0.31–0.39 mm). In addition, the new species was found in the Palaearctic region, but C. novaeguineae is distributed in Oceanian region. Consequently, we considered they are different species. Baker and Vaucher (1984) reported C. ornata from three species of frogs in Paraguay. Later, Moravec and Baruš (1990) considered that the material of C. ornata in Baker and Vaucher (1984) represented a new species (instead of C. ornata), and named Baker and Vaucher’s (1984) material as C. uruguayensis Moravec and Baruš 1990. However, the name of C. uruguayensis has been pre-occupated by C. uruguayensis Lent and Freitas, 1948. Thus Moravec and Kaiser (1994) proposed a new name, C. paraguayensis for it. Cosmocerca paraguayensis can be easily distinguished from the new species by having weakly sclerotized spicules slightly shorter than gubernaculum (vs well sclerotized spicules distinctly longer than gubernaculum), different morphology of tail in females (tail tapering abrumptly in about proximal fifth to a long caudal filament vs tail tapering abrumptly in bout proximal second to a long caudal filament) and different zoogeographical distribution (Neotropical region vs Palaearctic region).
The phylogenetic results based on the partial ITS and cox1 sequence data both supported that the new species and C. japonica have very close relationship. These results are consistent with the morphological data in which morphology and measurements of the new species are most similar to that of C. japonica (see the taxonomic remarks). However, the high level of interspecific nucleotide divergence was detected in ITS and cox1 sequences between C. simile n. sp. and C. japonica (2.74–3.37% and 10.2–13.5%, respectively), that indicated C. simile n. sp. and C. japonica represent different species.
The newly obtained genetic data of Cosmocerca species are very important for the further studies of the DNA-based taxonomy, population genetics and phylogenetics of the superfamily Cosmocercoidea.
References
Baker MR, Green DM (1988) Helminth parasites of native frogs (Leiopelmatidae) from New Zealand. Can J Zool 66:707–713
Baker MR, Vaucher C (1984) Parasitic helminths from Paraguay VI: Cosmocerca Diesing, 1861 (Nematoda: Cosmocercoidea) from frogs. Rev Suisse Zool 91:925–934
Bursey CR, Goldberg SR, Siler CD, Brown RM (2015) A new species of Cosmocerca (Nematoda: Cosmocercidae) and other helminths in Cyrtodactylus gubaot (Squamata: Gekkonidae) from the Philippines. Acta Parasitol 60:675–681
Chabaud AG, Campana-Rouget Y (1955) Helminths de la region de Banyule I. Nematode Parasites d’Amphibians Vie et Milieu 6:83–92
Chen H-X, Zhang L-P, Gibson DI, Lü L, Xu Z, Li H-T, Ju H-D, Li L (2018) Detection of ascaridoid nematode parasites in the important marine food-fish Conger myriaster (Brevoort) (Anguilliformes: Congridae). Parasite Vector 11:274
Falcón-Ordaz J, Windfield-Perez JC, Mendoza-Garfias B, Parra-Olea G, Perez-Ponce De Leon G (2007) Cosmocerca acanthurum n. sp. (Nematoda: Cosmocercidae) in Pseudoeurycea leprosa and Chiropterotriton orculus from the Transmexican Volcani Belt, Central Mexico, with a checklist of the helminth parasites of plethodontid salamanders. Zootaxa 1434:27–49
Floyd RM, Rogers AD, Lambshead PJD, Smith CR (2005) Nematode-specific PCR primers for the 18S small subunit rRNA gene. Mol Ecol Notes 5:611–612
González CE, Hamann MI (2008) Nematode parasites of two anuran species Rhinella schneideri (Bufonidae) and Scinax acuminatus (Hylidae) from Corrientes, Argentina. Rev Biol Trop 56:2147–2161
Kung CC, Wu HW (1945) Parasitic nematodes of amphibians from Pehpei, Szechwan, China. Sinensis 16:73–83
Lazarova SS, Malloch G, Oliveira CM, Hübschen J, Neilson R (2006) Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. J Nematol 38:404–410
Li L, Liu Y-Y, Zhang L-P (2012) Morphological and genetic characterization of Hysterothylacium zhoushanensis sp. nov. (Ascaridida: Anisakidae) from the flatfish Pseudorhombus oligodon (Bleeker) (Pleuronectiformes: Paralichthyidae) in the East China Sea. Parasitol Res 111:2393–2401
Li L, Du L-Q, Xu Z, Guo Y-N, Wang S-X, Zhang L-P (2014) Morphological variability and molecular characterisation of Dichelyne (Cucullanellus) pleuronectidis (Yamaguti, 1935) (Ascaridida: Cucullanidae) from the flatfish Pleuronichthys cornutus (Temminck & Schlegel) (Pleuronectiformes: Pleuronectidae) in the East China Sea. Syst Parasitol 87:87–98
Li L, Zhao J-Y, Chen H-X, Ju H-D, An M, Xu Z, Zhang L-P (2017) Survey for the presence of ascaridoid larvae in the cinnamon flounder Pseudorhombus cinnamoneus (Temminck & Schlegel) (Pleuronectiformes: Paralichthyidae). Int J Food Microbiol 241:108–116
Moravec F, Baruš V (1990) Some nematode parasites from amphibians and reptiles from Zambua and Uganda. Acta Soc Zool Bohemoslov 54:177–192
Moravec F, Kaiser H (1994) Description of Cosmocerca longispicula sp. nov. (Nematoda: Cosmocercidae), a parasite of a dendrobatid frog from Martinique, French Antilles. Parasitol Res 80:29–32
Moravec F, Sey O (1985) Some nematode parasites of frogs (Rana spp.) from North Viet Nam. Parasitol Hung 18:63–67
Moravec F, Sey O (1990) Some nematode parasites of frogs from Papua New Guinea and Australia. Acta Soc Zool Bohemoslov 54:268–286
Nadler SA, Hudspeth DSS (1998) Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): implications for morphological evolution and classification. Mol Phylogenet Evol 10:221–236
Rizvi AN, Bursey CR, Bhutia PT (2011) Cosmocerca kalesari sp. nov. (Nematoda, Cosmocercidae) in Euphlyctis cyanophlyctis (Amphibia, Anura) from Kalesar Wildlife Sanctuary, Haryana, India. Acta Parasitol 56:202–207
Rodrigues HO, Fabio SP (1970a) Nova especie do genero Cosmocera Diesing, 1861 (Nematoda, Oxyuroidea). Atas Soc Biol Rio de Janeiro 13:179–180
Rodrigues HO, Fabio SP (1970b) Contribuicao ao estudo do genero Cosmocera Diesing, 1861 (Nematoda, Oxyuroidea). Atas Soc Biol Rio de Janeiro 14:5–6
Sato A, Hasegawa H, Sekiya K, Tsubouchi T (2015) Is Cosmocerca (Nematoda: Cosmocercidae) parasitic in Japanese amphibians a single species? Japn J Vet Parasitol 14:7–12
Skrjabin KI, Shikhobalova NP, Lagodovskaya EA (1961) Oxyurata of animals and man. Part two, Translated from Russian. Israel Program for Scientific Translations, Jerusalem, 1974, pp. 460
Sou SK, Nandi AP (2015) On a new species of Cosmocerca (Nematoda; Cosmocercidae) from Microhyla rubra (Anura: Microhylidae) from West Bengal, India. Acta Parasitol 60:261–265
Sou SK, Sow KK, Nandi AP (2018a) Cosmocerca bengalensis sp. nov. (Nematoda: Cosmocercidae) in Hoplobatrachus tigerinus (Daudin, 1803) (Amphibia, Anura, Dicroglossidae) from West Bengal, India. Acta Parasitol 63:715–720
Sou SK, Sow KK, Nandi AP (2018b) Redescription of Cosmocerca ornata (Dujardin, 1845) Diesing, 1861 (Nematoda: Cosmocercidae) from Ranid frogs of West Bengal. India Proc Zool Soc 72:372–379. https://doi.org/10.1007/s12595-018-0279-6
Yamaguti S (1938) Studies on the helminth fauna of Japan. Part 23. Two new species of amphibian nematodes. Japn J Zool 7:603–607
Zhang K, Xu Z, Chen H-X, Guo N, Li L (2018) Anisakid and raphidascaridid nematodes (Ascaridoidea) infection in the important marine food-fish Lophius litulon (Jordan) (Lophiiformes: Lophiidae). Int J Food Microbiol 284:105–111
Zhao J-Y, Zhao WT, Ali AH, Chen H-X, Li L (2017) Morphological variability, ultrastructure and molecular characterisation of Hysterothylacium reliquens (Norris & Overstreet, 1975) (Nematoda: Raphidascarididae) from the oriental sole Brachirus orientalis (Bloch & Schneider) (Pleuronectiformes: Soleidae). Parasitol Int 66:831–838
Zhao W-T, Xu Z, Li L (2018) Morphological variability and molecular characterization of Mawsonascaris australis (Johnston & Mawson, 1943) (Nematoda: Ascaridoidea: Acanthocheilidae) from the brown guitarfish Rhinobatos schlegelii Müller & Henle (Rhinopristiformes: Rhinobatidae). J Helminthol 92:760–764
Zhu X, D’Amelio S, Paggi L, Gasser RB (2000) Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitol Res 86:677–683
Acknowledgements
The authors are grateful to Dr. František Moravec (Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Czech Republic), professor Hideo Hasegawa (Faculty of Medicine, Oita University, Japan), for providing important literature.
Funding
This study was supported by the National Natural Science Foundation of Hebei Province (C2019205094), the Support Program for 100 Excellent Innovative Talents of Hebei Province (SLRC2019033), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB26000000) and the Youth Top Talent Support Program of Hebei Province for Dr. Liang Li.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of Hebei Normal University as complying with the Animal protection law of the People’s Republic of China.
Additional information
Section Editor: Hiroshi Sato
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, HX., Zhang, LP., Feng, YY. et al. Integrated evidence reveals a new species of Cosmocerca (Ascaridomorpha: Cosmocercoidea) from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura). Parasitol Res 119, 1795–1802 (2020). https://doi.org/10.1007/s00436-020-06687-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-020-06687-3