Abstract
Main conclusion
Analysis of 180 accessions of Miscanthus using a DArT platform revealed high diversity. The phylogenetic analysis revealed that M. × giganteus accessions fall into two genetically distinct groups.
Miscanthus is a genus of perennial rhizomatous grasses that has emerged in last 20 years as a feedstock for bioenergy and biofuel production. Currently, the most widely used accession for bioenergy purposes is Miscanthus × giganteus, a sterile triploid hybrid between Miscanthus sinensis and Miscanthus sacchariflorus. However, previous reports have shown that genetic diversity of Miscanthus × giganteus is limited. Here, we report development of Diversity Arrays Technology platform for the analysis of genetic structure of a Miscanthus collection of 180 accessions. A total of 906 markers were obtained of which around 25.5 % exhibited polymorphism information content value in the range of 0.40 and 0.50 and are considered particularly informative. Newly developed marker system will serve as an additional resource to assist crop improvement, germplasm preservation and genetic studies. Three types of analysis indicated that 180 accessions from the collection were well differentiated and presented high diversity. Interestingly, the analysis revealed that there are two separate groups of plants, significantly differing in genetic diversity, that are commercially available as M. × giganteus. We suggest that one of these groups is most likely mutants or somaclonal variants of original M. × giganteus. The other group is recent hybrids of Miscanthus of higher genetic diversity. This study indicates that the diversity of commercially available M. × giganteus is higher than commonly assumed. Development of the new marker system can significantly assist breeding of new commercial cultivars of Miscanthus for bioenergy use.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Miscanthus is a genus of perennial rhizomatous grasses that originate from Eastern Asia and the Pacific (Greef and Deuter 1993). Since their introduction to Europe in early twentieth century, they have been primarily utilised as ornamental grasses. In the last 20 years, Miscanthus sp. emerged as potential feedstock for bioenergy and biofuel production (Heaton et al. 2008a, b; Jeżowski 2008; Sang and Zhu 2011; Mos et al. 2013). Various Miscanthus accessions have been assessed as a potential bioenergy crops due to a number of advantageous features (Jones et al. 2015) such as highly energy-efficient C4 photosynthesis, high water-use efficiency, reallocation of minerals and nutrients to rhizomes during winter senescence, and high biomass productivity, even on low-quality soils. Currently, the most widely used accession for bioenergy purposes is Miscanthus × giganteus Greef et Deu ex Hodkinson et Renvoize, a sterile triploid interspecific hybrid between the parent species Miscanthus sinensis and Miscanthus sacchariflorus. Miscanthus × giganteus was first introduced to Europe by Danish botanist Aksel Olsen in 1935. Since then, a number of similar crosses between the parental species have been produced and made available for commercial sale as M. × giganteus (Głowacka et al. 2015). Although not all of these accessions are triploids, they are all characterised by higher biomass yields than either of the parental species, a senescence profile that allows for optimal nutrient remobilisation in the Central European climate, low moisture content of biomass during winter harvest, and very low probability of producing viable seeds. However, it is widely believed that the genetic diversity of these crosses is limited (Głowacka et al. 2015) and their performance, in terms of biomass yield and stress resistance, is similar (Clifton-Brown et al. 2001; Lewandowski et al. 2003). Additionally, the sterility of some of these crosses makes further breeding of novel Miscanthus varieties from Miscanthus × giganteus very difficult or even impossible. It is therefore essential to go back to the genetic diversity of the Miscanthus genus to be able to breed new Miscanthus hybrids that will outperform Miscanthus × giganteus while maintaining its favourable characteristics as a non-invasive sterile bioenergy crop. New hybrids of Miscanthus should be bred for functional sterility to minimise their invasive potential in addition to typical agronomic features such as yield and resistance to biotic and abiotic stresses (Quinn et al. 2010; Matlaga and Davis 2013). One of the most important features of such functionally sterile new Miscanthus hybrids should be the inability to produce viable seeds at the location of planting. This could be achieved by targeting crop varieties to appropriate latitudes that lack the day length necessary to trigger the reproductive growth (Heaton et al. 2008a, b).
In light of limited information provided by morphological analyses of Miscanthus accessions, various molecular methods have been employed to characterise the genetic diversity of Miscanthus including AFLP (Greef et al. 1997), RAPD (Atienza et al. 2002), RFLP (Hernández et al. 2001), ISSR (Hodkinson et al. 2002), RAD-seq (Głowacka et al. 2015), and other PCR-based techniques (Chiang et al. 2001; Hodkinson et al. 2002). These studies revealed low genetic diversity of Miscanthus × giganteus accessions used for bioenergy. More recently, Diversity Array Technology (DArT) has been successfully applied in genetic diversity analyses, linkage mapping, and surveying population structure of various crop species (Oliver et al. 2011; Castillo et al. 2013; Von Mark et al. 2013). DArT is microarray-based method using DNA–DNA hybridisation (Jaccoud et al. 2001). Its advantages include rapid high-throughput characterisation, identification of single base changes and indels, and in particular, independence of any prior knowledge of sequence data (Jaccoud et al. 2001). DArT provides means of simultaneous genotyping of hundreds to thousands of dominant DArT markers obtained from nucleotide polymorphisms within restriction enzyme recognition sites. Therefore, DArT can be applied to minor crops such as Miscanthus, due to its potential to accelerate gene discovery and initiate molecular breeding without prior sequence data information (Ovesná et al. 2013; Grzebelus et al. 2014).
We present in this paper the development of a DArT microarray prepared from sorghum and sugarcane polymorphic markers and use of this microarray in genotyping a Miscanthus diversity collection of 180 accessions. By screening the DArT microarray with reduced complexity restriction fragment subsets from each accession, we were able to assess the genetic structure of the Miscanthus collection. This new molecular marker system for Miscanthus will serve as an additional resource to assist crop improvement efforts, germplasm preservation, and genetic studies. The genetic structure and clustering can also provide valuable information about potential development of polyculture Miscanthus plantations, composed of various Miscanthus accessions. This approach can not only improve biodiversity and disease resistance of the resulting plantation but also significantly improve biomass quality for downstream processing such as pelleting.
Materials and methods
Plant materials
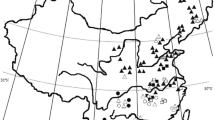
The plant material used for analysis comprised samples of 180 Miscanthus genotypes (Supplementary Table S1) originating from four different sources. A private collection of 88 horticultural Miscanthus accessions from Ogrody Traw (Zgierz, Poland) was used as the main source of M. sinensis genotypes. The plants were propagated from rhizomes and planted in a field site in southern Poland (50°39′59.91′′N, 18°07′50.29′′E). Thirty eight commercial accessions of Miscanthus were provided and grown in the aforementioned field by Energene sp. z o.o. (Energene, Lodz, Poland). Eight of these accessions were isolated from a commercially established plantation of Miscanthus × giganteus in northern Poland (54°06′18.67′′N, 19°16′19.08′′E), and three accessions were purchased from commercial vendors as Miscanthus × giganteus, but differed in their appearance and properties such as canopy structure, frost and cold tolerance, stem density, and dry mass yield (unpublished data, M. Mos personal communication). The Institute of Plant Genetics of the Polish Academy of Sciences (IPG-PAS), Poznan, Poland, provided thirty-one accessions, mainly M. sinensis but also M. sacchariflorus and several other Miscanthus sp., obtained through breeding efforts. Crucially, three accessions commercially used in Poland were provided and two Miscanthus × giganteus crosses initially developed by Dr Martin Deuter of Tinplant Biotechnik (Wanzleben-Borde, Germany), i.e. M116 (“Nagara”) and M119, were included. These crops have been described in detail in other publications (Jeżowski 2008; Jeżowski et al. 2009, 2011) and were used as a reference for this work. Purified Miscanthus DNA samples of native Chinese accessions collected from Guangdong, eastern Inner Mongolia, Heilongjiang, and Jilin provinces were provided by the Peking University Shenzhen Graduate School, School of Environment and Energy (PKUSZ SEE).
DNA isolation
Leaf tissues from Miscanthus plants were collected and processed within 8 h of collection. Leaf tissue was homogenised in liquid nitrogen with a mortar and pestle. Approximately, 100 mg of powdered tissue was used for nucleic acid isolation. Total genomic DNA was isolated with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). DNA quality and quantity were determined by agarose gel electrophoresis and Nanophotometer (Impeln, Munich, Germany), respectively. The DNA concentration of each sample was adjusted to 50 ng μl−1 before further processing.
Preparation of DArT probes and the microarray
We used DArT clones from the sorghum and sugarcane libraries described by Mace et al. (2008) and Heller-Uszynska et al. (2011). The array contained 5760 sugarcane clones and 1920 sorghum clones. Two replicates per clone were spotted on SuperChip poly-l-lysine slides (Thermo Scientific, Scoresby, Australia) using a MicroGrid arrayer (Genomics Solutions, Huntingdon, UK). After printing, slides were processed as described by Sharma et al. (2014) to construct a DArT array. The clones were constructed using PstI and TaqI digestions according to principles described at http://www.diversityarrays.com/dart-application-microarray-process-complexity-reduction as described by Mace et al. (2008) and Heller-Uszynska et al. (2011). Sample complexity reduction for each Miscanthus accessions was performed with a combination of two restriction enzymes, a rare cutter (PstI) and a frequent cutter (TaqI), adaptor ligation and PCR amplification, essentially as reported by Wenzl et al. (2004). Approximately 50 ng of genomic DNA of each of the accessions was digested with 2 units of both PstI and TaqI (NEB, Beverly, MA, USA). Reactions were incubated at 37 °C for 2 h, followed by 2 h at 60 °C. A PstI adapter (5′-CACGATGGATCCAGTGCA-3′ annealed with 5′-CTGGATCCATCGTGCA-3′) was ligated with T4 DNA ligase (NEB). One microlitre of digestion/ligation reaction product was used as a template for PCR amplification in a 50 μl reaction medium containing DArT-PstI primer (5′-GATGGATCCAGTGCAG-3′) and REDTaq polymerase (Sigma–Aldrich) using following cycling parameters: 94 °C for 1 min, followed by 30 cycles of 94 °C for 20 s, 58 °C for 40 s, 72 °C for 1 min, and final extension at 72 °C for 7 min. As a result of PCR only fragments containing PstI adapters at both sides were amplified. The resultant reduced-representation fragments were labelled with fluorescent dyes (Cy3/Cy5-dUTP) using the exo-Klenow fragment of Escherichia coli polymerase I (NEB).
Genotyping and scoring
Labelled DArT probe sets were hybridised to printed DArT arrays for 16 h at 65 °C in a water bath. Slides were washed, dried initially by centrifugation, and later by a vacuum desiccator as described by Kilian et al. (2012). The slides were scanned using a Tecan LS300 scanner (Tecan Group Ltd, Mannedorf, Switzerland). Three images were generated per array for each of the fluorescent dyes at appropriate wavelengths (Cy3: 543 nm, Cy5: 633 nm, FAM as reference signal: 488 nm). All images were analysed as described by Von Mark et al. (2013). The markers were scored as binary data (1/0), indicating the presence or absence of a specific marker in the representation of a particular sample as described by Wenzl et al. (2004).
Data analysis
The following quality parameters were used for filtering: call rate >80 % (percentage of valid scores in all possible scores for a marker), P value >50 % (between-cluster variance in relative hybridisation intensity as a percentage of the total variance), and reproducibility >95 %. A subset of accessions (48) were analysed in technical replication (completely independent assays) for scoring reproducibility assessment. The polymorphism information content (PIC) was also calculated to determine the informativeness of the DArT markers according to Dominguez-Garcia et al. (2012).
Diversity analysis
Genetic diversity of DArT markers was analysed using GenAlEx 6.5 (Peakall and Smouse 2006), including effective number of alleles, Shannon’s index, unbiased genetic diversity, and occurrence of unique, rare (0.0< frequency ≤0.05), common (0.05< frequency <0.95) and frequent alleles (0.95≤ frequency <1.0). Unique alleles are those that are present in one accession or one group of accessions belong to same cluster but absent in other accessions or clusters. To resolve genetic relationships among all examined accessions, the 0/1 binary matrix of the markers was used to calculate genetic similarity using Dice’s coefficient (Dice 1945), and cluster analysis was performed using Unweighted Pair Group Method with Algorithmic Mean (UPGMA) complemented in NTSYS-pc v.2.10e (Rohlf 1997). Principal coordinate analysis (PCoA) was conducted to complement the output of the cluster analyses using GenAlEx 6.5. The amount of variation among clusters was assessed by partitioning genetic diversity using analysis of molecular variance (AMOVA).
Structure analysis
The genetic structure of the Miscanthus collection was inferred using a Bayesian clustering framework, STRUCTURE v2.3.4 (Pritchard et al. 2000), assuming an admixture model and correlated allele frequencies. Three independent Markov Chain Monte Carlo (MCMC) runs were performed for each value of K ranging from 2 to 9 using 100,000 burn-in and 500,000 sampling iterations. A value of K was selected on the basis of the ad hoc approach described in the software documentation. Structure results of the selected K were subsequently visualised by HARVESTER (Earl 2012).
Results
DArT array development and evaluation
A total of 180 Miscanthus accessions that represent wide genome diversity of Miscanthus species were used for developing the diversity array. Sample complexity reduction has been performed with combination of two restriction endonucleases, a rare cutter (PstI) and a frequent cutter (TaqI). Selection of these two enzymes allows targeting active chromatin/genic regions of the genome and methylation sensitive complexity reduction. Utilisation of PstI adapter ligated to the digested DNA fragment as primer binding site ensured that only fragments which do not have an internal cleavage site for the frequent cutter (TaqI) are amplified during subsequent PCR reducing complexity even further. The method was selected based on its good performance in related species, i.e. sugarcane (Heller-Uszynska et al. 2011) and sorghum (Mace et al. 2008). After quality control and filtering, 906 markers were obtained (Supplementary Table S2) and used for further analysis. The call rate of the markers varied from 82 to 100 % with an average of 97.3 % and a scoring reproducibility of 99.3 %. The markers differed in PIC value, ranging from 0.01 to 0.50 with an average value of 0.22. About 39.1 % of the markers were highly discriminating with PIC values above 0.25. The majority of the DArT markers have values in the range of 0.06–0.10, accounting for 41.5 % of the total markers, followed by 0.46–0.50 (17.4 %), 0.41–0.45 (8.1 %), 0.11–0.15 (6.7 %), 0.36–0.40 (6.1 %) and 0.16–0.20 (5.3 %) (Fig. 1). The other PIC value classes are under-represented at below 5 %. Overall, the distribution of PIC values was asymmetrical and skewed towards the lower values.
Genetic relationship among Miscanthus accessions
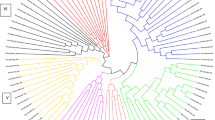
The 180 Miscanthus accessions were used to provide insights into genetic diversity. A UPGMA dendrogram was generated with the 180 Miscanthus accessions using the 906 DArT markers (Fig. 2). Two well-resolved clades were evident in the tree (Fig. 2a), each of which comprised three clusters (Fig. 2b, c), with 60, 29, 13, 12, 59 and 7 accessions clustered in clusters I to VI, respectively (Table 1). It is not surprising that accessions representing unspecified Miscanthus were widely distributed between clusters. Additionally, species-specific accessions grouping within clusters were also found.
Cluster analysis of 180 Miscanthus accessions based on 906 DArT markers. The dendrogram (a) was composed of Clade A (b) and Clade B (c). Scale bar indicates the dice coefficient of similarity (Dice 1945)
Within Clade A, Clusters I, II and III were mostly composed of M. sinensis accessions (Fig. 2b; Supplementary Table S1). Cluster I comprised fifty three M. sinensis accessions, M. floridulus M0528, M. transmorrisonensis M0507, and five unspecified Miscanthus sp., four of which have been annotated as M. sinensis on the basis of DArT and morphological analyses (guidelines described by Lee 1964a, b, c, d). Clusters II and III were exclusively composed of M. sinensis, accounting for 26 and 14 % of the total number of genotypes M. sinensis from the collection. Five initially unspecified Miscanthus sp. in Cluster II were shown to be M. sinensis during the analyses. As for Clade B, all Miscanthus × giganteus were distributed within either Cluster IV or VI grouped with unspecified isolates from commercial plantations, suggesting that these accessions also belong to Miscanthus × giganteus lineages. Interestingly, although both Clusters IV and VI are within the same clade of the dendrogram, separated from Cluster V, they are also clearly separated from each other, indicating the presence of two distinct types of Miscanthus × giganteus. Finally, all known M. sacchariflorus accessions were grouped in Cluster V. The same cluster also contained unclassified Miscanthus sp. what indicates that the initially unclassified plants also belong to M. sacchariflorus lineage. These several Miscanthus accessions appeared to fall into two categories on the basis of the analyses. Forty four of the accessions appeared to be Miscanthus sacchariflorus and fifteen accessions that were most likely the Miscanthus crosses with significant content of M. sacchariflorus genome admixture. In addition, M. oligostachyus M0002 has been allocated in Cluster V albeit with complex pattern of admixtures and large genetic distance from the rest of the Cluster V (Fig. 2).
PCoA analysis was conducted as a complement of cluster analysis to visualise the patterns of genetic relationship. The PCoA analysis (Fig. 3) confirmed the six major clusters obtained from the UPGMA dendrogram (Fig. 2). The two dimensions of PCoA accounted for 24.94 and 5.10 % of the genetic variations, respectively. The right-hand groups (in the upper and lower quadrates) appeared to be clustered around the X-axis with accessions representing Cluster I, II and III of the dendrogram tree that is different clusters of Miscanthus sinensis accessions. The left-hand groups were spread widely along both the X-axis and Y-axis, including the upper group representing accessions from Cluster IV, one of the Miscanthus × giganteus clusters and the lower group representing accessions from Cluster V. Interestingly, the middle group was formed by accessions from both Cluster V and VI of the dendrogram tree. These accessions represented several Miscanthus sp., M. sacchariflorus M0993 and M. oligostachyus M0002. These accessions together with an outlier (M. sinensis M0212) from Cluster I are most likely various horticultural crosses obtained by the breeders as analysis of allele frequencies for these accessions (see below) indicated a high proportion of genetic admixture and, therefore, more complex genetic history of these crops. On the other hand, another accession M. transmorrisonensis M0507, an outlier from Cluster I, has different characteristics. It shows low proportion of genetic admixture confirmed by the large genetic distance of M0507 from other members of Clade A observed on a dendrogram tree (Fig. 2).
Principal coordinate analysis (PCoA) of 180 Miscanthus accessions based on 906 DArT markers. Accessions were coloured by clusters of the dendrogram tree, respectively. Accession numbers were added for accessions mentioned in the text. Accessions belonging to Clusters I to VI are represented by: closed rhombi (I), closed squares (II), closed triangles (III), open circles (IV), open rhombi (V) and closed circles (VI), respectively
Population structure of Miscanthus accessions
The genetic structure of the Miscanthus collection was evaluated using different K ranging from 2 to 9 as described in “Materials and methods”. Data for multiple STRUCTURE runs using the admixture model with correlated allele frequencies indicated that K = 6 was optimal based on criteria described in Methods, suggesting that there are six clusters contributing significant genetic information in the Miscanthus collection. The bar plot of the population assignment when K = 6 is shown in Fig. 4 and the cluster probabilities assigned to each accessions were listed in Supplementary Table S3. Clusters I to VI comprised 60, 29, 13, 12, 59 and 7 accessions, respectively. The six clusters inferred by STRUCTURE were all corresponding to the six clusters in the UPGMA dendrogram tree (Fig. 2).
Population structure of 180 Miscanthus accessions as inferred by STRUCTURE. Accessions belonging to Clusters I to VI are represented by a colour block, whose width is proportional to the number of accessions assigned to it: I (red; 60), II (green; 29), III (yellow; 13), IV (dark blue; 12) and V (light blue; 59) V (pink; 7). Vertical lengths of each strain are proportional to each of the six inferred lineages
A majority of accessions (111 out of 180 accessions, 62 %) representing Clusters I to IV were assigned to one of the six clusters with a high probability ranging from 90 to 100 %, 88 (49 %) of which have close to homogeneous genetic background (>98 % probability). Of the remaining 69 Miscanthus accessions, 58 accessions (32 %) representing Clusters I, II, III, V and VI are heterogeneous showing intermediate (70–90 % probability) and/or highly (50–70 % probability) mixed composition, a majority (47 accessions) of which belong to M. sinensis. Strikingly, 11 accessions (6 %) representing M. sinensis (10 accessions) and M. oligostachyus M0002 from Cluster I, V or VI exhibited the highest probability lower than 50 % and up to five cluster assignments, suggesting that these accessions might have a highly complex genetic background. This supports prior observations regarding abundant flowering of M. sinensis plants and their abilities to easily cross within the same species and to produce viable seeds. M. floridulus has shown more common phenotypic characteristics with M. sacchariflorus, but the results of molecular analysis of the population structure indicates its closer genetic relationship with M. sinensis (Cluster I) than M. sacchariflorus (Cluster V). Cluster IV (one of Miscanthus × giganteus lineages) presented a relatively simple assignment, whereas the other five clusters have myriad cluster assignments, indicating the greater diversity and mixed genetic background within these clusters as well as higher rate of gene flow among clusters. Interestingly, all the M. sinensis accessions with mixed compositions within Cluster I, II and III revealed genetic admixture only from M. sinensis lineage (Cluster I, II or III), except for M. sinensis M0009, M0023, M0055, M0212 and M0611, which showed a very small proportion of admixture (below 10 % of Clusters IV, V or VI). This result indicated a once again high rate of gene flow among sub-lineages of M. sinensis due to their species compatibility and easy flowering synchronisation. As many as forty eight accessions were assigned to M. sacchariflorus lineage (Cluster V), as evidenced by phylogenetic and STRUCTURE analyses (Figs. 2, 3). Thirteen of these accessions had complex patterns of admixture from various lineages, suggesting that frequent interlineage gene flow had taken place. Although these accessions showed certain or high proportions of genetic admixture from M. sinensis lineage, there is no evident cluster that is predominant in the cluster assignment. Therefore, these accessions are most likely interspecific crosses between different M. sacchariflorus and various M. sinensis. Remaining thirty six accessions had relatively simple admixture profile. Although M. oligostachyus M0002 also occurs in Cluster V, closer analysis of its phylogenetic relations with other members of Cluster V reveals significant differences from other members of Cluster V, i.e. high admixture from M. sinensis Clusters I and III as well as significant distance from other members of Clade B on dendrogram tree (Fig. 2). One possible cause of this allocation is that morphology of this accession is similar to M. sinensis, resulting from the genetic materials obtained from M. sinensis lineage. Further investigations are necessary to verify the speculation.
Genetic variation among Miscanthus clusters
The analysis of molecular variance (AMOVA) was used to hierarchically partition genetic variation among the six clusters as revealed by a combination of phylogenetic, PCoA and STRUCTURE analysis. As shown in Table 2, there was a higher proportion of genetic variation within clusters (61 %) than among clusters (31 %) in the Miscanthus collection, although genetic differentiation was evident at all levels of analysis.
Clusters of M. sinensis (Cluster I, II and III) showed a relatively low genetic distance among each other, and were well differentiated from cluster of M. sacchariflorus (Cluster V) and clusters of Miscanthus × giganteus (Cluster IV and VI) (Table 3), indicating that M. sinensis was divergent from M. sacchariflorus and Miscanthus × giganteus and that there is a closer genetic link between Miscanthus × giganteus linage and M. sacchariflorus than it is with M. sinensis. The genetic distances between clusters of M. sinensis and cluster IV containing Miscanthus × giganteus was higher than the distance for Clusters V (M. sacchariflorus) and VI (other accessions of Miscanthus × giganteus) suggesting that there might be genetically different sub-lineages within Miscanthus × giganteus and that two clusters of Miscanthus × giganteus contain genetic material of different origin that results in high biomass yielding phenotype. Also, Cluster IV exhibited a higher genetic distance to Cluster VI than to Cluster V, which further supports this hypothesis. Deeper genotyping with DArT-seq or related platform of more Miscanthus × giganteus accessions especially representing Clusters IV and VI is necessary to validate this inference.
Genetic diversity of the Miscanthus collection
The statistics of genetic diversity for the Miscanthus collection was summarised in Table 1. The Miscanthus population comprised a lower proportion of rare and frequent alleles. Further, cluster-specific unique alleles were identified for Cluster I (24 alleles), Cluster IV (1 allele) and Cluster V (29 alleles). Cluster I exhibited the highest diversity as evidenced by number of effective alleles, Shannon’s index and unbiased gene diversity, while Cluster IV showed the lowest diversity at all the three indexes, in consistent with their clustering in PCoA analysis (Fig. 3) and levels of admixture in STRUCTURE analysis (Fig. 4). The diversity of Cluster III, IV and VI appeared to be underestimated, as the number of accessions assigned to the three clusters was at least twice lower than the other three clusters. However, the number of effective alleles and gene diversity of Cluster III (13 accessions) were even higher than that of Cluster V (59 accessions), indicating a greater genetic variations within Cluster III than within Cluster II, IV, V and VI. Indirectly, this result is in support of AMOVA analysis that a higher proportion of genetic variation existed within clusters than among clusters in the Miscanthus collection (Table 2).
Discussion
The importance of understanding genetic diversity in Miscanthus species is critical for their effective genetic resources management and any further utilisation of this promising energy crop in breeding program. Although a large number of molecular markers have been developed for Miscanthus (Greef et al. 1997; Zhao et al. 2011; Chouvarine et al. 2012), reliable and cost-efficient marker platform is still lacking. To the best of our knowledge, this is the first trial to develop DArT marker system for Miscanthus and utilise these makers in investigating genetic diversity of numerous Miscanthus accessions collected from various sources. The DArT technology is very suitable for high-throughput work and has been determined to have clear advantages in time and cost aspects of genotyping, in that it allows simultaneously type hundreds or thousands loci in a single array without prior knowledge genome sequence. The DArT technology has been successfully applied to various plants (Dominguez-Garcia et al. 2012; Simko et al. 2012; Castillo et al. 2013; Grzebelus et al. 2014), but no information is available on Miscanthus till date. In the present study, the DArT platform for Miscanthus collection were found both acceptable and provided robust information about the genetic variation in this collection. The 906 DArT markers were also found to be useful in providing a complete picture of genetic diversity in the Miscanthus collection of 180 accessions. Overall, the average PIC (0.22) of the Miscanthus microarray DArT markers was found to be lower than that observed in other plant species where similar markers were developed, such as wheat (0.44) (Raman et al. 2010), cassava (0.42) (Xia et al. 2005), sorghum (0.41) (Mace et al. 2008), and carrot (0.30) (Grzebelus et al. 2014), but comparable to that observed in Asplenium fern (0.21) (James et al. 2008), Lesquerella (0.21) (Von Mark et al. 2013) and sugar beet (0.28) (Simko et al. 2012). Around 25.5 % of the DArT markers exhibited a PIC value in the range of 0.40 and 0.50 and these markers might be considered particularly informative. The distribution of these new developed DArT markers remains to be determined. However, we could speculate that these DArT markers will be distributed throughout the genome with marker density highly correlated with gene density, in light of the empirical data from many other organisms whose DArT system was applied more broadly including genetic mapping and/or sequence-based physical mapping (Kilian et al. 2012; Grzebelus et al. 2014).
DArT markers developed in this study effectively profiled the diversity among the Miscanthus accessions. As the genotypes we studied represent both deep sampling of the single Miscanthus species and shallow sampling of relatively diverged species, a combination of phylogenetic and population genetic analysis were employed. All the three types of DArT diversity analysis (cluster analysis, PCoA and population structure) indicated that the 180 accessions were successfully differentiated by the maker system and presented high genetic diversity. The phylogeny of the Miscanthus accessions inferred by cluster analysis suggested six clusters among those accessions, in consistent with the six clusters inferred by STRUCTURE. However, 15 accessions from Cluster V appeared to group closer to accessions from Cluster VI than to the other accessions from Cluster V in PCoA analysis (Fig. 3), incongruent with the phylogeny in Fig. 2. This could be attributed to the fact that PCoA is more informative regarding distances among major groups whereas phylogenetic analysis is more sensitive to relationships between related individuals (Hauser and Crovello 1982). The relatively close relationship among these accessions was reflected by the branch positions of these accessions in the UPGMA tree (Fig. 2b) and by a high proportion of genetic admixture for these genotypes Supplementary Table S3). Accessions identified as M. sinensis were distributed among Clusters I, II, III with relatively high content of admixture form one another, especially in clusters II and III. This confirms prior observations that M. sinensis are compatible within the species and capable of crossing between one another due to earlier and extended flowering period (Jones et al. 2015). This translates into relatively synchronised flowering time of most M. sinensis plants, wide temperature and photoperiod profiles that promote flower development, ability to cross pollinate within the species and formation of good seed set. Most of the material collected as unspecified (almost 80 %) could be allocated to one of the species on the basis of the genetic structure and phylogenetic analyses (Supplementary Table S1), yet 20 % should be considered as either natural or bred inter specific hybrid containing admixture of both M. sinensis and M. sacchariflorus genomes on the basis of our analysis indicating breeding efforts of scientists, breeders and horticulturist to develop new varieties of Miscanthus.
Since Miscanthus × giganteus is already an important biomass crop that is increasingly deployed for bioenergy purposes throughout Europe and USA, special consideration should be given to Clusters IV and VI. These clusters contain plants sold commercially as Miscanthus × giganteus as well as plants isolated from a commercial plantation in northern Poland (M. species # in Supplementary Table S1). PCoA and UPGMA tree revealed that there are actually two separate groups of plants that are commercially available as Miscanthus × giganteus. Plants representing these clusters differ in appearance and properties (canopy structure, frost and cold tolerance, stem density and dry mass yield). These observations were further confirmed at the genetic level showing significant differences between Clusters IV and VI. Cluster IV contained plants that had almost identical genetic profiles (Supplementary Table S2 and Figs. 2, 4). We suggest that these plants are most likely mutants and/or somaclonal variants created through vegetative and tissue culture propagation of original Miscanthus × giganteus Greef et Deu ex Hodkinson et Renvoize hybrid. Cluster VI, on the other hand, contains crops with much higher genetic diversity than these within cluster IV, which is evident at each level of analysis, i.e. dendrogram tree (Fig. 2), PCoA (Fig. 3), and genetic diversity indices (Table 1). Also, Miscanthus × giganteus plants from Cluster IV contain much higher admixture of genetic material from other groups (Fig. 4) as opposed to the near homogeneity of Cluster VI.
Similar findings regarding two sub-lineages of Miscanthus × giganteus have been recently observed by Głowacka et al. (2015). The study using RAD-seq on the collection of 33 so-called legacy cultivars and their colchicine-induced polyploid variants revealed very little genetic difference between these crops, all of which were probably mutants and/or somaclonal variants of the plant introduced to Europe by Aksel Olsen. We believe that these so-called legacy cultivars correspond to accessions in Cluster IV of this study. In the study by Głowacka et al. (2015) the other group is composed of eight Miscanthus × giganteus accessions that contained much higher genetic diversity. This diverse population would probably correspond to Cluster VI in the present study.
Interestingly, representatives of Cluster IV and VI were observed and isolated from the same plantations, suggesting that in early stages of establishing Miscanthus plantations in Europe many crops of unknown origin were mixed and planted as Miscanthus × giganteus. Based on our observations, vast majority of plantations in Europe now contains genetic material originating from both original Miscanthus × giganteus Greef et Deu ex Hodkinson et Renvoize hybrid and more recent crosses that are representative of Cluster VI of this study.
There are both positive and negative outcomes of such situation. On the positive note, the increased biodiversity resulting from the mixing the crops of at least two groups increases the biodiversity of plantations in case of outbreak of diseases or pests. Moreover, industrial practice have shown that some of the genotypes from Cluster VI like “Nagara” show better properties than “legacy cultivars” of Miscanthus × giganteus during biomass pelleting (personal communication with Blankney Estates Limited, Blankney, UK). This feature has significant impact on biofuel production from the point of view of biomass processers. Conversely, it appears that genetic materials originating from different sources have been mixed during the establishment of early Miscanthus plantations in Europe. Although it has not been confirmed by our dataset, there exists a risk that other Miscanthus genotypes phenotypically similar to Miscanthus × giganteus might have been introduced at these plantations as well. Some of them might not be functionally sterile and may pose a risk of spreading the material beyond the intended borders of a plantation. Genotyping of much larger dataset containing numerous plants from the same plantation is required to verify this hypothesis. If this hypothesis proved positive, the development of molecular diagnostic tools may help to identify the plantations that pose such risk and would allow mitigating this risk to minimum.
To the best of our knowledge, this is the first report regarding different classes of allele frequency among the polymorphic markers in Miscanthus. Hundreds of common alleles were detected in the present study, and these will be useful for understanding genetic diversity and molecular profiling of Miscanthus accessions from diverse origins. Further, cluster-specific alleles detected will be very useful for molecular profiling of a particular cluster, such as M. sinensis, M. sacchariflorus and two separate Miscanthus × giganteus clusters.
Conclusions
In this study, the DArT marker platform for Miscanthus was successfully developed and utilised in genotyping of a Miscanthus collection, suggesting that this technology is a powerful tool for investigating genetic diversity in Miscanthus populations. The high number of DArT markers allowed a great resolution of genetic differences among Miscanthus accessions and enabled us to examine the extent of variability in the Miscanthus collection in the present study, as well as provide support to ambiguous taxonomy of certain Miscanthus species. Further utilisation of these DArT markers in developing a linkage map in Miscanthus will assist breeding efforts and future genetic mapping studies. The acquired genetic diversity information of Miscanthus accessions will facilitate better germplasm management and conservation of the species. Finally two separate sub-linages of Miscanthus × giganteus were identified and showed different genetic variance, admixture profiles and properties. This indicates that the genetic diversity of commercially established Miscanthus plantations may be significantly higher that currently assumed.
Author contribution
Michal Mos, Maurycy Daroch and Andrzej Kilian conceived and designed research. Michal Mos, Maurycy Daroch, Stanislaw Jezowski and Marta Pogrzeba provided plant material. Maurycy Daroch and Michal Mos processed plant material. Jie Tang performed analyses and interpreted data. All authors participated in preparation of the manuscript. All authors read and approved the final manuscript.
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- DArT:
-
Diversity arrays technology
- PCoA:
-
Principal coordinate analysis
- PIC:
-
Polymorphism information content
- RAD-seq:
-
Restriction site associated DNA sequencing
- UPGMA:
-
Unweighted pair group method with algorithmic mean
References
Atienza S, Satovic Z, Petersen K, Dolstra O, Martin A (2002) Preliminary genetic linkage map of Miscanthus sinensis with RAPD markers. Theor Appl Genet 105:946–952
Castillo A, Ramírez MC, Martín AC, Kilian A, Martín A, Atienza SG (2013) High-throughput genotyping of wheat-barley amphiploids utilising diversity array technology (DArT). BMC Plant Biol 13:87
Chiang YC, Chou CH, Lee PR, Chiang TY (2001) Detection of leaf-associated fungi based on PCR and nucleotide sequence of the ribosomal internal transcribed spacer (ITS) in Miscanthus. Bot Bull Acad Sinica 42:39–44
Chouvarine P, Cooksey AM, McCarthy FM, Ray DA, Baldwin BS, Burgess SC, Peterson DG (2012) Transcriptome-based differentiation of closely-related Miscanthus lines (transcriptome-based differentiation of Miscanthus). PLoS One 7:e29850
Clifton-Brown JC, Lewandowski I, Andersson B, Basch G, Christian DG, Kjeldsen JB, Jorgensen U, Mortensen JV, Riche AB, Schwarz KU, Tayebi K, Teixeira F (2001) Performance of 15 Miscanthus genotypes at five sites in Europe. Agron J 93:1013–1019
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Dominguez-Garcia MC, Belaj A, De la Rosa R, Satovic Z, Heller-Uszynska K, Kilian A, Martin A, Atienza SG (2012) Development of DArT markers in olive (Olea europaea L.) and usefulness in variability studies and genome mapping. Sci Hortic 136:50–60
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Głowacka K, Clark LV, Adhikari S, Peng J, Ryan Stewart J, Nishiwaki A, Yamada T, Jørgensen U, Hodkinson TR, Gifford J (2015) Genetic variation in Miscanthus × giganteus and the importance of estimating genetic distance thresholds for differentiating clones. GCB Bioenergy 7:386–404
Greef J, Deuter M (1993) Syntaxonomy of Miscanthus × giganteus Greef et Deu. Angew Bot 67:87–90
Greef J, Deuter M, Jung C, Schondelmaier J (1997) Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genet Resour Crop Ev 44:185–195
Grzebelus D, Iorizzo M, Senalik D, Ellison S, Cavagnaro P, Macko-Podgorni A, Heller-Uszynska K, Kilian A, Nothnagel T, Allender C (2014) Diversity, genetic mapping, and signatures of domestication in the carrot (Daucus carota L.) genome, as revealed by Diversity Arrays Technology (DArT) markers. Mol Breeding 33:625–637
Hauser LA, Crovello TJ (1982) Numerical analysis of generic relationships in Thelypodieae (Brassicaceae). Syst Bot 7:249–268
Heaton EA, Dohleman FG, Long SP (2008a) Meeting US biofuel goals with less land: the potential of Miscanthus. Glob Change Biol 14:2000–2014
Heaton EA, Flavell RB, Mascia PN, Thomas SR, Dohleman FG, Long SP (2008b) Herbaceous energy crop development: recent progress and future prospects. Curr Opin Biotech 19:202–209
Heller-Uszynska K, Uszynski G, Huttner E, Evers M, Carlig J, Caig V, Aitken K, Jackson P, Piperidis G, Cox M, Gilmour R, D’Hont A, Butterfield M, Glaszmann JC, Kilian A (2011) Diversity Arrays Technology effectively reveals DNA polymorphism in a large and complex genome of sugarcane. Mol Breed 28:37–55
Hernández P, Dorado G, Laurie D, Martin A, Snape J (2001) Microsatellites and RFLP probes from maize are efficient sources of molecular markers for the biomass energy crop Miscanthus. Theor Appl Genet 102:616–622
Hodkinson TR, Chase MW, Lledó DM, Salamin N, Renvoize SA (2002) Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J Plant Res 115:381–392
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25–e25
James KE, Schneider H, Ansell SW, Evers M, Robba L, Uszynski G, Pedersen N, Newton AE, Russell SJ, Vogel JC (2008) Diversity arrays technology (DArT) for pan-genomic evolutionary studies of non-model organisms. PLoS One 3:e1682
Jeżowski S (2008) Yield traits of six clones of Miscanthus in the first 3 years following planting in Poland. Ind Crops Prod 27:65–68
Jeżowski S, Glowacka K, Kaczmarek Z (2009) Wstępna ocena głównych parametrów wymiany gazowej związanych z fotosyntezą w odniesieniu do plonowania traw energetycznych z rodzaju Miscanthus w pierwszym roku uprawy. Acta Agrophysica 14:73–81
Jeżowski S, Głowacka K, Kaczmarek Z (2011) Variation on biomass yield and morphological traits of energy grasses from the genus Miscanthus during the first years of crop establishment. Biomass Bioenergy 35:814–821
Jones MB, Finnan J, Hodkinson TR (2015) Morphological and physiological traits for higher biomass production in perennial rhizomatous grasses grown on marginal land. GCB Bioenergy 7:375–385
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity Arrays Technology: a generic genome profiling technology on open platforms. In: Pompanon F, Bonin A (eds) Data production and analysis in population genomics. Humana Press, New York, pp 67–89
Lee NY (1964a) Taxonomic studies on the genus Miscanthus: anatomical patterns of leaves. Bot Mag Tokyo 77:122–130
Lee NY (1964b) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 1. J Jpn Bot 39:196–205
Lee NY (1964c) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 2. J Jpn Bot 39:257–268
Lee NY (1964d) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 3. J Jpn Bot 39:289–298
Lewandowski I, Clifton-Brown JC, Andersson B, Basch G, Christian DG, Jorgensen U, Jones MB, Riche AB, Schwarz KU, Tayebi K, Teixeira F (2003) Environment and harvest time affects the combustion qualities of Miscanthus genotypes. Agron J 95:1274–1280
Mace ES, Xia L, Jordan DR, Halloran K, Parh DK, Huttner E, Wenzl P, Kilian A (2008) DArT markers: diversity analyses and mapping in Sorghum bicolor. BMC Genom 9:26
Matlaga DP, Davis AS (2013) Minimizing invasive potential of Miscanthus × giganteus grown for bioenergy: identifying demographic thresholds for population growth and spread. J App Ecol 50:479–487
Mos M, Banks SW, Nowakowski DJ, Robson PRH, Bridgwater AV, Donnison IS (2013) Impact of Miscanthus × giganteus senescence times on fast pyrolysis bio-oil quality. Bioresour Technol 129:335–342
Oliver R, Jellen E, Ladizinsky G, Korol A, Kilian A, Beard J, Dumlupinar Z, Wisniewski-Morehead N, Svedin E, Coon M (2011) New Diversity Arrays Technology (DArT) markers for tetraploid oat (Avena magna Murphy et Terrell) provide the first complete oat linkage map and markers linked to domestication genes from hexaploid A. sativa L. Theor Appl Genet 123:1159–1171
Ovesná J, Kučera L, Vaculová K, Milotová J, Snape J, Wenzl P, Huttner E, Kilian A, Martelli G, Milella L (2013) Analysis of the genetic structure of a barley collection using DNA Diversity Array Technology (DArT). Plant Mol Biol Rep 31:280–288
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quinn LD, Allen DJ, Stewart JR (2010) Invasiveness potential of Miscanthus sinensis: implications for bioenergy production in the United States. GCB Bioenergy 2:310–320
Raman H, Stodart B, Cavanagh C, Mackay M, Morell M, Milgate A, Martin P (2010) Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop Pasture Sci 61:222–229
Rohlf F (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system Exeter Software. Exeter publishing Setauket, NY
Sang T, Zhu W (2011) China’s bioenergy potential. GCB Bioenergy 3:79–90
Sharma M, Nagavardhini A, Thudi M, Ghosh R, Pande S, Varshney RK (2014) Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genom 15:454
Simko I, Eujayl I, van Hintum TJ (2012) Empirical evaluation of DArT, SNP, and SSR marker-systems for genotyping, clustering, and assigning sugar beet hybrid varieties into populations. Plant Sci 184:54–62
Von Mark VC, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop Lesquerella and related species. PLoS One 8:e64062
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA 101:9915–9920
Xia L, Peng K, Yang S, Wenzl P, De Vicente MC, Fregene M, Kilian A (2005) DArT for high-throughput genotyping of cassava (Manihot esculenta) and its wild relatives. Theor Appl Genet 110:1092–1098
Zhao H, Yu JY, You FM, Luo MC, Peng JH (2011) Transferability of microsatellite markers from Brachypodium distachyon to Miscanthus sinensis, a potential biomass crop. J Integr Plant Biol 53:232–245
Acknowledgments
This study was mainly funded by Energene sp. z o.o., and partially supported by Peking University Shenzhen Graduate School start up grant 0068 to MD and a Key Project of Shenzhen Emerging Industries (No. JC201104210118A). MM holds executive position and has financial interest in Energene.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Tang and M. Daroch were contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, J., Daroch, M., Kilian, A. et al. DArT-based characterisation of genetic diversity in a Miscanthus collection from Poland. Planta 242, 985–996 (2015). https://doi.org/10.1007/s00425-015-2335-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2335-z