Abstract
Canine Hepatozoon species from Brazil was molecular identified and characterized for the first time. From 31 dogs, 7 were positive for blood smear examination and 21 positive for PCR. Partial sequences of the 18S rRNA gene from eight naturally infected dogs were analyzed. Sequences revealed that Brazilian Hepatozoon is closely related with the Japanese Hepatozoon, that has 99% nucleotide identity with Hepatozoon canis from Israel, and different from Hepatozoon americanum. These results indicate that the canine Hepatozoon species from Brazil is H. canis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Canine hepatozoonosis is a tick-borne disease caused by the protozoan Hepatozoon. There are two described species: Hepatozoon canis, transmitted by the dog tick Rhipicephalus sanguineus and Hepatozoon americanum transmitted by Amblyomma maculatum (Vicent-Johnson et al. 1997; Baneth et al. 2003). In most cases, the clinical presentation of H. canis infection is a mild disease but a severe illness, characterized by extreme lethargia, cachexia and anemia, that may occur in dogs with high parasitaemia. In contrast, H. americanum infection is a debilitating and often fatal disease, characterized by generalized pain, muscle atrophy, weakness and bone proliferative lesions (Baneth et al. 2003). Mathew et al. (2000) demonstrated that H. canis and H. americanum are closely related. Nevertheless, in addition to the biological and clinical differences between the two species, Baneth et al. (2000) achieved the distinction between them using genetic and antigenic analysis. Inokuma et al. (2002) determined, using the PCR, that the species from Japan might be a strain variant of H. canis.
In Brazil, the canine Hepatozoon infection has been reported and has being diagnosed during laboratory examinations (Mundim et al. 1992, 1994; Gondim et al. 1998) or during epidemiological studies in urban or rural areas (Massard 1979; O’Dwyer et al. 2001, 2004; Paludo et al. 2003). O’Dwyer et al. (2001) observed high prevalence (39.2%) in dogs from rural areas from Rio de Janeiro State and low prevalence (5.9%) in stray dogs from São Paulo State, Brazil (O’Dwyer et al. 2004). The canine Hepatozoon species from Brazil was not determined but the clinical signs and the low pathogenity of the Brazilian species indicated that we were dealing with H. canis or a close related species and not with H. americanum (O’Dwyer et al. 2001, 2004; Paludo et al. 2003).
Thus, the objective of this study was to perform the molecular characterization of canine Hepatozoon species from Brazil, by the analysis of the partial 18S rRNA gene sequences from Hepatozoon detected in naturally infected dogs.
Material and methods
Hepatozoon isolates
Investigation of dogs naturally infected by Hepatozoon sp. was performed by blood smears taken from the ear margin capillary bed, fixed with methanol and stained with Giemsa. Blood was collected by puncture of the cephalic vein to perform DNA extraction. A total of 31 dogs were examined. Positive EDTA-anticoagulated peripheral blood was kept frozen at −80°C until used.
DNA extraction
DNA extraction of Hepatozoon protozoa was performed pre-incubating the blood with proteinase K (digestion) by 4 h at 56°C. Afterwards, DNA was isolated from 200 μl aliquots of the blood using a QIAamp DNA mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Each DNA sample was eluted in 100 μl of TE buffer. Five microliter portions of these DNA extracts were used for PCR amplification.
PCR assay
The primers set HepF (5′ ATA-CAT-GAG-CAA-AAT-CTC-AAC 3′) and HepR (5′ CTT-ATT-CCA-TGC-TGC-AG 3′) was designed to amplify a partial 18S rRNA gene sequence of Hepatozoon spp. based upon alignment data from the H. canis from a dog in Israel (Genbank accession number AF176835), H. americanum from a dog in the US (AF176836) and H. catesbianae from a frog (AF176837) as described by Inokuma et al. (2002).
The amplification reaction was carried out in a total of 25 μl containing 1X Taq polymerase buffer (Pharmacia), 2 mM MgCl2, 0.2 mM dNTPs, 1.5 U of Taq DNA polymerase (Pharmacia) and 1 μM of each primers Hep F and Hep R (Inokuma et al. 2002).
PCR reactions were performed with the thermal profile consisted of a hot start of 3 min at 94°C and 35 repetitive cycles of 1 min at 94°C, 2 min at 57°C, and 2 min at 72°C followed by a 7 min extension at 72°C for one cycle. All amplifications were performed on thermocyler Biometra (T gradient).
Aliquots of amplified products (8 μl) were analyzed in ethidium bromide-stained 1% agarose gel by electrophoresis at 100 V for 30 min in TAE buffer and visualized under UV transluminator. The total remaining reaction products were purified by purification Kit MontageTM PCR Centrifugal Filter Devices (Millipore). The purified products were dissolved in 20 μl of TE prior to sequencing. Selected products results were confirmed by sequencing.
Sequencing
Sequencing of PCR products amplified from dog blood samples was carried out in both directions using the “ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit” (PE Applied Biosystems, Foster City, CA, USA). Approximately, 10 ng of purified DNA, for each sequencing reaction, was combined with 3.2 pmol of primer (sense and/or reverse) used in the amplification reaction. Nucleic acid sequence analysis was performed on an automated Applied Biosystems 377 DNA sequencer.
Nucleic acid sequence analysis
The computer analysis of nucleic acid sequence data was performed using MERGER package software. The multiple sequence alignment method and a neighbor joining phylogenetic tree (Saitou and Nei 1987) was constructed using the Clustal W program (Thompson et al. 1994). A phylogenetic tree was visualized using the TREEVIEW 1.4 program (Page 1996). The bootstrap test was applied to estimate the confidence of branching patterns of the neighbor-joining tree (Felsenstein 1985).
Divergences were estimated by the two-parameter method using the MEGA “Molecular evolutionary Genetics” software in the final documentation.
Results and discussion
From the 31 dogs examined, 7 were positive by blood smear examination (22.6%) and 21 were positive by the PCR (67.7%) (Fig. 1). All the positive animals by blood smear examination were also positive by PCR. From the 21 PCR positive blood, 8 were selected for perform the sequencing.
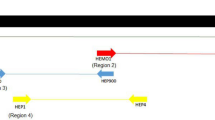
The nucleotide sequences of the 625 bp PCR product excluding the primer regions revealed that Brazilian isolates were closely related with the Japanese Hepatozoon species, which was considered a strain variant of H. canis, but significantly different from H. americanum (Fig. 2).
Neighbor-Joining tree based on Hepatozoon rDNA gene. Individual sequences (HEP) were used to construct a Neighbor-Joining tree. Numbers on branches are bootstrap values (only values above 50 are shown). Fukuoka-sequence from Inokuma et al. (2002)
Inokuma et al. (2002) obtained Hepatozoon DNA from only two dogs, and both sequences were identical to each other. The sequence of the Japanese Hepatozoon was similar to that of H. canis from Israel with 99% nucleotide identity and distantly related with H. americanum with 94% identities. Our sequences showed the presence of one polymorphic site with a transversion (T↔G).
These results indicate that the canine Hepatozoon species from Brazil is H. canis but with the possibility of the occurrence of variant strains in the same geographic region. O’Dwyer et al. (2004) already suggested that the canine Hepatozoon species from Brazil was H. canis. The infection is very prevalent in some regions of Brazil, has low pathogenicity and is frequently associated with other hemoparasites (O’Dwyer et al. 2001). Also, the tissue stages observed were similar to the ones of H. canis (O’Dwyer et al. 2004).
The clinical, epidemiological and biological significance of the Brazilian isolates should be investigated.
References
Baneth G, Barta JR, Shkap V, Martin DS, Macintire DK, Vincent-Johnson N (2000) Genetic and antigenic evidence supports the separation of Hepatozoon canis and Hepatozoon americanum at the species level. J Clin Microbiol 38:1298–1301
Baneth G, Mathew JS, Shkap V, Macintire DK, Barta JR, Ewing SA (2003) Canine hepatozoonosis: two disease syndromes caused by separate Hepatozoon spp. Trends Parasitol 19:27–31
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gondim LFP, Konayagawa A, Alencar NX, Biondo AW, Takahira RF, Franco SRV (1998) Canine hepatozoonosis in Brazil: description of eight naturally occurring cases. Vet Parasitol 74:319–323
Inokuma H, Okuda M, Ohno K, Shimoda K, Onishi T (2002) Analysis of the 18S rRNA gene sequence of a Hepatozoon detected in two Japanese dogs. Vet Parasitol 106:265–271
Massard CA (1979) Hepatozoon canis (James, 1905) (Adeleida: Hepatozoidae) de cães do Brasil, com uma revisão do gênero em membros da ordem carnívora. Seropédica: UFRRJ, Departamento de Parasitologia (Tese, Mestrado) 121 p
Mathew JS, Van Den Bussche RA, Ewing AS, Malayer JR, Latha BR, Panciera RJ (2000) Phylogenetic relationships of Hepatozoon (Apicomplexa: Adeleorina) based on molecular, morphologic and life-cycle characters. J Parasitol 86:366–372
Mundim AV, Jacomini JO, Mundim MJS, Araújo SF (1992) Hepatozoon canis (James, 1905) em cães de Uberlândia, Minas Gerais. Relato de dois casos. Braz J Vet Res Anim Sci 29:259–261
Mundim AV, Mundim MJS, Jensen NMP, Araújo SF (1994) Hepatozoon canis: estudo retrospectivo de 22 casos de infecção natural em cães de Uberlândia, MG. Rev Cent Ciênc Bioméd Univ Fed Uberlândia 10:89–95
O’Dwyer LH, Massard CL, Pereira De Souza JC (2001) Hepatozoon canis infection associated with dog ticks of rural areas of Rio de Janeiro State, Brazil. Vet Parasitol 94:143–150
O’Dwyer LH, Saito ME, Hasegawa MY, Kohayagawa A (2004) Tissue stages of Hepatozoon canis in naturally infected dogs from São Paulo State, Brazil. Parasitol Res 94:240–242
Page RDM (1996) Tree view: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Paludo GR, Dell’Porto A, Castro e Trindade AR, Mcmanus C, Friedman H (2003) Hepatozoon spp.: report of some cases in dogs in Brasília, Brazil. Vet Parasitol 118:243–248
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Acids Res 22:4673–4680
Vincent-Johnson NA, Macintire DK, Lindsay DL, Lenz SD, Baneth G, Shkap V, Blagburn BL (1997) A new Hepatozoon species from dogs: description of the causative agent of canine hepatozoonosis in North America. J Parasitol 83:1165–1172
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rubini, A.S., Paduan, K.d.S., Cavalcante, G.G. et al. Molecular identification and characterization of canine Hepatozoon species from Brazil. Parasitol Res 97, 91–93 (2005). https://doi.org/10.1007/s00436-005-1383-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-005-1383-x