Abstract
Nematodes of the genus Cosmocerca are commonly found in various amphibians in South Africa and in most cases are identified as C. ornata. However, after detailed morphological studies and molecular approaches, three new species of the genus were recently described from three different frogs in South Africa. In present study, we describe another new species – Cosmocerca goroensis parasitising the Northern Pygmy Toad Poyntonophrynus fenoulheti in Soutpansberg mountains, Limpopo province, South Africa. The new species is characterised by prominent sex dimorphism, wide lateral alae, numerous somatic papillae in both sexes, and wide triangularly shaped gubernaculum and simple prominent spicules in males. Cosmocerca goroensis n. sp. distinguished from congeners, previously reported in Southern Africa by the shape of the gubernaculum and arrangement of somatic papillae in males. Morphological differences were confirmed by molecular analysis based on fragments of the 28S gene. Phylogenetic analysis based on the 28S gene fragments, including C. goroensis n. sp. and newly obtained sequence of C. ornata from Pelophylax lessonae from Ukraine, supported previously known data of closer relationships between species of Cosmocerca and Aplectana and more distant with Cosmocercoides spp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cosmocercid nematodes are one of the most common parasites of anuran amphibians worldwide, yet one of the most understudied. In southern Africa, representatives of two genera namely Aplectana Railliet and Henry, 1916 and Cosmocerca Diesing, 1861 were previously reported. Until recently, the species C. ornata Dujardin, 1845 was the only representative of the genus reported from the Afrotropical realm (Baker, 1981). Harnoster et al. (2022) described three new species of Cosmocerca from three different frog species in South Africa, all of which were distinguished based on morphological differences (mainly structure of the gubernaculum) and molecular data of ITS-28S region.
Since wide usage of the molecular approaches for nematodes, representatives of Cosmocercidae were scarcely involved in phylogenetic studies of other groups with no specific studies on the interrelationships within the family. Recently, Chen et al. (2020, 2021a, 2021b) and Harnoster et al. (2022) provided phylogenetic analyses of several species of Cosmocercidae (including up to six species of Cosmocerca and three species of Aplectana) based on partial 28S and ITS sequences. Alcantara et al. (2022) provided phylogenetic analysis of six species of Cosmocerca and two species of Cosmocercoides based on partial cox1 sequences. Rebêlo et al. (2023) described a new species of Cosmocercoides from Brazil and also included it in the phylogenetic analysis based on cox1 gene marker. It should be noted that in most of those studies, authors used sequences of C. ornata collected from Hylarana spinulosa (Smith) (Anura: Ranidae) (GenBank MW326675 for 28S) and from Bufo gargarizans Cantor (Anura: Bufonidae) (GenBank MT108304 for cox1 and MT108302 for ITS) in China (Chen et al., 2020, 2021a, 2021b).
In the present study, we collected several specimens of Cosmocerca from the Northern Pygmy Toad Poyntonophrynus fenoulheti (Hewitt and Methuen) (Anura: Bufonidae) in South Africa which appeared to be clearly different from all previously known species within the genus. Additionally, we collected specimens of C. ornata from the Pool Frog Pelophylax lessonae (Camerano) (Anura: Ranidae) from Ukraine and obtained genetic markers of ITS region and partial 28S and cox1 gene for both species. The description of the new species, followed by phylogenetic analysis based on 28S gene fragments is presented herein.
Materials and methods
Specimens of P. fenoulheti were captured on a rocky North face mountain slope in one locality, namely Goro Nature Reserve in the Soutpansberg mountains (Limpopo Province, South Africa). Toads were collected in February 2020 (nine specimens) and February 2022 (five specimens). Specimen of P. lessonae was collected in the outskirts of Kyiv (Ukraine) in 2018. Amphibians were anaesthetised with tricaine ethyl-4-amin-obenzoate (MS222) and subsequently euthanised through cutting the spine and destroying the brain according to the standard operating procedure (NWU-00492–16-A5) and dissected. Nematodes were removed from intestine and rectum, washed in saline and fixed with hot 70% ethanol. For morphological examination nematodes were placed in distilled water for 10-30 minutes and then cleared in lactophenol for about 30 minutes. Cleared nematodes were studied as temporary mounts in lactophenol under the Zeiss Axio Imager M1 microscope equipped with a digital imaging system. Line drawings were made based on the series of photomicrographs.
In total, 69 nematode specimens were collected, of which 20 of C. goroensis n. sp. were measured. All measurements in the text are given in micrometers, unless stated otherwise and presented as ranges followed by the mean values in parentheses and measurements of holotype or allotype in square brackets.
For the DNA extraction anterior fragments of male specimens were used while posterior part was preserved for morphological identification. DNA was extracted using the ZYMO ZR tissue and insect DNA miniprep extraction kit (Zymo Research, USA) following the protocol recommended by the manufacturer. The ITS-28S region was amplified using the primer pair rift (5′-GCG GCT TAA TTT GAC TCA ACA CGG-3′) and 1500R (5′-GCT ATC CTG AGG GAA ACT TCG-3′) and the thermocycling profile following Tkach et al. (2014). Unpurified PCR products were sent to a commercial sequencing company, Inqaba Biotechnical Industries (Pty) Ltd (Pretoria, South Africa). PCR products were sequenced in both directions using pairs of PCR primers and additional internal primers: ITS4 (5′-TCC TCC GCT TATTGA TAT GC-3′), 300R (5′-CAA CTT TCC CTC ACGGTA CTT G-3′), ITS5 (5′- GGA AGT AAA AGT CGTAAC AAG G-3′) and ECD2 (5′-CTT GGT CCG TGT TTCAAG ACG GG-3′). The cox1 amplicons were obtained using the primer pair LCO1490 (50-GGT CAA CAA ATC ATAAAG ATA TTG G-30) and HCO2198 (50-TAA ACTTCA GGG TGA CCA AAA AAT CA-30) (Folmer et al., 1994) with the thermocycling profile as follows: 3 min denaturation at 94°C, 20 cycles of 94°C for 30 s, 45°C for 30 s, 72°C for 60 s and 40 cycles at 94°C for 30 s, 51°C for 60 s, 72°C for 60 s for amplification, 72°C for 10 min for final extension. Contiguous sequences were assembled and edited using Geneious Prime software (https://www.geneious.com) and submitted to GenBank.
For the phylogenetic analysis, the newly obtained sequences as well as those retrieved from GenBank were aligned using the ClustalW tool in the MEGA 11 software (Tamura et al., 2021) and trimmed. Prior to Bayesian Inference (BI) analysis, the GTR + G + I nucleotide substitution model was estimated as the best-fitting model using the same software. BI analysis was run using MrBayes v. 3.2.2 software with the Markov Chain Monte Carlo (MCMC) algorithm run for 8 million generations and using the default parameters. Maximum Likelihood (ML) was performed in Mega 11 with nodal support assessed using 1000 bootstrap pseudoreplicates.
Family Cosmocercidae Railliet, 1916
Subfamily Cosmocercinae Railliet, 1916
Genus Cosmocerca Diesing, 1861
Cosmocerca goroensis n. sp.
Type-host: Northern Pygmy Toad Poyntonophrynus fenoulheti (Hewitt and Methuen) (Anura: Bufonidae).
Type-locality: Goro Nature Reserve, Waterpoort, Limpopo Province, South Africa Coordinates: 22°57.522S, 29°25.68E.
Type-material: Holotype (NMBP 954), allotype (NMPB 955) and paratypes (NMBP 956-968) stored in the Parasitic Worm Collection, National Museum, Charles Street, Bloemfontein, South Africa.
Site in host: Intestine, rectum.
Etymology: The species is named after its type locality.
Infection parameters: Intensity – 1-33 (5.8), Prevalence – 92%, Abundance –5.3.
Representative DNA sequences: GenBank cox1 OR349766, ITS-28S OR211678.
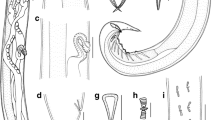
Cosmocerca goroensis n. sp., photomicrographs. A – male, ventral view; B – female, lateral view; C – anterior end of female, lateral view; D – posterior region of male, lateral view; E – male, transverse section at mid-body region; F – region of vulva, lateral view; G – posterior end of female, lateral view. Scale bars: A, B – 1 mm; C–G – 100 µm.
Description
General. Small stout nematodes with prominent sexual dimorphism, males at least twice shorter than females. Lateral alae in both sexes wide, prominent, beginning at level of anterior quarter of oesophagus and ending at level of anus in females and slightly anterior to cloaca in males (Fig. 2A, B, E). Numerous somatic papillae on body cuticle. Apical: small triangular mouth surrounded by three lips of which dorsal one with two prominent cephalic papillae and two subventral ones each with one cephalic papilla and prominent amphid; six minute outer papillae (Fig. 1B). Oesophagus with anteriorly differentiated pharynx, elongated cylindrical corpus, narrow undifferentiated isthmus and wide rounded bulb (Figs. 1A, 2C). Nerve ring encircling oesophagus at level of its anterior third. Excretory pore wide, prominent, opening at level of oesophageal bulb. Deirids not observed. Intestine and rectum straight, narrow.
Males. Measurements based on seven specimens. Body 1.4–2.2 (1.9) [2.0] mm long, 124–195 (167) [186] wide. Lateral alae 39–73 (55) [50] maximum wide, beginning at 76–232 (145) 92 from anterior end, ending at 336–517 (420) [388] from tail tip. Oesophageal pharynx 22–33 (30) [30] long, 19–24 (21) [22] wide. Oesophageal corpus and isthmus 224–308 (263) [271] long, 21–35 (26) [29] and 16–24 (21) [20] wide at corpus and isthmus level, respectively. Oesophageal bulb 58–83 (69) [78] long, 63–92 (76) [83] wide. Total oesophagus 309–421 (362) [379] long, spanning 16.8–21.5 (18.8) [18.6] % of body length. Nerve ring and excretory pore at 72–110 (86) [84] and 227–391 (317) [333] from anterior end of body. Tail 134–182 (155) [140] long, narrowing with blunt tip lacking any additional processes. Gubernaculum (Fig. 1E) triangular, with wide well sclerotized edges and less sclerotized plate in middle of proximal end. Edges of gubernaculum with uneven inner surface. Gubernaculum 102–138 (116) [114] long, comprising 5.5–7.3 (6.1) 5.6 % of body length, 27–57 (39) [57] wide in lateral projection. Spicules well visible, 57–71 (64) 64 long, simply-shaped, equal, located in gubernaculum groove (Fig. 1E). Ten identical plectanes arranged in two irregular rows present at posterior end of body (Figs. 1C, 2D). Each plectane bearing seven tubercles directed posteriorly (Fig. 1D). Two almost equal parallel rows, each of around 25 somatic papillae beginning at level of posterior third of body and extending to tail tip on ventral side of body (Fig. 1C). One pair of adcloacal papillae and one pair of enlarged papillae posterior to cloaca also present on ventral side (Fig. 1C).
Females. Measurements based on 13 specimens. Body 3.6–4.6 (4.1) [4.1] mm long, 328–421 (364) [370] wide. Lateral alae 47–79 (63) [55] wide, beginning at 122–289 (180) [235] from anterior end, ending at 282–699 (558) [594] from tail tip. Oesophageal pharynx 51–81 (62) [55] long, 34–48 (42) [34] wide. Oesophageal corpus and isthmus 347–470 (401) [405] long, 39–50 (46) [43] and 30–43 (36) [35] wide at corpus and isthmus level, respectively. Oesophageal bulb 106–133 (122) [118] long, 116–156 (139) [131] wide. Total oesophagus 544–658 (586) [578] long, spanning 13.0–15.6 (14.5) [14.2] % of body length. Nerve ring and excretory pore at 156–270 (201) [213] and 424–501 (451) [455] from anterior end of body. Tail needle-shaped (Fig. 2G), 556–763 (637) [645] long, 13.8–19.9 (15.9) [15.9] % of body length. Vulva transverse slit without prominent lips (Fig. 2F), located at 1.7–2.3 (2.0) [2.0] mm from anterior end of body, 45.7–50.1 (48.2) [48.2] % of body length. Both uteri directed posteriorly, filled with 4–57 (25) [24] eggs. Eggs (N=20) 154–199 (174) × 75–123 (100) in size, containing developed larvae.
Remarks
The new species belongs to the genus Cosmocerca due to the possession of plectanes and two weakly sclerotized spicules in males, presence of numerous papillae along the body and prominent sexual dimorphism with males usually less than a half the size of females (Baker 1981; Gibbons, 2010). Cosmocerca goroensis n. sp. can be easily distinguished from three recently described congeners from South Africa by the shape of the gubernaculum. Cosmocerca goroensis n. sp. has gubernaculum with wide Y-shaped well sclerotized edges and less sclerotized part on proximal end while C. monicae has narrow Y-shaped gubernaculum without less sclerotized plate, C. makhadoensis has V-shaped gubernaculum, and C. daly has club-shaped one. Additionally, the new species possesses 7 tubercles on each plectane (counted on plectanes on three specimens) while all three mentioned species have 5-6 tubercles on their plectanes. Moreover, males of C. goroensis have two parallel rows of somatic papillae that were not reported in other African Cosmocerca spp.
Cosmocerca ornata (Dujardin, 1845)
Fig. 3.
Cosmocerca ornata from Pelophylax lessonae photomicrographs. A – male, ventral view; B – male, lateral view; C – posterior end of male, ventral view; D – posterior region of male, ventral view, arrows indicate plectanes; E – anterior end of female, lateral view; F – posterior end of female, lateral view. Scale bars: A, B – 1 mm; C–F 100 µm.
Remarks
Dujardin (1845) described the species based on material from the green frog of the genus Pelophylax (reported as Rana esculenta) and the European Common Frog Rana temporaria (L.) (Anura: Ranidae) from France. Subsequently, C. ornata was reported from more than 50 amphibian and reptile hosts in Europe, Asia and Africa (Yildrimhan et al., 2009; Sou et al., 2019). The initial description of C. ornata was rather vague and no detailed redescription based on the type material or material collected from type host and type locality were made ever since. In present study, we collected nematodes from the pool frog P. lessonae in Ukraine. Morphologically, these nematodes correspond to the description of C. ornata by Dujardin (1845) and Travassos (19311931). Since our specimens are from the frog that is very closely related to the type host and are collected from Europe, we identify them as C. ornata. Representative DNA sequences of the collected specimens can be accessed under following GenBank numbers: cox1 OR350241; 28S OR211679.
Molecular analysis
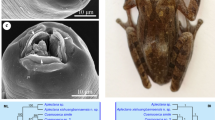
For the phylogenetic analysis, the final alignment composed of 738 nucleotides of 15 species of Cosmocerca, Aplectana and Cosmocercoides and Cruzia americana as an outgroup. The resulted tree (Fig. 4) based on both Bayesian Inference (BI) and Maximum Likelihood (ML) analyses showed three distinct clades: one composed by two species of Cosmocercoides plus Cosmocerca longicauda (most likely misidentified, see below), one composed by two species of Aplectana from China and one paraphyletic clade composed of nine species of Cosmocerca plus Aplectana macintoshii. In the latter clade C. ornata GenBank MW326675 (probably misidentified, see below) located basal to other paraphyletic clade formed by C. goroensis n. sp. as basal branch to seven other congeners and A. macintoshii. Cosmocerca ornata from Kyiv located in the same clade with three African Cosmocerca spp., Cosmocerca sp.1 from Hoplobatrachus rugulosus from China and A. macintoshii.
Pairwise analysis (Table 1) of the partial 28S gene showed that the new species differs in 2.5 % (18 nt.) – 5.0 % (37 nt.) from all available congeners. Pairwise comparison of C. ornata showed significant differences from sequences of C. ornata available in GenBank, comprising 2.8 % in 28S (MW326675) and 17.9 % (173 nt.) in ITS (MT108302) fragments.
Discussion
Harnoster et al. (2022) described three new species of Cosmocerca from South Africa and supposed that previous records of C. ornata in that region might be misidentified species. The morphological differences between C. goroensis n. sp. and other three South African species are mostly based on the shape of the gubernaculum. In our opinion, detailed studying of the gubernaculum (preferably dissected out of nematode) might reveal additional characters for morphological differentiation. Unfortunately, as same as for many other cosmocercids, we did not observe significant differences between females of C. goroensis n. sp. and other congeners from southern Africa.
To the date, in studied region all studied Cosmocerca spp. were found only in their type hosts. Expectedly, same was also observed in the present study, as the Northern Pygmy Toads were only found in rock pools on a rocky mountain slop, a habitat not shared with any other observed amphibians. In our opinion, host specificity of Cosmocerca might be much stricter than it was previously assumed. In the present study we obtained sequence of C. ornata from the green frog closely related to its type host and the sequence appeared clearly different from specimens obtained from H. spinulosa in China. We believe that Chinese specimens might belong to another species while specimens from the present study most likely belong to C. ornata. It is also possible that the real C. ornata may be specific to Ranids from Western Palaearctic, which can be illuminated after detailed studies of Cosmocerca from different amphibians from other regions on both morphological and molecular fronts.
In light of the available literature, the phylogenetic relationships between different genera of Cosmocecrcidae were studied based on concatenated 18S and 28S gene fragments, partial ITS and cox1 markers. Except for the cox1, all trees showed similar topologies with distinct clade of Cosmocercoides and closer relationships between Aplectana and Cosmocerca. In the present study we could use 15 different species that overlap obtained sequences in region of the 28S gene. Similar to previous studies, resulted tree showed three distinct clades composed of Cosmocerca, Aplectana and Cosmocercoides. The presence of Cosmocerca longicauda (GenBank OL468682) in the same clade with Cosmocercoides can probably be explained as a misidentification of the species. The sequence authors did not indicate the host and the paper was unpublished, though entitled “Morphological and Molecular Characterization of slug and snail parasitic nematodes”. Since Cosmocerca are rarely found in snails (unlike commonly found Cosmocercoides), we believe that the sequence most likely belongs to some misidentified species of Cosmocercoides. The two species of Aplectana from China form a clearly distinct clade while A. chameleonis from African frog located closed to two African species of Cosmocerca. Position of A. chameleonis close to Cosmocerca was also shown in previous studies (Chen et al., 2021b). It is clear that only further analyses based on longer sequences of more species of cosmocercids from different continents can resolve taxonomic and phylogenetic issues within this family.
References
Alcantara, E. P., Ebert, M. B., Müller, M. I., Úngari, L. P., Ferreira-Silva, C., Emmerich, E., Santos, A.L.Q., O’Dwyer, L. H. & Da Silva, R. J. (2022). First molecular assessment on Cosmocerca spp. from Brazilian anurans and description of a new species of Cosmocerca (Ascaridomorpha: Cosmocercoidea) from the white-spotted humming frog Chiasmocleis albopunctata (Boettger, 1885) (Anura: Microhylidae). Journal of Helminthology, 96 (e64), 1–8. https://doi.org/10.1017/S0022149X22000517
Baker, M. Rr. (1981). Cosmocercid nematode parasites from frogs of Southern Africa. Museum national d'Histoire naturelle, 24(1), 25–32. https://doi.org/10.4102/koedoe.v24i1.616
Chen, H.-X., Zhang, L.-P., Feng, Y.-Y. & Li, L. (2020). Integrated evidence reveals a new species of Cosmocerca (Ascaridomorpha: Cosmocercoidea) from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura). Parasitology Research, 119, 1795–1802. https://doi.org/10.1007/s00436-020-06687-3
Chen, H.-X., Gu, X.-H., Ni, X.-F. & Li, L. (2021a). Description of a new species of Aplectana (Nematoda: Ascaridomorpha: Cosmocercidae) using an integrative approach and preliminary phylogenetic study of Cosmocercidae and related taxa. Parasites & Vectors, 14:165. https://doi.org/10.1186/s13071-021-04667-9
Chen, H.-X., Ni, X.-F., Gu, X.-H., Sinch, U. & Li, L. (2021b). Morphology, genetic characterization and phylogeny of Aplectana dayaoshanensis n. sp. (Nematoda: Ascaridida) from frogs. Infection, Genetics and Evolution, 96, 1–9. https://doi.org/10.1016/j.meegid.2021.105123
Dujardin, D. N. (1845). Histoire naturelle des helminthes ou vers intestinaux. Paris. 658.
Folmer, O., Black, M., Hoeh, W., Lutz, R., & Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299.
Gibbons, L. (2010). Keys to the nematode parasites of vertebrates. Supplementary volume. CAB International, Oxon, 416. https://doi.org/10.1079/9781845935719.0003
Harnoster, F., Du Preez, L. & Svitin, R. (2022). Three new species of Cosmocerca Diesing, 1861 (Nematoda: Cosmocercidae) parasitising frogs Cacosternum boettgeri Boulenger, 1882, Kassina senegalensis Dumeril and Bibron, 1841 and Phrynomantis bifasciatus Smith, 1847 from South Africa. Parasitology Research, 121, 563–571. https://doi.org/10.1007/s00436-021-07390-7
Rebêlo, G. L., Santos, A. N., Travares-Costa, L. F. S., Dias-Souza, M. R., Müller, M. I., Jesus, R. F., Costa-Campos, C. E., Dos Santos, J. N. & Melo, F. T. V. (2023). Morphological and molecular characterization of Cosmocercoides amapari n. sp. (Nematoda: Cosmocercidae), parasitic in hylid frogs from the Brazilian Amazon. Parasitology, 150(3), 286–296. https://doi.org/10.1017/S0031182022001767
Sou, S. K., Sow, K. K. & Nandi, A. P. (2019). Redescription of Cosmocerca ornata (Dujardin, 1845) Diesing, 1861 (Nematoda: Cosmocercidae) from Ranid Frogs of West Bengal, India. Proceedings of the Zoological Society, 72, 372–379. https://doi.org/10.1007/s12595-018-0279-6
Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis version 11. Molecular Biology and Evolution, 38, 3022–3027.
Tkach, V. V., Kuzmin, Y., & Snyder, S. D. (2014). Molecular insight into systematics, host associations, life cycles and geographic distribution of the nematode family Rhabdiasidae. International Journal for Parasitology, 44(5), 273–284. https://doi.org/10.1016/j.ijpara.2013.12.005
Travassos, L. (1931) Note préliminaire sur les Cosmocercidae d’Europe. Compte rendu des Séances de la Société de Biologie, 107, 175–176.
Yildirimhan, H. S., Bursey, C. R. & Goldberg, S. R. (2009). Helminth parasites of the Caucasian parsley frog, Pelodytes caucasicus, from Turkey. Comparative Parasitology, 76, 247–257. https://doi.org/10.1654/4376.1.
Acknowledgements
The study was financially supported by Grant of the National Academy of Sciences of Ukraine for research laboratories/groups of young scientists of the National Academy of Sciences of Ukraine for conducting research in the priority areas of development of science and technology in 2022-2023. Roman Svitin and Yuriy Kuzmin wish to express their sincere thanks to the Armed Forces of Ukraine for the ability to continue research in Ukraine even during the full-scale Russian aggression.
Author information
Authors and Affiliations
Contributions
RS, TN, FH, YK and LdP collected amphibians and nematodes. RS, FH, TN and YK completed the morphological part of the species study. FH, TN and RS completed the molecular part of the study. All authors reviewed the text of the manuscript and images.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed. North-West University ethics approval no NWU-00380-16-A5.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Svitin, R., Kuzmin, Y., Harnoster, F. et al. Cosmocerca goroensis n. sp. (Nematoda: Cosmocercidae) from South Africa and its phylogenetic relationships with other cosmocercids based on partial 28S sequences. Syst Parasitol 100, 601–610 (2023). https://doi.org/10.1007/s11230-023-10109-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-023-10109-0