Abstract
Chagas disease, caused by the protozoan Trypanosoma cruzi, is a major parasitic disease that affects millions of people in America. However, despite the high impact of this disease on human health, no effective and safe treatment has been found that eliminates the infecting parasite from human patients. Among the possible chemotherapeutic targets that could be considered for study in T. cruzi are the DNA polymerases, in particular DNA polymerase beta (polß), which previous studies have shown to be involved in kinetoplast DNA replication and repair. In this paper, we describe the expression, purification, and biochemical characterization of the Miranda clone polß, corresponding to lineage T. cruzi I (TcI). The recombinant enzyme purified to homogeneity displayed specific activity in the range described for a highly purified mammalian polß. However, the trypanosome enzyme exhibited important differences in biochemical properties compared to the mammalian enzymes, specifically an almost absolute dependency on KCl, high sensitivity to N-ethylmaleimide (NEM), and low sensitivity to ddTTP. Immuno-affinity purification of T. cruzi polymerase beta (Tcpolß) from epimastigote extracts showed that the native enzyme was phosphorylated. In addition, it was demonstrated that Tcpolß interacts with some proteins in a group of about 15 proteins which are required to repair 1–6 bases of gaps of a double strand damaged DNA. It is possible that these proteins form part of a DNA repair complex, analogous to that described in mammals and some trypanosomatids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trypanosoma cruzi, the etiological agent of Chagas disease, causes pathologies affecting millions of people, principally in America. Transmission is mainly through hematophagous insect vectors (Brisse et al. 2000; Teixeira et al. 2006), although other transmission mechanisms exist, such as congenital infection (Jercic et al. 2010) and by ingestion of food contaminated with infective forms of the parasite (Ramírez et al. 2013). Due to the migration of infected persons, this disease is spread to non-endemic regions through transfusion and congenital transmission (Jannin and Villa 2007). The current etiological treatment is effective only during the acute phase and recently infected cases, but important side effects are produced (Coura and Castro 2002; Jannin and Villa 2007); the results of drug treatment during the chronic phase are controversial and very limited (Coura and Castro 2002; Jannin and Villa 2007). For this reason, the search for new chemotherapeutic targets to develop more efficient drugs is one of the strategies to control this disease. An important consideration in the study of this parasite is the particular biological and biochemical characteristics of its single and peculiar mitochondrion (kinetoplast), which is composed of a complex network of minicircles and maxicircles of DNA that are replicated by a unique mechanism (Ferguson et al. 1991). According to several studies, the kinetoplast plays a critical role in parasite locomotion, suggesting that its biochemical and molecular mechanisms would be favored as specific chemotherapeutic targets (Schamber-Reis et al. 2012). In kinetoplast replication, the DNA polymerase beta (polß) has a key function in DNA repair (Torri and Englund 1995; Saxowsky et al. 2003; Schamber-Reis et al. 2012). T. cruzi has a complex life cycle in both the vertebrate and the invertebrate hosts, where different parasitic forms exist, the epimastigote, amastigote, and trypomastigote, which are exposed to different oxidative stresses. In the invertebrate hosts, epimastigotes and metacyclic trypomastigotes coexist with a high Fe2+/Fe3+ burden, sufficient to generate radical oxygen stress (ROS) by the Fenton reaction (Graca-Souza et al. 2006). Amastigotes, on the other hand, coexist in the cytoplasm of colonized vertebrate cells of macrophages where ROS and reactive nitrogen stress (RNS) are frequent (Gupta et al. 2009; Piacenza et al. 2009).
T. cruzi has been divided into two major phylogenetic lineages, T. cruzi I (TCI) and T. cruzi II (TCII) (Souto et al. 1996; Momen 1999). Later, based on multilocus enzyme electrophoresis and random amplified polymorphic DNA (RAPD), TCII was further subdivided into five discrete phylogenetic clusters called discrete typing units (DTU) 2a–2e (Brisse et al. 2000). Several authors have postulated that the lineages DTU 2d and 2e are hybrids generated probably by ancestral genetic exchanges between lineages DTU 2b and DTU 2c (Westenberger et al. 2005; De Freitas et al. 2006). At present, the main lineage TCI is called TcI, and DTU 2a, DTU 2b, DTU 2c, DTU 2d, and DTU 2e are called TcIV, TcII, TcIII, TcV, and TcVI, respectively (Zingales et al. 2012). The T. cruzi CL Brener clone used in previous studies of T. cruzi polß belongs to the hybrid lineage TcVI (Schamber-Reis et al. 2012; Lopes et al. 2008). It is noteworthy that the genome sequencing of this clone confirmed not only that this parasite is predominantly diploid but also that the nuclear genome is formed by genes similar to those described for the T. cruzi Esmeraldo clone belonging to the TcII lineage (Esmeraldo haplotype) and similar to those described for TcIII clones (non-Esmeraldo haplotype) (El-Sayed et al. 2005a). Several studies have shown that the six lineages (TcI–TcVI) have different biological and genetic properties (Souto et al. 1996; Momen 1999; Brisse et al. 2000; Westenberger et al. 2005; De Freitas et al. 2006).
Polß of mammals has been extensively studied (Burgers 1998; Lindhl and Wood 1999; Wilson 2000). This enzyme has a very important role in the base excision repair system of nuclear DNA (Jannin and Villa 2007; Ramírez et al. 2013). Also, the tertiary structure of rat and human polß as well as their different biochemical functions have been thoroughly investigated (Davies et al. 1994; Pelletier et al. 1994; Sawaya et al. 1994; Lindhl and Wood 1999; Wilson 2000). Although the enzyme is composed of only one low molecular mass polypeptide chain, it has four domains and several enzymatic activities: DNA-dependent DNA synthesis, single-stranded DNA binding activity, 5′-phosphate recognition activity in gapped DNA structures, and a 5′-deoxyribose phosphate (dRP) lyase activity (Matsumoto and Kim 1995; Burgers 1998; Prasad et al. 1998; Lindhl and Wood 1999). Other homologues to mammalian polß have been found in recent years, stimulating great interest in the real physiological function of these enzymes (Torri and Englund 1992, 1995; Taladriz et al. 2001). In lower eukaryotes, however, a very particular situation has been described. Some years ago, a mitochondrial polß was described in the kinetoplastid protozoan Crithidia fasciculata (Torri and Englund 1992, 1995). A nuclear polß enzyme was characterized in another trypanosomatid, Leishmania infantum, suggesting that it may be involved in parasite drug resistance (Taladriz et al. 2001; Ramiro et al. 2002). Two mitochondrial polßs were described subsequently in Trypanosoma brucei (Saxowsky et al. 2003). More recently, the genome projects of the three most medically relevant trypanosomatids, T. cruzi, T. brucei, and Leishmania major were published (El-Sayed et al. 2005a; b; Ivens et al. 2005). All of the deduced amino acid sequences for these polßs have putative mitochondrial presequences. These genomic studies confirmed the previous facts revealed from the analysis of T. brucei, showing that the paralog sequence named DNA polß-PAK is found near the polß gene (Lopes et al. 2008; El-Sayed et al. 2005a). In addition to polß and polß-PAK, five other mitochondrial DNA polymerases (pols) have been identified in trypanosomatids (Klingbeil et al. 2002; Chandler et al. 2008; Rajao et al. 2009; Bruhn et al. 2010). Among these enzymes, there are four DNA pol homologues of the bacterial DNA pol I (POLIA, POLIB, POLIC, and POLID) and one homologue to the eukaryotic DNA pol kappa (POLK). Studies using RNA interference methodology (iRNA) performed in T. brucei have shown that these enzymes play essential roles in different stages of kinetoplast DNA (kDNA) replication (Klingbeil et al. 2002; Chandler et al. 2008; Bruhn et al. 2010). In relation to POLK, studies performed in T. cruzi cells overexpressing the recombinant gene Tcpolκ have shown that this enzyme has the capacity to overcome oxoguanine lesions of DNA, and the parasites increase their resistance to hydrogen peroxide exposition (Rajao et al. 2009).
The polß gene from the T. cruzi TcVI lineage CL Brener clone (Lopes et al. 2008) was expressed as fusion peptides of polß with maltose-binding protein (MBP); this study demonstrated that the enzyme has DNA polymerase and dRPlyase activities, and it is inhibited by NaCl (0–100 mM) and has an optimal pH of about 8.0 (Lopes et al. 2008). A later report based on transfection of T. cruzi parasites with an expression vector containing the full-length polß gene from the T. cruzi CL Brener clone demonstrated that parasites overexpressing polß showed increased survival in a range of hydrogen peroxide concentrations, suggesting that polß contributed to the repair of oxidative lesions of kDNA and improved the efficiency of base excision repair (Schamber-Reis et al. 2012). However, on those in vitro studies with the fusion peptide of MBP-T. cruzi polymerase beta (Tcpolß), the recombinant enzyme could not perform base excision repair or overcome DNA lesion with 8-oxodG (Lopes et al. 2008).
To obtain more information about the biochemical and physiological role of T. cruzi DNA polymerases, some years ago, we carried out the purification and characterization of DNA polymerases (Rojas et al. 1992; Venegas and Solari 1995; Venegas et al. 2000) and the cloning and characterization of a polß gene from the TcI Miranda clone of this parasite (Venegas et al. 2009). In the present manuscript, we present the expression of this gene in a highly efficient bacterial expression vector, purification of the enzyme, and a study of its biochemical features.
Materials and methods
Cloning of recombinant polß from T. cruzi
The complete ORF was amplified by PCR using a high-fidelity polymerase (Invitrogen, USA). The template was the recombinant plasmid pUC-7-2 containing the Tcpolß Miranda clone, described previously (Venegas et al. 2009). The primers used in the PCR were F: 5′ CCCATATGTTTCGTCGCACGTTCTGG 3′ and R: 5′ CCGGATCCTTAGGGGTCGCGGTTTTCCG 3′. The PCR amplification was performed according to the following program: initial denaturation, 5 min at 94 °C; 30 cycles of 30 s of denaturation at 94 °C, annealing of 30 s at 55 °C, and extension period of 1 min at 72 °C; and final extension of 10 min at 72 °C. The PCR products were inserted in pGEM-T Easy vector (Promega, USA). The cloned sequence was evaluated by sequencing. The cloned ORF was excised by NdeI/BamHI digestion, and the product was purified from the gel and inserted into NdeI/BamHI sites of the expression vector pET15b (Novagen, USA). Bacterial DH5α cells were transformed with the expression vector containing the ORF of polß (pET15b-pol), and positive colonies were examined by PCR screening and sequencing.
Expression and purification of recombinant Tcpolß
To produce recombinant protein, bacterial BL21 (DE3) cells were transformed with pET15b-pol vector and grown in Luria-Bertani agar plates overnight at 37 °C. One single colony was inoculated overnight at 37 °C in 5 ml of Terrific broth medium supplemented with 1 μg/ml ampicillin. The next day, the pre-inoculum was added to 500 ml of Terrific Broth medium and incubated to an A600 nm of 0.8. Protein induction was performed by adding 0.5 mM isopropyl 1-thio-ß-d-galactopyranoside (IPTG); cells were cultured for 3 h at 37 °C. Cells were harvested by centrifugation and suspended in 20 ml STE buffer (100 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA). The buffer was supplemented with 0.35 mg/ml lysozyme and incubated at room temperature for 30 min. The cell suspension was sonicated and insoluble bodies with the recombinant protein were processed as previously described (Tamayo et al. 2004). Briefly, insoluble bodies were solubilized and suspended in 40 ml of 6 M guanidinium hydrochloride, 10 mM HEPES pH 7.9, and 1 mM 2-mercaptoethanol and incubated overnight. Then denatured proteins were diluted with 160 ml of 10 mM HEPES pH 7.9, 5 mM 2-mercaptoethanol, and 20 % glycerol. The next day, the proteins were dialyzed against buffer containing 20 mM Tris-HCl pH 7.6, 10 % glycerol, 0.5 M KCl, 0.01 % Triton X-100, 20 mM imidazole, 1 mM 2-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The induced protein was first purified by Ni-NTA, but binding to the resin was null. Recombinant protein solution was dialyzed again to reduce imidazole and KCl and to replace 2-mercaptoethanol by dithiothreitol (DTT), using the same buffer but with 1 mM DTT, 50 mM KCl, and no imidazole. Two steps of ionic exchange were used to purify the recombinant polymerase. First, the extracts containing the recombinant proteins were loaded onto phosphocellulose (P11) resin (Whatman, USA), equilibrated with BC50 buffer (20 mM Tris-HCl pH 7.6, 50 mM KCl, 1 mM EDTA, 0.1 mM PMSF). Protein fractions were eluted with BC500 (as BC50 but with 500 mM KCl). Positive fractions were dialyzed against hydroxyapatite (HY) buffer (20 mM sodium phosphate pH 7.5, 50 μM EGTA, 50 μM EDTA, 1 mM DTT, 20 % glycerol, 0.1 mM PMSF) and loaded onto hydroxyapatite resin equilibrated with HY buffer. Proteins were eluted with a linear gradient from 50 to 500 mM sodium phosphate prepared in HY buffer. The yield of Tcpolß purification was 45–50 mg protein obtained from 500 ml of recombinant bacteria culture transformed with the respective recombinant plasmid.
Protein fractions were evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot using a monoclonal mouse his-tag antiserum (Invitrogen, USA) and colorimetric development using nitroblue tretrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Bio-Rad, USA). SDS-PAGE gels were analyzed using the conventional methods of Coomassie blue and silver staining. Fractions containing recombinant protein were dialyzed against polymerase storage buffer (20 mM Tris-HCl, pH 8.0, 80 mM KCl, 1 mM EGTA, 50 μM EDTA, 1 mM DTT, 20 % glycerol, 0.1 mM PMSF). Polß aliquots were stored at −80 °C until used in kinetic assays.
DNAse I-activated calf thymus DNA preparation
One milliliter of a reaction mix (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 5 mM KCl, 10 mg bovine serum albumin (BSA)) containing 500 mg calf thymus DNA and 4 μg pancreatic DNAse (Worthington) was incubated for 30 min at 30 °C. DNAse inactivation was performed by incubation for 15 min at 75 °C. The reaction mix was slowly cooled to 20 °C over a period of 30 min. DNA was precipitated by adding 0.1 M sodium acetate and four volumes of cold ethanol. The reaction mix was incubated for 2 h at −80 °C. The DNA was recovered by centrifugation and washed with 80 % ethanol. Finally, the activated DNA was suspended in 500 μl 30 mM Tris-HCl pH 7.5 and 30 mM NaCl and stored at −20 °C until used. The quality of the activated DNA was determined using DNA synthesis assays with Klenow fragment and αdATP32 incorporation.
Enzyme assays
Polymerase assays were performed by adding 5 μl of the hydroxyapatite fractions to a mix containing reaction buffer (40 mM Tris-HCl pH 8.0, 10 mM DTT, 1 mM MnCl2, 40 mM KCl, 1 mM EGTA, 0.2 mg/ml BSA, 40 mg/ml DNAse I-activated calf thymus DNA, 500 μM dNTP mix, 5 μM dATP, and 3.32 nM of α-dATP32) (3000 Ci/mmol, Perkin Elmer, USA) obtaining a specific activity of 438.24 cpm/pmol in a total volume by assay of 50 μl. Incubations were performed at 37 °C for 30 min. Total reaction volumes were put onto pieces of DE81 paper (Whatman, USA) and incubated for 15 min at 25 °C. Papers were washed three times with 0.3 M Na2HPO4 pH 8.0 to eliminate non-incorporated radionucleotide and dried at 25 °C, and labeled nucleotide incorporation was measured in a scintillation counter. Data were plotted and enzymatic activity was determined. In titration polymerase assays, different amounts of enzyme were added to the reaction mix using 80 mM KCl and 40 mg/ml activated DNA or polydT-oligodA as substrate. Titration assays were performed using the same conditions. In N-ethylmaleimide (NEM) assays, the recombinant polymerase was dialyzed in the storage buffer without DTT. Specific enzymatic activity was defined as units/mg protein; one unit was defined as the incorporation of 1 nmol of deoxynucleoside triphosphate into acid-insoluble material in 60 min at 37 °C.
Mass spectrometry analysis of T. cruzi polß
The putative recombinant protein was analyzed by mass spectrometry. After SDS-PAGE, the polymerase band was excised from the gel and sent to the Pasteur Institute (Montevideo, Uruguay) for analysis. The resulting peptides were sequenced and identified by liquid chromatography coupled to mass spectrometry. All the peptides identified correspond to T. cruzi polß.
Antibody generation
Two rabbits were immunized subcutaneously with 1 mg of the recombinant protein prepared in complete Freund’s adjuvant; after 21 days, they were inoculated again with 500 μg of the recombinant protein prepared in incomplete Freund’s adjuvant. After 14 days, rabbits were inoculated again with 500 μg of the recombinant protein prepared in PBS buffer by intraperitoneal injection. Blood and serum were obtained from these animals 1 week later and stored at −20 °C until antiserum purification. For antibody purification, a Ni-NTA resin (Invitrogen, USA) was packed and equilibrated with S buffer (6 M guanidinium hydrochloride, 50 mM Tris-HCl pH 8.0). All the procedures with the rabbits and molecular methods were approved by the bioethics committee of our institution.
Recombinant DNA ß polymerase was prepared from inclusion bodies and solubilized with S buffer. Six milligram of this protein fraction was incubated with 3 ml of the resin suspension. Unbound material was washed first with a washing buffer (8 M urea and 20 mM Tris-HCl pH 8.0) and then with the same buffer but pH = 5.9. Finally, the resin was equilibrated with Tris-buffered saline (TBS) buffer (10 mM Tris-HCl pH 7.5, 200 mM NaCl). Three milliliter of serum was loaded onto the column and unbound material eliminated with TBS buffer washing steps until no protein was detected by the Bradford method. Purified antibodies were eluted with 5 M MgCl2 and collected fractions of 750 μl were analyzed to detect proteins. Positive fractions were dialyzed against PBS buffer supplemented with 10 % glycerol and 2 mM ß-mercaptoethanol overnight at 4 °C. Purified antibody yield was 0.5 mg/ml of serum. Western blots were performed with the purified antibody using 0.1 μg/ml as working final protein concentration.
The specificity of affinity-purified antibodies was assessed by Western blot against homologous and heterologous extracts (Fig. S1). As seen in the figure, these antibodies detected a band of approximately 50 kDa only in the extract of epimastigote and pure recombinant Tcpolß. No reaction against other heterologous extracts or against T4 DNA pol (lane 6) or the Klenow fragment (lane 7) was detected.
Preparation of T. cruzi protein extracts
T. cruzi epimastigote cells (2 × 108) were washed three times with PBS and suspended in lysis buffer (50 mM Tris-HCl pH 7.8, 1 mM DTT, 1 mM EDTA, 1 M KCl, 1.5 % NP-40, 0.1 % Triton X-100, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 1 mM PMSF, 10 % glycerol, and phosphatase inhibitor cocktail (Roche)). The cells were incubated for 10 min at 4 °C and then were centrifuged at 12,000g for 15 min at 4 °C. Protein extracts were dialyzed against 20 mM Tris-HCl pH 7.8, 50 mM KCl, 1 μg/ml pepstatin A, 1 μl/ml leupeptin, 0.5 mM PSMF, 20 % glycerol, 1 mM EDTA, 1 mM EGTA, and 2.5 mM DTT.
Purification of Tcpolß-associated proteins
Purified Tcpolß antibody was covalently bound to Sepharose 4B resin activated with cyanogen bromide (Sigma-Aldrich, Germany) to a final concentration of 1 mg/ml resin according to manufacturer instructions. The resin was incubated overnight with the antibody and then was washed and blocked by the addition of 0.5 M Tris-HCl pH 7.8 during 3 h at 4 °C. Later, the resin was washed with 0.2 M glycine pH 2.5 and quickly neutralized by the addition of 0.5 M Tris-HCl pH 7.8. The resin was washed with equilibrium buffer (20 mM Tris-HCl pH 7.8, 50 mM KCl, 0.5 mM DTT, 10 % glycerol, 0.1 mM EDTA, 0.15 % NP-40). A volume of 200 μl of resin was incubated for 3 h at 4 °C with 600 μl of T. cruzi protein extract (3 mg) prepared in equilibrium buffer supplemented with protease inhibitor cocktail (Roche). The resin was then washed with 4 ml of equilibrium buffer supplemented with 150 mM KCl. The resin was eluted with the addition of 600 μl of equilibrium buffer with the salt replaced by 1.2 M NaCl. After elution, the resin was washed with 4 ml of same elution buffer, and then 600 μl of 0.2 glycine pH 2.5 was added to elute native Tcpolß.
Phosphoprotein analysis
The native Tcpolß fraction eluted in a volume of 600 μl by 0.2 M glycine pH 2.5 from the purified Tcpolß antibody covalently bound to Sepharose 4B column was precipitated by adding 600 μl 20 % trichloroacetic acid. Proteins were collected by centrifugation and washed with 80 % ethanol. The dried pellet was resuspended in 20 μl of Laemmli sample buffer supplemented with 1 μl of 2 M Tris base. Precipitated proteins were subjected to SDS-PAGE and transferred to a polyvinylidene fluoride membrane, which was methanol activated and blocked with boiled 3 % gelatin. Membranes were incubated for one night with rabbit anti-phosphoSer-Thr-Tyr (Abcam, USA) in a dilution of 1/100. Alkaline phosphatase-conjugated secondary antibody (Promega, USA) was used at a 1/7000 dilution, incubating for 30 min. The membranes were developed using NBT/BCIP (Bio-Rad, USA) according to the manufacturer’s instructions.
DNA repair assays
Protein fractions eluted with 1.2 M NaCl were concentrated fourfold by dialyzing against 20 mM Tris-HCl pH 7.8, 50 mM KCl, 0.1 mM EDTA, 0.1 mM PMSF, 60 % glycerol, and 0.5 mM DTT. This fraction was assayed with or without the addition of Tcpolß using the DNA polymerase assay short gap (1–6 bases) filling kit (ProFoldin, USA) according to the manufacturer’s instructions. The fluorescence measurements were carried out in a nanoquant plate (Tecan, Infinite 200 PRO NanoQuant).
Statistical analysis
All assays were repeated three times. In inhibition assays, data were analyzed by two-way ANOVA and Sidak’s multiple comparison test using Prism 6.0 (Graph Software, USA). Differences were considered significant at p < 0.05.
Results
Expression and purification of DNA polymerase ß from T. cruzi
Recombinant polß was purified from insoluble bodies accumulated in BL21 (DE3) bacterial cells transformed with pET15b-pol vector and induced with IPTG. The lysate was diluted and dialyzed to eliminate guanidinium hydrochloride in order to refold the enzyme. Enzymatic activity of this protein fraction was null. The proteins were fractioned by phosphocellulose (P11) and HY resins for further purification. The purification scheme is shown in Fig. 1a. The fractions containing recombinant protein were identified by SDS-PAGE and immunoblotting using his-tag antiserum (Fig. 1b, c). DNA polymerase activity of the P11 fraction was null despite the purity of the fraction (Fig. 1B). The HY fraction showed clearly the presence of a protein band with the expected molecular weight (50 kDa) (Venegas and Solari 1995) and a low molecular weight contaminant (12 kDa) (Fig. 1b, c). This contaminant corresponds to endonuclease A (end A), according to our experience which appears to be a common contaminant present in inclusion body recombinant proteins. The putative recombinant protein band was excised from an SDS-PAGE stained with Coomassie blue and further analyzed by MALDI-TOF-TOF. The peptides produced achieved 49 % coverage of the polß sequence (Venegas et al. 2009) was the best match. The polymerase activity of the HY fractions was evaluated, but only two fractions showed high activity (fractions 14 and 16), while another fraction with high amounts of recombinant proteins showed low activity (fraction 12) (Fig. 1d). Fractions 10, 18, and 20 showed low activity compared to fractions 14 and 16. The polymerase activity observed in the HY fractions was performed by the recombinant protein and was not due to other polymerase activities such as DNA polymerase I from E. coli, which is a 100-kDa molecular weight polypeptide; no such protein appeared in the silver-stained gel (Fig. 1c). In addition, recombinant proteins in native conditions were unable to bind to the Ni-NTA resin despite the his-tag of the pET15b expression vector, as shown by immunoblotting (Fig. 1b, bottom). In urea-denaturing conditions, the recombinant enzyme was able to bind to the Ni-NTA resin but did not achieve appropriate folding (not shown). The recombinant polß with the highest enzymatic activity (fraction 14) was used in further assays.
Purification of recombinant DNA polymerase ß from T. cruzi. a Sequential steps of recombinant polymerase purification, which begin with BL21 whole cell extracts (WCE) containing expressed recombinant protein. WCE was purified using a phosphocellulose resin and the 500 mM KCl eluate was passed through a hydroxyapatite resin. The eluates were obtained by linear gradient of sodium phosphate (50 to 500 mM) and analyzed to identify the recombinant polymerase fractions. b Coomassie blue-stained SDS-PAGE showing the hydroxyapatite fractions (1 to 22). Arrow indicates recombinant DNA polymerase ß. NI indicates non-induced protein extract, IN indicates induced protein extract, and I indicates the phosphocellulose eluate. Asterisk indicates a DNAse contaminant. Bottom shows a Western blot against his-tag epitope. c Silver-stained SDS-PAGE showing the purification steps of recombinant ß polymerase. Arrow indicates the migration of recombinant DNA ß polymerase. NI indicates non-induced extracts, S corresponds the soluble phase, P corresponds the insoluble phase, P11 corresponds to the phosphocellulose eluate, and HY indicates fraction 14 of hydroxyapatite purification step. d Activity graph of hydroxyapatite fractions. Polymerase activity is indicated as picomoles corresponding to the αdATP radionucleotide incorporation to a DNAse-activated DNA (see “Materials and methods”). Klenow fragment activity is shown as positive control
Recombinant Tcpolß enzymatic activity with two substrates
DNA and synthetic polydT-oligodA nucleic acid were used to evaluate the enzymatic activity of the recombinant Tcpolß enzyme. The first substrate was prepared using DNAse I partially digested calf thymus DNA. Increasing amounts of recombinant protein were added to the assays and radiolabeled nucleotide incorporation was quantified. The increase of polymerase activity was proportional to the increase of recombinant protein and this activity was not dependent upon the substrate. The magnitude of the activity was high if activated DNA (data not shown) was used as substrate instead of polydT-oligodA.
As positive control, the activities of recombinant Tcpolß and Klenow fragments (KF) were compared. With activated DNA, the KF showed an increase of 17-fold, whereas with synthetic DNA, KF showed an increase of only 1.4-fold, in relation to the activity found with the recombinant Tcpolß fraction. These results, together with the silver-stained SDS-PAGE (Fig. 1c), confirm that the enzymatic activity observed in all the assays is due to recombinant Tcpolß and is not an intrinsic bacterial DNA polymerase activity.
KCl, manganese, and magnesium ions modulate the enzymatic activity of Tcpolß
The enzyme activity of Tcpolß is affected by many factors, including salt and bivalent cations. To study the effect of salt and cations, several assays were performed adding increasing amounts of KCl (Fig. 2a), MnCl2 (Fig. 2b), and magnesium acetate (MgAc) (Fig. 2c). In all cases, the enzymatic activity was modulated, showing a peak activity interval between 80 and 160 mM KCl, 1–4 mM MnCl2, and 10–15 mM magnesium acetate, using activated DNA as substrate.
Salts and pH modulate recombinant DNA polymerase ß activity. Polymerase activity graphs of different assays with recombinant polß (500 ng) and DNAse-activated DNA as template in the presence of increased amounts of a KCl (0, 10, 20, 40, 80, 160, and 320 mM), b MnCl2 (0, 0.5, 1, 2, 4, 8, and 16 mM), and c magnesium acetate (MgAc) (0, 2.5, 5, 10, 15, 20, and 40). d Polymerase assays were performed at different pH conditions: 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, and 9.0. In all graphs, the αdATP radionucleotide incorporation is expressed as picomoles and each point is the result of three different experiments
pH dependence of DNA polymerase activity
The recombinant Tcpolß enzyme was evaluated in different pH values from acidic (pH = 5.0) to alkaline (pH = 9.0). The peak of its activity was near pH = 7.5 (Fig. 2d).
ddTTP and NEM inhibit the enzymatic activity of recombinant Tcpolß
DNA polymerases are inhibited by the incorporation of dideoxy-nucleotide triphosphates (ddNTP) in the active sites, stopping polymerization. We asked whether recombinant polß could be inhibited by the addition of ddTTP to the assays compared to the Klenow fragment. We observed that the inhibitory effect was greater for Tcpolß; 25 μM ddTTP inhibited more than 50 % of the initial activity (Fig. 3a).
Inhibition of the polymerase activity of recombinant DNA ß polymerase. Polymerase activity graphs of different assays with recombinant DNA ß polymerase (500 ng) and DNAse-activated DNA as template in presence of increased amounts of a dTTP and b NEM, classical polymerase inhibitors. As control, Klenow fragment activity is shown in each graph. In all graphs, the αdATP radionucleotide incorporation is expressed as percentage of total incorporation of dNTP without any inhibitor, and each is the result of three different experiments. Data were analyzed by two-way ANOVA and Sidak’s multiple comparison test. Double asterisk indicates significant differences (p < 0.05)
Other compound important to characterize biochemically the DNA polymerases is NEM, used to alkylate and covalently modify nucleophilic thiol residues on proteins, which was used at 0–40 mM (Fig. 3b). The Tcpolß fraction was dialyzed to eliminate DTT from the medium. With 5 mM, the activity of the recombinant enzyme was reduced to 25 % of the initial activity (Fig. 3b). The Klenow fragment was insensitive to the effect of NEM. These results show the effect of NEM on the thiol residues present in Tcpolß that are involved in catalytic activity. Additionally, these results confirm that the DNA polymerase activity described in this study is due to recombinant Tcpolß and not to a bacterial-fragmented DNA polymerase I activity, which is not inhibited by NEM.
Native polß is detected in T. cruzi extracts with recombinant protein antiserum
Recombinant protein was used as immunogen in rabbits and the antibody obtained was evaluated. As is shown in Fig. 4, purified antibody is able to recognize two protein bands in epimastigote extracts with high sensitivity, since detection is observed with 0.5 μg of extract or more (Fig. 4). One of the protein bands detected by the antibody migrated with higher molecular weight than the recombinant protein, indicating the possibility of post-translational modifications. The other protein band could be a proteolysis product.
Native Tcpolß is found as phosphorylated form
We purified the native enzyme to identify post-translational modification of Tcpolß using an affinity resin. For this, we generated a resin containing anti-Tcpolß bound covalently (see “Materials and methods”). The resin was incubated with acidic buffer to elute Tcpolß, which was dialyzed and analyzed by immunoblot. As seen in Fig. 5, using an anti-phosphoprotein antibody, we detected a phosphorylated form of Tcpolß compared to the enzyme treated with alkaline phosphatase.
T. cruzi polß in a phosphorylated form. a Immunoblot using anti-phospho protein. Lane 1: Recombinant Tcpolß. Lane 2: Eluate from affinity column constructed with IgG (control). Lane 3: Native Tcpolβ eluted from affinity column constructed with anti-Tcpolß antibody. Lane 4: Native Tcpolß from affinity column treated with alkaline phosphatase. Lane 5: Native Tcpolß from affinity column treated only with alkaline phosphatase buffer. b Immunoblot using anti-Tcpolß. Lane 1: Recombinant Tcpolß. Lane 2: Eluate from affinity column constructed with IgG (control). Lane 3: Native Tcpolß eluted from affinity column constructed with anti-Tcpolß. Lane 4: T. cruzi protein extract
T. cruzi polß needs additional factors to repair DNA gaps
To investigate DNA repair features of Tcpolß, we performed in vitro assays using a polß assay kit (ProFoldin). The substrate was a DNA with gaps and repair was measured by fluorescence. T. cruzi protein extract showed repair activity but recombinant Tcpolß alone did not. However, we observed repair activity in a dose-dependent manner when purified Tcpolß-associated proteins were mixed with recombinant Tcpolß. In contrast, Tcpolß-associated proteins alone did not repair DNA gaps. As control, we mixed the recombinant Tcpolß with a control protein eluted from a resin containing IgG instead of anti-Tcpolß. The Klenow fragment was also assayed and did not repair DNA gaps (Fig. 6).
Tcpolß requires additional factors to repair gaps in DNA templates. a Plot showing the incorporation of fluorescence-labeled nucleotides in a repair assay. Affinity eluate corresponds to proteins from the elution with high salt of an affinity column constructed with anti-Tcpolß. Control eluate corresponds to proteins from the elution of an affinity column constructed with control IgG. Polß indicates the recombinant enzyme. b Silver-stained SDS-PAGE showing the eluted extracts from affinity columns. Lane 1: Molecular weight standard. Lanes 2–7: Tc protein extract (30, 20, 10, 5, 2.5, and 1.25 μg). Lane 8: High-salt elution from anti-Tcpolß affinity column. Lane 9: High-salt elution from IgG control affinity column
Phylogenetic analysis of recombinant T. cruzi Miranda polß
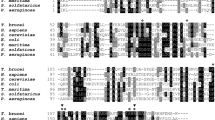
A phylogenetic tree is shown in Fig. 7, based on the nucleotide sequence of the ORF encoding polß of various species of organisms. It is observed that the sequences of T. cruzi are grouped into three monophyletic nodes corresponding to TcI lineage, the TcI non-Esmeralda haplotype, and TcII. It is noteworthy that the TcI group, where the recombinant polß described in this manuscript is located, has a high bootstrap support (90 %). Both sequences of the CL Brener clone (TcVI) grouped in other nodes. The sequences are grouped according to topologies previously described in the literature (File et al. 2002).
a Phylogram and b cladogram of DNA polymerase ß orthologue genes. The phylogenetic trees were constructed using the open reading frame nucleotide sequences published in the GenBank. The sequence codes for the T. cruzi polß sequences are Tc-Dm28c (TcI Dm28cclone, gb|AYLP01000037.1), Tc-Miranda (TcI Miranda clone, gi 71666010), Tc-JRcl4 (JR-cl4 clone, gb|AODP01000717.1), Tc-CLB-N-Hap (TcVI CL Brener clone haplotype non-Esmeraldo, gb|AAHK01000070.1|:20532–21826), Tc-Tula (TcVI Tula clone, gb|AQHO01018169.1|:123–1417), Tc-CLB-E-Hap (TcVI CL Brener clone haplotype Esmeraldo, gb|AAHK01000480.1|:1629–2925), Tc-Esm (TcII Esm clone, gb|ANOX01000503.1|:1035–2330), and Tc-marinkellei (gb|AHKC01018446.1|:1295–2596). The GenBank codes for the other polß sequences are C-fasciculata (gi|33356562), X-laevis (gi|2661841), Human (gi|4505930), rat (gi|206277), S-cerevisiae (Saccharomyces cerevisiae pol4, YCR014C), and S-pombe (Schizosaccharomyces pombe pol4 SPAC2F7.06c.1). The phylogenetic reconstruction was performed by the neighbor-joining method with 1000 bootstrap replicas. Numbers in each node are the percentage of bootstrap support. The TcI, TcI-N-Hap, and TcII in front of each tree indicate the node grouping sequences of T. cruzi lineage TcI, the non-Esmeraldo haplotype, and TcII lineage, respectively
The phylogenetic differences found between the Miranda Tcpolß and the other T. cruzi counterpart sequences are also illustrated, analyzing the single nucleotide polymorphisms (SNP) among them (Table 1). As may be observed, among the Miranda and the other six polß ORF described, there are 20 SNPs, of which five are responsible for coding for the four amino acids that differentiate this enzyme from the other six. In addition, the differences between the Miranda Tcpolß and the two CL Brener polß haplotypes (N-Hap and E-Hap) are based on 12 and 13 different SNPs, respectively. Comparing Miranda with the Esm and Tula sequences, dissimilar 15 and 11 SNPs are observed, respectively. By contrast, with the other TcI group sequences, Dm28c and JRcl4, only four different SNPs were found.
The primary and secondary structures of a broad spectrum of polßs are illustrated in supplementary Fig. S2. The greater degree of conservation of the primary structure is correlated with the presence of different secondary structures described previously in the literature (Pelletier et al. 1994; Sawaya et al. 1994). Conservation is also observed with catalytic residues such as D190 and D191 (motif C) and the residue R256 (motif A) responsible for the DNA synthesis. Also in this figure, the four amino acids (vertical arrows pointing up) that are different in the T. cruzi Miranda clone enzyme compared to the other T. cruzi sequences are indicated.
Discussion
In this paper, we show the expression, purification, and biochemical characterization of the recombinant DNA polymerase ß encoded by the T. cruzi Miranda clone (Miranda Tcpolß), which belongs to the TcI lineage. The recombinant protein was expressed as a fusion peptide with an N-terminal histidine tag encoded in a pET15 vector and purified from the insoluble inclusion bodies by conventional column chromatography. Unexpectedly, the recombinant renatured protein did not bind to the Ni-NTA resin, despite the location of the histidine tag present in the N-terminus, which was confirmed by Western blotting. As a consequence, the purification was achieved by conventional column chromatography. It is possible that in the “native” conformation of recombinant protein, the histidine tag is not accessible to the external environment.
Importantly, although there are other recombinant Tcpolß described in the literature; they are from the CL Brener clone of the TcVI lineage which were expressed as fusion peptides bound to maltose-binding protein (Lopes et al. 2008; Schamber-Reis et al. 2012). As we mentioned in a previous paper, despite the similarity of the TcI polß Miranda clone to the CL Brener clone, they are not identical (Venegas et al. 2009). The greatest differences between these proteins are observed at the nucleotide sequence level, as shown by the phylogenetic reconstruction presented here and the analysis of SNPs. Although only four amino acids are different between the Miranda enzyme and the other T. cruzi counterparts, it cannot be ruled out that these changes could be responsible for the different biochemical characteristics observed. This possibility is reinforced by the very different lineages of the Miranda (TcI) and CL Brener (TcVI) clones, whose biological, genetic, and pathogenic differences have been extensively described by several groups of researchers (Brisse et al. 2000; Teixeira et al. 2006; Souto et al. 1996; Momen 1999; Westenberger et al. 2005; De Freitas et al. 2006; Zingales et al. 2012). In addition, the fact that in previous reports the recombinant Tcpolß was expressed as a fusion peptide with the maltose-binding protein (Lopes et al. 2008; Schamber-Reis et al. 2012), which is a protein of 387 amino acids and 42.5 kDa (Riggs 2000), about the same size as Tcpolß, could alter its biochemical and biological properties.
The pure Tcpolß recombinant protein described here presents several interesting biochemical characteristics. One is the absolute dependency on KCl, which increased its enzymatic activity by about 20,000 times in a concentration range of 100–150 mM. Although this feature has also been observed in most polß, the very high degree of stimulation is not common (Stalker et al. 1976; Hubschers 1983). Further investigation will be necessary to discover the exact cause which determines this behavior. At least an effect produced by truncated or partially proteolyzed form of this enzyme may be ruled out because the full length and intact polypeptide chain was confirmed by SDS-PAGE and protein sequencing by mass spectroscopy.
The optimum pH range observed in our recombinant Tcpolß is more acid than has been described for mammalian polß (Hubschers 1983; Davies et al. 1994; Pelletier et al. 1994; Sawaya et al. 1994). This may indicate another aspect of the catalytic differences between T. cruzi and mammal polßs. A similar situation may be mentioned with regard to the concentration range of Mn which produces the highest Tcpolß activity, which was found at 1–4 mM, higher than the range of 0.3–0.8 mM described in the literature (Hubschers 1983). The optimal range of Mg was similar to that described for mammalian enzymes, which is between 5 and 25 mM (Hubschers 1983).
Another important biochemical characteristic shown by our Tcpolß is its high sensitivity to the sulfhydryl-blocking reagent N-ethylmaleimide (NEM), with a residual enzymatic activity less than 20 % at 10 mM. Mammalian polßs are generally insensitive up to 10 mM (Hubschers 1983; Wang 1991). However, in some cases, the mammalian polßs are highly sensitive to NEM, such as the polß from the Novikoff hepatoma (Stalker et al. 1976). In that case, the authors purified a protein with a size of 32 kDa, smaller than the previously described mammalian polß of about 40 kDa; they suggested that the reactivity of cysteines is more dependent on the exposition to the sulfhydryl-blocking reagent rather than to cysteine abundance (Bollum 1975; Weissbach 1975). Interestingly, yeast DNA pol IV, an orthologous 68-kDa gene product of polß, is highly sensitive to NEM, with less than 2 % activity at 0.1 mM (Shimizu et al. 1993). The natural T. cruzi polß we characterized displayed partial inhibition by NEM; enzyme activity was greater than 50 % at 10 mM (Venegas and Solari 1995). This dissimilar behavior between these two enzymes could be related to their dissimilar purification level, in which unknown contaminant proteins could protect or hide the residues which are modified by NEM, taking into account that the natural enzyme was not purified to homogeneity (Venegas and Solari 1995). However, other causes cannot be ruled out, such as possible post-translational modification of the natural Tcpolß or other unknown causes.
The present recombinant Tcpolß protein showed sensitivity to the ddTTP reagent similar to that of the previous natural Tcpolß, only partially inhibited by this reagent, with about 50 % activity at a TTP/ddTTP ratio of 1:2 (Venegas and Solari 1995). Thus both enzymes showed atypical resistance to this reagent, since the majority of polß are almost completely inhibited by a dTTP/ddTTP ratio of 1:1 (Hubschers 1983; Wang 1991).
Two proteins of about 35 and 55 kDa were detected in epimastigote extracts by Western blot using a pure anti-polß polyclonal antibody produced here. The small protein possibly corresponds to the known proteolytic product without the 8-kDa N-terminal described earlier (Venegas and Solari 1995). Alternatively it could be the form that is found inside the mitochondrion. The second protein had greater molecular mass than its recombinant counterpart; it maybe a post-translational modification of Tcpolß, such as acetylation (Hasan et al. 2002), methylation (El-Andaloussi et al. 2007), or ubiquitination (Parsons et al. 2008), previously described for mammalian polßs. Further studies will be required to determine this matter.
In vitro studies carried out in the rat polß identified two residues (Ser-44 and Ser-55) which were phosphorylated by protein kinase C (Tokui et al. 1991). The phosphorylation produced the inhibition of DNA polymerase activity. Later, a study of cerebral ischemia experiments in rat models showed that polß was phosphorylated “in vivo” and is correlated with increased levels of inhibition (Lou et al. 2007). In this study, we show for the first time also that the high molecular weight form Tcpolß is phosphorylated in vivo in epimastigote cells. It is possible that the same homologous residues as those identified in rat polß are phosphorylated in Tcpolß because both residues are highly conserved in a broad spectrum of species including trypanosomatids (Fig. S2). However, since there are at least 16 other hypothetical phosphorylation sites for various kinases (Fig. S2), further studies should be undertaken to identify exactly which sites are phosphorylated in this T. cruzi enzyme.
Regarding the additional factors that Tcpolß required in order to repair short gaps in double strand DNA, in mammals, at least 16 proteins have been described that form a complex with polß; among them it is important to mention XRCC1 and PARP1, which interact directly with polß and appear to be responsible for forming a kind of molecular skeleton for complex stability (Almeida and Sobol 2007). However, in trypanosomatids, a counterpart of the XRCC1 gene has not been found, suggesting that perhaps other proteins such as XAB2 could play this role (Passos-Silva et al. 2010). In our study, the eluted fraction that interacted with Tcpolß is required for its activity. They are a group of at least 15 proteins with a range of 15 to about 150 kDa, highlighting one major protein of approximately 40 kDa. Further studies will be required to identify these proteins and to determine whether any of them correspond to proteins that have previously been suggested to have a role in DNA repair in trypanosomatids.
In this manuscript, we describe the expression, purification to homogeneity, and biochemical characterization of Tcpolß of the TcI lineage. This study confirms that the previously described differences between the T. cruzi enzyme and its mammalian counterparts based on primary structure and functional properties are also reflected in its biochemical properties, such as its high dependence on and stimulation by KCl, high sensitivity to NEM, and low sensitivity to ddTTP. These results, together with the detection of phosphorylation in the native form of the enzyme and the requirement of additional proteins to repair short DNA gaps, indicate that this enzyme forms a protein complex involved in DNA repair, as has been described in mammals and some trypanosomatids (Almeida and Sobol 2007; Passos-Silva et al. 2010). The exact importance of these results require future studies to know exactly what are the role of these enzyme characteristics in the biology of this parasite and which could be useful as potential chemotherapeutic targets for the development of better drugs for Chagas disease treatment.
References
Almeida KH, Sobol RW (2007) A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair 6:695–711
Bollum FJ (1975) Mammalian DNA polymerases. Prog Nucleic Acid Res Mol Biol 15:109–144
Brisse S, Barnabé C, Tibayrenc M (2000) Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol 30:34–44
Bruhn DF, Mozeleski B, Falkin L, Klingbeil MM (2010) Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Mol Microbiol 75:1414–1425
Burgers PMJ (1998) Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107:218–227
Chandler J, Vandoros AV, Mozeleski B, Klingbeil MM (2008) Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplasto DNA replication in trypanosomes. Eukaryot Cell 7:2141–2146
Coura JR, Castro SL (2002) A critical review on Chagas disease chemotherapy. Mem Inst Oswaldo Cruz 97:3–24
Davies JF, Almassy RJ, Hostomska Z, Ferre RA, Hostomsky Z (1994) 2.3 A crystal structure of the catalytic domain of DNA polymerase ß. Cell 76:1123–1133
De Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Vanessa F, Gonçalves VF, Teixeira SMR, Chiari E, Junqueira ACV, Fernández O, Macedo AM, Machado CR, Pena SDJ (2006) Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog 2:0226–0235. doi:10.1371/journal.ppat.0020024
El-Andaloussi N, Valovka T, Toueille M, Hassa PO, Gehrig P, Covic M, Hubscher U, Hottiger MO (2007) Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. Faseb J 21:26–34
El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, Da Silveira JF, De Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, Mcculloch R, Mckenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B (2005a) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415
El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, Ghedin E, Peacock C, Bartholomeu DC, Haas BJ, Tran AN, Wortman JR, Alsmark UC, Angiuoli S, Anupama A, Badger J, Bringaud F, Cadag E, Carlton JM, Cerqueira GC, Creasy T, Delcher AL, Djikeng A, Embley TM, Hauser C, Ivens AC, Kummerfeld SK, Pereira-Leal JB, Nilsson D, Peterson J, Salzberg SL, Shallom J, Silva JC, Sundaram J, Westenberger S, White O, Melville SE, Donelson JE, Andersson B, Stuart KD, Hall N (2005b) Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409
Ferguson M, Torri AF, Ward DC, Englund PT (1991) In situ hybridization to the Crithidia fasciculate kinetoplast reveals two antipodal sites involved in kinetoplast DNA replication. Cell 70:621–629
File J, Forterre P, Sen-Lin T, Laurent J (2002) Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol 54:763–773
Graca-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GRR, Paes MC, Sorgine MHYF, Oliveira MF, Oliveira PL (2006) Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol 36:322–335
Gupta S, Wen J-J, Garg NJ (2009) Oxidative stress in Chagas disease. Interdisc Perspect Infect Dis. doi:10.1155/2009/1903540
Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, Hottiger MO (2002) Acetylation regulates the DNA end-trimming activity of DNA polymerase β. Mol Cell 10:1213–1222
Hubschers U (1983) DNA polymerases in prokaryotes and eukaryotes: mode of action and biological implications. Experientia 39:1–25
Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O’neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442
Jannin J, Villa L (2007) An overview of Chagas disease treatment. Mem Inst Oswaldo Cruz 102(I):95–97
Jercic MI, Mercado R, Villarroel R (2010) Congenital Trypanosoma cruzi infection in neonates and infants from two regions of Chile where Chagas’ disease is endemic. J Clin Microbiol 48:3824–3826
Klingbeil MM, Motyka SA, Englund PT (2002) Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol Cell 10:175–186
Lindhl T, Wood RD (1999) Quality control by DNA repair. Science 286:897–1905
Lopes DO, Schamber-Reis BL, Regis-da-Silva CG, Rajao MA, Darocha WD, Macedo AM, Franco GR, Nardelli SC, Schenkman S, Hoffmann J-S, Cazaux C, Pena DJ, Teixeira SMR, Machado CR (2008) Biochemical studies with DNA polymerase beta and DNA polymerase beta-PAK of Trypanosoma cruzi suggest the involvement of these proteins in mitochondrial DNA maintenance. DNA Repair 7:1882–1892
Lou Y, Ji X, Ling F, Li W, Zhang F, Cao G, Chen J (2007) Impaired DNA repair via the base-excision repair pathway after focal ischemic brain injury: a protein phosphorylation-dependent mechanism reversed by hypothermic neuroprotection. Front Biosci 12:1852–1862
Matsumoto Y, Kim K (1995) Excision of deoxyribosephophate residues by DNA polymerase ß during DNA repair. Science 269:699–702
Momen H (1999) Taxonomy of Trypanosoma cruzi: a commentary on characterization and nomenclature. Mem Inst Oswaldo Cruz 94(1):181–184
Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL (2008) CHIP mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell 29:477–487
Passos-Silva DG, Andrade M, Nascimento de Aguiar PH, Vieira-da-Rocha JP, Machado CR, Carolina Furtado C (2010) Overview of DNA repair in Trypanosoma cruzi, Trypanosoma brucei and Leishmania major. J Nucleic Acids. doi:10.4061/2010/840768
Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J (1994) Structure of ternary complexes of rat DNA polymerase ß a DNA template-primer, and ddCTP. Science 264:1891–1903
Piacenza L, Alvarez MN, Peluffo G, Radi R (2009) Fighting the oxidative assault: the Trypanosoma cruzi journey to infection. Curr Opin Microbiol 12:415–421
Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH (1998) Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase ß as revealed by site directed mutagenesis. J Biol Chem 273:11121–11126
Rajao MA, Passos-Silva DG, DaRocha WD, Franco GR, Macedo AM, Pena SD, Teixeira SM, Machado CR (2009) DNA polymerase kappa from Trypanosoma cruzi localizes to the mitochondria, bypasses 8-oxoguanine lesions and performs DNA synthesis in a recombination intermediate. Mol Microbiol 71:185–197
Ramírez JD, Montilla M, Zulma M, Cucunuba ZM, Floréz AC, Zambrano P, Guhl F (2013) Molecular epidemiology of human oral Chagas disease outbreaks in Colombia. PLoS Negl Trop Dis 7(2):e2041. doi:10.1371/journal.pntd.0002041
Ramiro MJ, Hanke T, Taladriz S, Larrage V (2002) DNA polymerase beta mRNA determination by relative quantitative RT-PCR from Leishmania infantum intracellular amastigote. Parasitol Res 88:760–767
Riggs P (2000) Expression and purification of recombinant proteins by fusion to maltose-binding protein. Mol Biotechnol 15:51–63
Rojas C, Venegas J, Litvak S, Solari A (1992) Two DNA polymerases from Trypanosoma cruzi: biochemical characterization and the effect of inhibitors. Comp Biochem Physiol 101:27–33
Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J (1994) Crystal structure of rat DNA polymerase ß: evidence for a common polymerase mechanism. Science 264:1930–1935
Saxowsky T, Choudhary G, Klingbeil MM, Englund PT (2003) Trypanosoma brucei has two distinct mitochondrial DNA polymerase ß enzymes. J Biol Chem 278:49095–49101
Schamber-Reis BLF, Nardellib S, Régis-Silva CG, Carneiro P, Goncalves P, Almeida S, Franco GR, Macedo AM, Pena SDJ, Cazaux C, Hoffmann J-S, Machado MC, Schenkman S, Ribeiro SM, Machado CR (2012) DNA polymerase beta from Trypanosoma cruzi is involved in kinetoplast DNA replication and repair of oxidative lesions. Mol Biochem Parasitol 183:122–131
Shimizu K, Santocanale C, Ropp PA, Longhese MP, Plevani P, Lucchini G, Sugino A (1993) Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae. A probable homolog of mammalian DNA polymerase ß. J Biol Chem 268:27148–27153
Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B (1996) DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol 83:141–152
Stalker DM, Mosbaugh DW, Meyer RR (1976) Novikoffhepatoma deoxyribonucleic acid polymerase. Purification and properties of a homogeneous ß polymerase. Biochemistry 15:3114–3121
Taladriz S, Hanke T, Ramiro MJ, García-Díaz M, García De Lacoba M, Blanco L, Larraga V (2001) Nuclear DNA polymerase beta from Leishmania infantum. Cloning, molecular analysis and developmental regulation. Nucleic Acids Res 29:3822–3834
Tamayo E, Bernal G, Teno U, Maldonado E (2004) Mediator is required for activated transcription in a Schizosaccharomyces pombe in vitro system. Eur J Biochem 271:2561–2572
Teixeira ARL, Nascimento RJ, Sturm NR (2006) Evolution and pathology in Chagas disease—a review. Mem Inst Oswaldo Cruz 101:463–491
Tokui T, Inagaki M, Nishizawa K, Yatani R, Kusagawa M, Ajiro K, Nishimoto Y, Date T, Matsukage A (1991) Inactivation of DNA protein kinase C. J Biol Chem 266:10820–10824
Torri AF, Englund PT (1992) Purification of a mitochondrial DNA polymerase from Crithidia fasciculata. J Biol Chem 267:4786–4792
Torri AF, Englund PT (1995) A ß-like DNA polymerase from mitochondrion of the trypanosomatid Crithidia fasciculate. J Biol Chem 270:3495–3497
Venegas J, Solari A (1995) Purification and characterization of a ß-like DNA polymerase from Trypanosoma cruzi. Mol Biochem Parasitol 73:53–62
Venegas J, Salas J, Gonzalez C, Zulantay I, Díaz E, Gajardo M, Sanchez G, Solari A (2000) Isolation and partial characterization of three DNA polymerases from Trypanosoma cruzi. Comp Biochem Physiol B B1(27):11–19
Venegas J, Åslund L, Solari A (2009) Cloning and characterization of a DNA polymerase beta gene from Trypanosoma cruzi. Parasitol Int 58:187–192
Wang TS-F (1991) Eukaryotic DNA polymerases. Annu Rev Biochem 60:513–552
Weissbach A (1975) Vertebrate DNA polymerases. Cell 5:101–108
Westenberger SJ, Barnabé C, Campbell DA, Sturm NR (2005) Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171:527–543
Wilson SH (2000) Structural design of eukaryotic DNA repair polymerase. Mutat Res 460:231–244
Zingales B, Miles M, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12:240–253. doi:10.1016/j.meegid.2012.04.024
Acknowledgments
We thank Dr. Catherine Connelly for critically reading the manuscript and for her valuable suggestions. We thank Dr. Rodrigo Contreras from Plant Physiology and Biotechnology Laboratory, Chemistry and Biology Faculty in the University of Santiago of Chile (USACH), for the use of the fluorimeter equipment and his technical advice in fluorescence measurement.
This study was supported by FONDECYT Grant 1120507 (E.M.) and CONICYT-COLCIENCIAS Grant PCCIC2006, N°018/DRI/644 (J.V.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edio Maldonado and Diego A. Rojas contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 2764 kb)
Rights and permissions
About this article
Cite this article
Maldonado, E., Rojas, D.A., Moreira-Ramos, S. et al. Expression, purification, and biochemical characterization of recombinant DNA polymerase beta of the Trypanosoma cruzi TcI lineage: requirement of additional factors and detection of phosphorylation of the native form. Parasitol Res 114, 1313–1326 (2015). https://doi.org/10.1007/s00436-014-4308-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-4308-8