Abstract
Miscanthus genetic resources are widely distributed throughout China. However, genetic studies on Miscanthus lagged far behind other crops (e.g., sorghum, maize). To establish the comprehensive genetics knowledge of Miscnathus in China, here we report the genetic and phylogenetic diversity of 174 domestic Miscanthus accessions, along with an external Miscanthus × giganteus control. Cytological observations and flow cytometry analyses indicated that there were two major Miscanthus cytotypes in China: diploid (86.86%) and tetraploid (12.57%) without triploid. A total of 108 polymorphic loci generated from 25 SSR primers were used to evaluate the genetic variation. Large variations in genetic similarity coefficients (GSCs), ranging from 0.08 to 0.97 with a mean value of 0.39, were observed between these Miscanthus accessions. Our phylogenetic data revealed that these accessions were clustered into four main clades: M. section Miscanthus, M. section Diandranthus, M. section Triarrhena, and hybrids. The average percentage of polymorphic loci (P), gene diversity (H), and Shannon’s diversity index (I) among Miscanthus species are 70.93%, 0.22, and 0.34, respectively. These were consistent with the analysis of molecular variance (AMOVA) results, showing that 85% of genetic variation was found within clades. This study investigated the clear phylogenetic relationship of Miscanthus species in China, which will be valuable for further utilization of the germplasm in genetic improvement and hybrid breeding of Miscanthus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Miscanthus, a perennial C4 crop with rich lignocellulose, has received increasing attention worldwide for that it is potentially the next-generation renewable bioenergy resource. This biofuel crop is closely related to sorghum and sugarcane and is a member of the subtribe Saccharinae, tribe Andropogoneae, subfamily Panicoideae, and grass family of Poaceae (Hodkinson et al. 2002a). Of the 25 Miscanthus species worldwide, 14 of them are found in southern and eastern Asia as well as the Pacific Islands (Hodkinson et al. 2002b).
As one of the countries where Miscanthus originated, there is a wide range of genetically diversified germplasm in China (Sun et al. 2010; Zhang et al. 2013; Clark et al. 2014). To date, disagreements on how to classify Miscanthus species are still present. Early classifications were mainly based on morphological differences, resulting in two sections (Miscanthus and Triarrhena), along with a subsection (Kariyasua). Miscanthus sinensis, Miscanthus sacchariflorus, and Miscanthus oligostachyus were believed to belong to sections of Miscanthus, Triarrhena, and Kariyasua, respectively, and were further divided into 11 to 12 species (Hodkinson et al. 2002b, c; Sun et al. 2010). Chen and Renvoize (2006) identified seven species, M. sinensis, M. sacchariflorus, Miscanthus lutarioriparius, Miscanthus floridulus, Miscanthus paniculatus, Miscanthus nepalensis, and Miscanthus nudipes based on their paniculate inflorescences of racemes in mainland China. While M. sinensis and M. sacchariflorus are widely distributed in various locations in China, the remaining species are predominately identified in some specific ecological regions. For example, M. paniculatus, M. nepalensis, and M. nudipes are rarely found at high altitude areas (Chen and Renvoize 2006). Compared with M. sacchariflorus, M. lutarioriparius is endemic to central China where it can naturally grow vigorously up to 7 m in height with much thicker stems (Yan et al. 2012).
In general, Miscanthus can almost grow under any kind of environment (e.g., dry and unproductive mountainous regions, low wetlands, field footpaths, or seashores) due to its high water and nutrient use efficiency (Lewandowski and Schmidt 2006). Once established, it would show its high tolerance to various adverse conditions through the well-developed rhizome system. These favorable traits make Miscanthus an ideal lignocellulosic energy crop for most marginal land which are not ideal for grain or any crops of high economic value (Sang and Zhu 2010). Additional benefits include avoiding competition with food crops for fertile land and improving soil conditions and biological ecosystems.
Unlike many agronomic grain crops, most of Miscanthus germplasm remain uncharacterized and underutilized. Attention has greatly increased recently due to its huge potential as an important bioenergy crop (Chou 2009). The focus of Miscanthus studies over the last few years has been mainly limited to a very specific Miscanthus (e.g., Miscanthus × giganteus, M. sinensis). Cytologically, the base chromosome number of Miscanthus is x = 19, constituting its diploid 2n = 2x = 38, triploid 2n = 3x = 57, tetraploid 2n = 4x = 76, and hexaploid 2n = 6x = 114, respectively (Lafferty and Lelley 1994). Most Miscanthus species are self-incompatible and have a large complex genome with about 2.5 Gbp (Rayburn et al. 2009). Such genome size is equivalent to that of the maize genome but 6- and 20-fold of the Oryza sativa and Arabidopsis thaliana genomes, respectively.
Originated from Japan, M. × giganteus is recognized as a sterile triploid (2n = 3x = 57), and this vegetatively propagated Miscanthus is a hybrid of an allotetraploid M. sacchariflorus (2n = 4x = 76) × a diploid M. sinensis (2n = 2x = 38) (Linde-Laursen 1993; Lafferty and Lelley 1994; Hodkinson et al. 2002c). Compared with other types of Miscanthus, M. × giganteus has a number of advantages such as hybrid vigor, high yield, and wide range of adaptation to adverse environments (Cosentino et al. 2007; Christian et al. 2008). Currently, this is almost the only hybrid that has been widely grown in Europe and the USA for its biofuel purpose (Danalatos et al. 2007; Heaton et al. 2008). Clearly, the use of such narrow germplasm, i.e., only one hybrid, could result in a potential threat to the Miscanthus industry, due to its vulnerability to crop insects, diseases, and many other issues (Głowacka et al. 2015). Furthermore, the lack of genetic diversity studies on a wide range of Miscanthus germplasm may prevent the full use of other Miscanthus potentials.
Clark et al. (2014) recently studied the diversity and population structure on 703 M. sinensis, 12 M. sacchariflorus, 4 M. floridulus, 1 M. oligostachyus, as well as some hybrids by using SNP markers, and found south-eastern China to be the origin of M. sinensis populations. Studies on Miscanthus genetic diversity in South Korea (Qin et al. 2013; Yook et al. 2014), Japan (Shimono et al. 2013; Huang et al. 2014), and Europe (Slavov et al. 2013; Tang et al. 2015) have been mainly focused on a single species, M. sinensis, using limited samples from a restricted geographical distributions, while the similar results can also be found in China (Xu et al. 2013; Zhao et al. 2013; Nie et al. 2014).
Several studies on the ploidy level of Miscanthus have been reported. Lafferty and Lelley (1994) studied on four Miscanthus accessions and found diploid M. sinensis, triploid M. × giganteus, and both diploid and tetraploid M. sacchariflorus. Moon et al. (2013) studied ploidy diversity on 216 accessions collected from South Korea, including 49 M. sinensis, 164 M. sacchariflorus, and 3 M. × giganteus. In these accessions, diploid M. sinensis (21.29%), triploid M. × giganteus (1.39%), and both diploid (20.37%) and tetraploid (55.56%) M. sacchariflorus were identified. Li et al. (2013) studied 36 Chinese accessions including 12 M. sinensis, 14 M. sacchariflorus, and 10 M. lutarioriparius, and found that 2 M. sacchariflorus and 1 M. lutarioriparius were tetraploid (8.33%) and all of the others were diploid. A comprehensive genetic diversity study on a wide range of Miscanthus samples in China will be valuable for genetic improvement, breeding, and fundamental research in Miscanthus worldwide.
In this study, we collected 175 wild Miscanthus germplasm, including various species, varieties, and hybrids from different distinct geographic and ecological locations in China to reveal genetic diversity and understand the evolutionary relationships. We reported the results of cytological analyses on the germplasm to determine their ploidy levels and elucidated their phylogenetic relationships using the simple sequence repeat (SSR) markers. In contrast with previous work, our findings suggested that M. lutarioriparius was in fact a variant of M. sacchariflorus in China. The objectives of this study were to characterize the germplasm pool for wild Miscanthus germplasm in China, detect the ploidy levels of Miscanthus, and explore genetic diversity and phylogenetic evolution relationship for Miscanthus. This knowledge is essential for promoting genetic improvements and developing new germplasm and novel varieties of Miscanthus.
Materials and Methods
Miscanthus Germplasm Collections
According to the distribution of the genus Miscanthus which is recorded in Flora of China (Chen and Renvoize 2006), we collected 164 wild Miscanthus including 39 M. sinensis, 20 M. floridulus, 83 M. sacchariflorus, 16 M. lutarioriparius, 2 M. nudipes, and 4 hybrids that were widely distributed across the country. Miscanthus wild germplasm, naturally growing in various ecosystems of the most provinces with the exception of Xinjiang, Tibet, Qinghai, and Inner Mongolia in mainland China, were collected. Of all wild Miscanthus genetic resources collected, M. sinensis and M. sacchariflorus were widely distributed in China and exhibited strongest ecological adaptabilities. M. sinensis was found from latitude of 43.82° N of northeast China (e.g., Jilin Province) where temperature is very low to that of 19.45° N of southeast China where the temperature is very high (e.g., Hainan Province), and from longitude of 104° E to that of 127° E (altitude 27–1255 m). M. sacchariflorus was discovered from latitude of the northeast China 47.20° N (e.g., the north of Heilongjiang Province) to that of the southeast China 30.30° N (e.g., Hubei Province) and from longitude of 107° E to that of 128° E (altitude 3–1119 m). M. floridulus and M. lutarioriparius, on the other hand, were spread only within the region of southeast China. M. lutarioriparius is unique to China, which was found only in the middle and lower parts of Yangtze River. M. nudipes grows only within the districts of Yunnan, Guizhou, and Chongqing where the altitude is about 1600–1700 m above the sea level. Interestingly, several hybrids derived from naturally crossing between M. sinensis and M. lutarioriparius and between M. sinensis and M. sacchariflorus had been identified and collected. Figure 1 and Online Resource 1 (ESM 1) illustrate all Miscanthus accessions collected, each of which with the detailed information on sample name, species, geographical location and altitude, etc. These locations represent many distinct ecological environments in China.
Native distribution map of wild Miscanthus germplasm collected in China. Filled triangles indicate Miscanthus sacchariflorus, empty triangles indicate Miscanthus sinensis, filled squares indicate Miscanthus lutarioriparius, and empty squares indicate Miscanthus floridulus. Miscanthus nudipes and hybrids are marked by filled circles and empty circles, respectively
An external triploid M. × giganteus (kindly provided by the University of Illinois at Urbana-Champaign, USA) and ten domestic man-made interspecific hybrids (crossed between different Miscanthus species in China, Online Resource 1 (ESM 1)) were added into this study as controls. The collections of Miscanthus germplasm were planted in a 2.0 × 2.0 m plot, one replicate/genotype, on the Experimental Farm of Shandong Agricultural University, Tai’an, China, in March, 2011, and each genotype was also planted in the university greenhouse as a backup. No fertilizer was applied at any time during this study.
Ploidy Levels Determined by Cytological Observations and Flow Cytometry
Mitotic observations were performed based on the recent improved cytological techniques described by Kim et al. (2010). Specifically, root tips (3–4 cm long) from healthy Miscanthus accessions were taken at 8:30–9:30 am. Roots were then fixed with fresh fixative containing ethanol and acetic acid with 3:1 ratio (v/v) for 20 h and transferred to 70% ethanol to store at 4 °C. After the hydrolytic process, the roots were stained with carbol fuchsin. Active dividing cells with C-metaphase chromosomes were visualized and counted under an Olympus BX60 microscope (Olympus America Inc., Melville, NY, USA) and cells with clear chromosome images were recorded using a Nikon DIGITAL SIGHT DS-U3 camera (Nikon Instruments Inc., Tokyo, Japan).
Ploidy levels of various Miscanthus samples were determined based on the cytological data and nuclear DNA contents were measured by flow cytometry (PA model, Germany Partec) according to Rayburn et al. (2009). Briefly, 1~2 cm2 fresh leaf tissue of each sample was submerged in 800 μl extraction buffer (13% hexylene glycol, 10 mM Tris-HCL, 10 mM MgCl2) and 12 μl 25% Triton X-100 in a petri dish. The tissue was chopped into homogenized mixture and filtered. For DNA staining, 400 μl 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/ml) was added into the centrifuged nuclear pellet suspension. DNA contents of the diploid Miscanthus and triploid M. × giganteus were used as standards. Each sample was tested three times. Cytological results of all 175 Miscanthus samples examined in this study are listed in Table 1.
Genetic Diversity and Phylogeny Detected by SSR
To investigate genetic diversity and phylogenetic relationship, genomic DNA of 175 Miscanthus accessions were isolated using sodium dodecyl sulfate (SDS) method and diluted into 5 ng/μl as a working solution.
SSR markers were identified by searching the published genome database and EST sequences of Miscanthus (Ma et al. 2012; Barling et al. 2013) using MIcroSAtellite identification tool (MISA) and Primer5. Primers were synthesized by Dingguo Biotech Company (Beijing, China).
SSR markers were amplified in 10 μl PCR mixtures, each of which contained 10 ng of template DNA, 1× Green GoTaq buffer (Promega Corporation, Madison, USA) consisting of 2.0 mM MgCl2, 0.15 μM of both reverse and forward primers (Beijing Dingguo Changsheng Biotechnology Co. LTD, Beijing, China), and 0.5 units of Taq polymerase (Thermo Scientific, Scoresby, Australia). The PCR reactions were performed on a Veriti 96-well thermal cycler (Applied Biosystems, USA). The reaction mixture of each sample was incubated at 94 °C for 5 min, followed by 35 cycles of 45 s denaturation at 94 °C, 30 s annealing at 55 °C, 45 s extension at 72 °C, and 7 min final extension at 72 °C. PCR products were separated on 8% polyacrylamide gels (PAG) (Amresco, Solon, OH, USA) and visualized by ethidium-bromide staining using Bio-Rad Gel Doc™ XR+ System (Bio-Rad Laboratories, Inc., California, USA).
Data Analysis
Out of the 40 SSRs, 25 primers generated scorable polymorphic amplicons in Miscanthus. Sequence information of the SSRs is listed in Online Resource 2 (ESM 1). These polymorphic SSR bands were converted to haplotype profiles as the required format of the program. Polymorphism information content (PIC) was calculated according to the method of Smith et al. (1997):
where P ij is the frequency of the jth allele for the ith marker and is summed over n alleles. A binary matrix generated from coding 1 or 0 depending on presence or absence of each polymorphic SSR allele was used for calculating the genetic similarity coefficients (GSCs) by a Jaccard’s similarity coefficient (Chae et al. 2014). The GSCs of these Miscanthus germplasm were employed for Miscanthus cluster classifications using neighbor joining (NJ). The NTSYS-pc version 2.02 (Rohlf 2000) was used to conduct both the phylogenetic tree construction with bootstrapping 1000 times and principal coordinate analysis (PCoA). The mean GS estimates of the inter- and intra-groups were calculated among all possible pairs of the collections belonging to each group.
To analyze phylogenetic relationships among various Miscanthus germplasm collected, 40 pairs of polymorphic SSR primers were initially used. Analysis of molecular variance (AMOVA) was performed using GenAlEx 6.5 (Peakall and Smouse 2012) to assess the genetic differentiation in the Miscanthus germplasm among and within species. Furthermore, the percentage of polymorphic loci (P), Nei’s (1978) gene diversity (H), Shannon’s information index of diversity (I), and FST (Wright 1965) among species was also calculated.
Results
Cytotypes of Various Miscanthus Collections
Confirmation of Ploidy Levels by Flow Cytometry
Figure 2 illustrates the flow cytometric G0/G1 DAPI-stained nuclear DNA histograms generated by relative fluorescent intensities of representatives of diploids, triploids, and tetraploids, respectively. As shown in Fig. 2, the dominant peaks of a known diploid and a triploid M. × giganteus used as controls appeared at their respectively expected channels 80 and 120 (Fig. 2a, c). Clearly, samples showed their DNA peaks on the same channel as the diploid control were the results of diploids (Fig. 2b). Conversely, those with DNA peaks on channel 160 were tetraploids as their peaks had shifted to the right by another 80 channels (Fig. 2d). Similarly, DNA peaks positioned in between the diploids and tetraploids on channel 120 were the results of triploids. Coefficients of variations (CVs) for the sample peaks in 175 Miscanthus germplasm were all within the normal range of 3 to 5% (Online Resource 1 (ESM 1)) (Doležel et al. 1998). The results of ploidy levels revealed by flow cytometry DNA data were consistent with those of cytological analyses.
Cytological Analyses
Chromosome counts revealed two major cytotypes, 2n = 2x = 38 (Fig. 3a) and 2n = 4x = 76 (Fig. 3c), along with M. × giganteus, 2n = 3x = 57 (Fig. 3b). These data showed that, of the 175 Miscanthus samples examined, 152 (86.86%) were diploids (2n = 2x = 38), including 68 M. sacchariflorus, 9 M. lutarioriparius, 39 M. sinensis, 20 M. floridulus, 2 M. nudipes, and 14 hybrids. The second major cytotype with a total of 22 (12.57%) was tetraploids (2n = 4x = 76), 15 of which were M. sacchariflorus, and 7 were M. lutarioriparius. The triploid M. × giganteus control chromosome number was further confirmed to be 2n = 3x = 57 (Table 1).
In the 83 M. sacchariflorus accessions, 15 (18.07%) were tetraploid, whereas 7 (43.75%) out of 16 M. lutarioriparius were tetraploids. No polyploid (neither triploid nor tetraploid) was identified among M. sinensis, M. floridulus, M. nudipes, and hybrids (Table 1). Tetraploids were found only from M. sacchariflorus and M. lutarioriparius. None of the accessions collected in this study was identified as triploid.
Genetic Diversity and Phylogeny Detection
Polymorphism of SSR Markers
Of 40 pairs of primers, only 25 resulted in amplified products showing clear abundant polymorphisms (Online Resource 2 (ESM 1)), and a total of 149 polymorphic bands identified from 175 accessions were amplified. The numbers of polymorphic bands varied with different SSRs, ranging from 3 to 9 and averaging 5.96 per locus. With the exclusion of those non-specific bands, a total of final 108 polymorphic bands (4.32/SSR) were used for the phylogenetic evaluation of the collected Miscanthus samples (Online Resource 2 (ESM 1)).
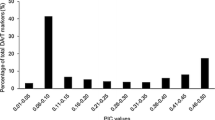
In general, PIC reflect the allele diversities and frequencies within the examined materials. In this study, ESSR13 showed the lowest PIC value of 0.41, whereas the highest PIC value for GSSR66 was 0.81, with an average of 0.63 among all loci (Online Resource 2 (ESM 1)).
Genetic Variations and AMOVA Analysis
The GSCs were calculated based on SSR polymorphisms present in these Miscanthus germplasm, ranging from 0.08 to 0.97, with an average GSC 0.39 (Online Resource 3 (ESM 1)). Two diploid genotypes M157 (M. sacchariflorus collected from Beijing, China) and M324 (M. sinensis collected from Sichuan, China) showed the lowest GSC 0.08, whereas two tetraploid cytotypes M049 and M050 (M. sacchariflorus collected from Henan, China) gave rise to the highest GSC 0.97 (Online Resource 3 (ESM 1)). These GSC data were the clear indications of the presence of abundant genetic variations in these Miscanthus in China.
As indicated in Online Resource 4 (ESM 1), both M. sinensis (GSCs from 0.26 to 0.90) and M. sacchariflorus/M. lutarioriparius (GSCs from 0.34 to 0.97) showed higher intra-species genetic diversity than that of other species. Similarly, hybrids also exhibited high genetic variations (GSC 0.34 to 0.93). However, little variation was detected within M. nudipes, perhaps largely due to the very limited numbers (only two) collected from only two locations. Medium intra-species genetic diversity with GSCs from 0.31 to 0.86 was detected in M. floridulus. The mean GSC (0.51) among M. floridulus was higher than that among M. sinensis (0.49) (Online Resource 4 (ESM 1)).
Data listed in Table 2 indicated that the highest interspecies GSC between M. sinensis and M. floridulus is 0.37 and the lowest interspecies GSC between M. floridulus and M. sacchariflorus/M. lutarioriparius is 0.22. These results not only revealed clear genetic variations but also showed evolutionary relationships among these germplasm resources. Interestingly, comparing the GSCs of the hybrids to those detected between Miscanthus species, the highest GSC (0.43) appeared in-between the hybrids and the two species M. sacchariflorus/M. lutarioriparius, demonstrating their close evolutionary relationship. The hybrids have close genetic distance to the M. sacchariflorus/M. lutarioriparius and this may be due to that M. sacchariflorus/M. lutarioriparius is the dominant population in China.
The AMOVA analysis (Table 3) showed that genetic variations were significant in both within and between Miscanthus species, and the genetic variation of within species (85%) was substantially greater than that of between species (15%). The global genetic differentiation between these species was relatively weak but statistically significant (FST = 0.16, P < 0.001). The highest genetic differentiation was between M. floridulus and M. sacchariflorus/M. lutarioriparius (FST = 0.40, P < 0.001), while the lowest was between M. sacchariflorus/M. lutarioriparius and hybrid (FST = 0.17, P < 0.001). The indirect estimator of gene flow (Nm) among species was 2.75. According to the three main genetic diversity parameters including Nei’s genetic distance (H), Shannon’s information index of diversity (I), and percentage of polymorphic bands (P), the Miscanthus germplasm genetic resources in China have a varied genetic diversity. Among the Miscanthus species, the P value ranged from 16.67% (M. nudipes) to 94.44% (M. sacchariflorus/M. lutarioriparius), with an average of 70.93%. The H value ranged from 0.08 to 0.30, with an average of 0.22 at the population level. While the variation trend of I value was similar to the other two parameters, with an average of 0.34 (Table 4).
Cluster Analysis
A phylogenetic tree was constructed based on the GSC data (Fig. 4). In this tree, 175 samples were divided into four major clades (I, II, III, and IV) (Fig. 4). The major clades (I, II, and III) represented M. section Miscanthus, M. section Diandranthus, and M. section Triarrhena, respectively, while clade IV included all hybrids. Clade I consists of two sub-branches, corresponding to the two species in M. section Miscanthus: M. sinensis and M. floridulus, clade II includes M. nudipes, clade III represents the two species (M. sacchariflorus and M. lutarioriparius) combined in M. section Triarrhena, and clade IV covers all hybrids, including the external triploid M. × giganteus control (Fig. 4 and Online Resource 1 (ESM 1)).
Dendrogram constructed with a neighbor-joining clustering algorithm from the pairwise matrix of genetic similarities among 175 Miscanthus germplasm accessions with bootstrapping 1000 times. Miscanthus floridulus (blue), Miscanthus sinensis (red), Miscanthus nudipes (yellow), hybrids (green), Miscanthus sacchariflorus (purple, black letters) and Miscanthus lutarioriparius (purple, orange letters)
To characterize the genetic variation of these Miscanthus germplasm, a three-dimensional PCoA (Fig. 5) were constructed. All of 175 materials were scattered in a specific determined space and clearly divided into four groups (I, II, III, and IV), which were corresponding to the groups in the phylogenetic tree (Figs. 4 and 5). Clusters I, II, III, and IV were occupied by M. sinensis and M. floridulus, M. nudipes, M. sacchariflorus and M. lutarioriparius, and hybrids, respectively. The first, second, and third principal coordinates, termed PC1, PC2, and PC3 explained 22.68, 4.43, and 3.54% of the total variations, respectively. Three species, M. sinensis, M. nudipes, and M. floridulus were clustered together along dimension of PC1. M. floridulus was genetically separated from M. sinensis and M. nudipes along dimension of PC2 and PC3. M. lutarioriparius were located at the lower position of the PC3 axis within the M. section Triarrhena group (Fig. 5).
The hybrids in cluster IV were distributed between M. section Triarrhena and M. section Miscanthus (Fig. 5). Triploid hybrid M. × giganteus was also included in cluster IV. M. × giganteus was believed to be a hybrid of a tetraploid M. sacchariflorus (2n = 4x = 76) × a diploid M. sinensis (2n = 2x = 38) (Linde-Laursen 1993).
Discussion
Miscanthus Genetic Diversity in China and Their Evolutionary Relationships
The data presented in this report are the result of comprehensive analyses using the techniques of cytology, flow cytometry, SSR molecular markers, and phylogenetics. Our cytological studies indicated that most of Miscanthus germplasm in China, including all natural hybrids, were diploid, along with some specific regional distributed tetraploids of M. sacchariflorus and M. lutarioriparius. Our results are in good agreement with those reported by Li et al. (2013), who studied 36 populations of three Miscanthus species from China: M. lutarioriparius, M. sacchariflorus, and M. sinensis by flow cytometry technique. Using the same method, similar conclusions from investigating the 216 South Korea Miscanthus germplasm with diploid (20.37%) and tetraploid (55.56%) M. sacchariflorus have recently been reported (Moon et al. 2013).
Our SSR and phylogenetic analyses revealed that there were relatively high genetic variations within each species and among various species in the Chinese genus Miscanthus. These data demonstrated that high genetic similarity is present in the Miscanthus germplasm in China. Early studies on 260 individuals of M. sinensis collected from 24 natural populations in Southwest China showed that inter- and intra-species genetic similarities ranged from 0.57 to 0.97 with an average of 0.66 (Nie et al. 2014). Similarly, genetic similarities of 69 M. sinensis and M. sacchariflorus/M. lutarioriparius derived from South Korea, Japan, and China were from 0.34 to 0.86 (Yook et al. 2014).
Our interspecific phylogenetic data showed that the least genetic similarity was between M. floridulus and M. sacchariflorus (GSC 0.22, Table 2), and the closest interspecific relationship was between M. sinensis and M. floridulus (GSC 0.37, Table 2). In the study of Clark et al. (2014), M. floridulus was also genetically close to M. sinensis, whereas M. sacchariflorus was clearly distinct from M. sinensis. Furthermore, the highest GSC (0.43, Table 2) appeared in between the hybrids and the two species M. sacchariflorus and M. lutarioriparius, demonstrating that hybrids were more evolutionarily related to M. sacchariflorus/M. lutarioriparius than to other species, which might be due to its self-incompatibility, high heterozygosity, and complex genome in the genus Miscanthus.
Classifications of Miscanthus Germplasm in China
By combining cytological and molecular biological techniques, we successfully evaluated the genetic evolutionary relationship of the 174 wild Miscanthus germplasm collected from various ecological districts in China. Our results suggested that the use of molecular phylogenetic analysis enabled us to distinguish different species and identify the naturally derived hybrids. The examined Miscanthus germplasm in this study could be divided into three sections: M. section Miscanthus, M. section Diandranthus, and M. section Triarrhena. These three sections include five main Miscanthus species, of which M. sinensis and M. floridulus belong to M. section Miscanthus, M. nudipes is part of M. section Diandranthus, and M. sacchariflorus and M. lutarioriparius are members of M. section Triarrhena. Additionally, our ten man-made interspecific hybrids, four naturally derived hybrids, and the external control M. × giganteus were gathered closely in the phylogenetic tree (Fig. 4). On the other hand, Chae et al. (2014) suggested that Miscanthus accessions fell into five taxonomic groups, including the existing taxonomic section Miscanthus, diploid and tetraploid Miscanthus sacchariflorus, and a fourth (M. × giganteus) and fifth group (Miscanthus “purpurascens”). Furthermore, AMOVA analysis agreed with the clustering pattern, the NJ tree, and the genetic distance measurements among the four clades includinge five main Miscanthus species and the hybrids, suggesting the accuracy and reliability of our phylogenetic analyses.
It is noteworthy that the two species M. sacchariflorus and M. lutarioriparius in M. section Triarrhena, could not be categorized into two independent sub branches. With regard to the considering morphological similarities (e.g., similar stamens and awnless spikelets per flower) between two species except the major difference in plant height (Chen and Renvoize 2006; Yan et al. 2012), it seems that M. lutarioriparius is just a variant of M. sacchariflorus species. This suggestion is not in agreement with the result of Chae et al. (2014) who reported that diploid and tetraploid M. sacchariflorus were taxonomically different, the latter more closely related to M. lutarioriparius. However, our speculation is similarity to that of Sun et al. (2010) who concluded that M. lutarioriparius was most likely to be a variant of the same species, M. sacchariflorus, since plant heights were the only difference between the two.
Miscanthus Tetraploids and Their Regional Distributions
Through analyzing distribution and localization of ploidy level of each accession, we found that tetraploids of M. sacchariflorus and M. lutarioriparius were geographically distributed in the Middle East of China (Latitude 28° 16′ N–37° 30′ N, Longitude 122° 18′ E–122° 18′ E, Online Resource 1 (ESM 1)). While both diploid and tetraploid coexisted in these regions, diploids were also identified in other parts of China.
Plant whole genome duplication (WGD) has been recognized to be one of the major driving forces for plant evolution and speciation, and almost all angiosperms had undergone one or several times of WGDs during their long evolutionary history (Mallet 2007; Renny-Byfield et al. 2010). Maize, one of the most important economic crops, is a paleopolyploid, a result of at least three times of WGDs (Gaut and Doebley 1997; Blanc and Wolfe 2004; Swigonova 2004). Miscanthus belongs to subtribe sugarcane, and the ancestor of the sugarcane subtribe was separated from the sorghum subtribe about 3.8–4.6 million years ago (Kim et al. 2014b). A recent study on genome comparison has demonstrated the high similarities between M. sinensis and Sorghum bicolor, suggesting that Miscanthus was a result of WGD of the ancestral Saccharinae chromosome number x = 10, with a single chromosome fusion, leading to the odd base chromosome number x = 19 (Swaminathan et al. 2012; Kim et al. 2014a). Tetraploidization of Miscanthus occurred most likely after its divergence from its closely associated sugarcane clade (Swaminathan et al. 2012). However, the mechanism underlying the occurrence of the tetraploids of M. sacchariflorus and M. lutarioriparius in China remains to be further elucidated.
In conclusion, this study described, for the first time, cytological analyses, the genetic diversity, and phylogenetic evolution of various Miscanthus species and hybrids located in China. Abundant genetic variations were detected within and among Miscanthus species. Another significant achievement of this study is the discovery of M. sacchariflorus and M. lutarioriparius tetraploids that are localized in several provinces where climates are similar. Furthermore, our phylogenetic data suggest that M. lutarioriparius is in fact a variant of M. sacchariflorus. These results will be undoubtedly valuable for further genetic breeding, fundamental research, and ultimate utilization of the rich Miscanthus genetic resources not only in China but also elsewhere in the world.
References
Barling A, Swaminathan K, Mitros T, James BT, Morris J, Ngamboma O, Hall MC, Kirkpatrick J, Alabady M, Spence AK, Hudson ME, Rokhsar DS, Moose SP (2013) A detailed gene expression study of the Miscanthus genus reveals changes in the transcriptome associated with the rejuvenation of spring rhizomes. BMC Genomics 14:864. https://doi.org/10.1186/1471-2164-14-864
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691. https://doi.org/10.1105/tpc.021410
Chae WB, Hong SJ, Gifford JM, Rayburn AL, Sacks EJ, Juvik JA (2014) Plant morphology genome size and SSR markers differentiate five distinct taxonomic groups among accessions in the genus Miscanthus. GCB Bioenergy 6:646–660. https://doi.org/10.1111/gcbb.12101
Chen SL, Renvoize SA (2006) Miscanthus. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, science press, Beijing, China. Missouri Botanical Garden Press, St Louis, pp 581–583
Chou CH (2009) Miscanthus plants used as an alternative biofuel material: the basic studies on ecology and molecular evolution. Renew Energy 34:1908–1912. https://doi.org/10.1016/j.renene.2008.12.027
Christian DG, Riche AB, Yates NE (2008) Growth yield and mineral content of Miscanthus giganteus grown as a biofuel for 14 successive harvests. Ind Crop Prod 28:320–327
Clark LV, Brummer JE, Głowacka K, Hall MC, Heo K, Peng JH, Yamada T, Yoo JH, Yu CY, Zhao H, Long SP, Sacks EJ (2014) A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Ann Bot 114:97–107. https://doi.org/10.1093/aob/mcu084
Cosentino SL, Copani V, Foti S, Patane C, Sanzone E (2007) Effects of soil water content and nitrogen supply on the productivity of Miscanthus ×giganteus Greef et Deu in a Mediterranean environment. Ind Crop Prod 25:5–88. https://doi.org/10.1016/j.indcrop.2006.07.006
Danalatos NG, Archontoulis SV, Mitsios I (2007) Potential growth and biomass productivity of Miscanthus × giganteus as affected by plant density and N-fertilization in central Greece. Biomass Bioenergy 31:145–152. https://doi.org/10.1016/j.biombioe.2006.07.004
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82:17–26. https://doi.org/10.1006/anbo.1998.0730
Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental Allotetraploid origin of maize. Proc Natl Acad Sci 94:6809–6814. https://doi.org/10.1073/pnas.94.13.6809
Głowacka K, Clark LV, Adhikari S, Peng J, Stewart JR, Nishiwaki A, Yamada T, Jorgensen U, Hodkinson TR, Gifford J, Juvik J, Sacks EJ (2015) Genetic variation in Miscanthus × giganteus and the importance of estimating genetic distance thresholds for differentiating clones. GCB Bioenergy 7:386–404. https://doi.org/10.1111/gcbb.12166
Heaton E, Dohleman FG, Long SP (2008) Meeting US biofuel goals with less land: the potential of Miscanthus. Glob Chang Biol 14:2000–2014. https://doi.org/10.1111/j.1365-2486.2008.01662.x
Hodkinson TR, Chase MW, Renvoize SA (2002a) Characterization of a genetic resource collection for Miscanthus (Saccharinae Andropogoneae Poaceae) using AFLP and ISSR PCR. Ann Bot 89:627–636. https://doi.org/10.1093/aob/mcf091
Hodkinson TR, Chase MW, Lledó MD, Salamin N, Renvoize SA (2002b) Phylogenetics of Miscanthus Saccharum and related genera (Saccharinae Andropogoneae Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J Plant Res 115:381–392. https://doi.org/10.1007/s10265-002-0049-3
Hodkinson TR, Chase MW, Takahashi C, Leitch IH, Bennett MD, Renvoize SA (2002c) The use of DNA sequencing (ITS AND TRNL-F) AFLP and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae). Am J Bot 89:279–286. https://doi.org/10.3732/ajb.89.2.279
Huang CL, Ho CW, Chiang YC, Shigemoto Y, Hsu TW, Hwang CC, Ge XJ, Chen C, Wu TH, Chou CH, Huang HJ, Gojobori T, Osada N, Chiang TY (2014) Adaptive divergence with gene flow in incipient speciation of Miscanthus floridulus/sinensis complex (Poaceae). Plant J 80:834–847. https://doi.org/10.5169/seals-268384
Kim S, Rayburn AL, Lee DK (2010) Genome size and chromosome analyses in prairie cordgrass. Crop Sci 50:2277–2282. https://doi.org/10.2135/cropsci2010.03.0140
Kim C, Lee TH, Guo H, Chung SJ, Paterson AH, Kim DS, Lee GJ (2014a) Sequencing of transcriptomes from two Miscanthus species reveals functional specificity in rhizomes and clarifies evolutionary relationships. BMC Plant Biol 14:57–60. https://doi.org/10.1186/1471-2229-14-134
Kim C, Wang X, Lee TH, Jakob K, Lee GJ, Paterson AH (2014b) Comparative analysis of Miscanthus and Saccharum reveals a shared whole-genome duplication but different evolutionary fates. Plant Cell 26:2420–2429. https://doi.org/10.1105/tpc.114.125583
Lafferty J, Lelley T (1994) Cytogenetic studies of different Miscanthus species with potential for agricultural use. Plant Breed 113:246–249
Lewandowski I, Schmidt U (2006) Nitrogen, energy and land use efficiencies of Miscanthus, reed canary grass and triticale as determined by the boundary line approach. Agric Ecosyst Environ 112:335–346. https://doi.org/10.1016/j.agee.2005.08.003
Li X, Hu D, Luo M, Zhu M, Li X, Luo F, Li JQ, Yan J (2013) Nuclear DNA content variation of three Miscanthus species in china. Genes Genom 35:13–20. https://doi.org/10.1007/s13258-013-0063-y
Linde-Laursen I (1993) Cytogenetic analysis of Miscanthus ‘Giganteus’ an interspecific hybrid. Hereditas 119:297–300. https://doi.org/10.1111/j.1601-5223.1993.00297.x
Ma X-F, Jensen E, Alexandrov N, Troukhan M, Zhang L, Thomas-Jones S, Farrar K, Clifton-Brown J, Donnison I, Swaller T, Flavell R (2012) High resolution genetic mapping by genome sequencing reveals genome duplication and tetraploid genetic structure of the diploid Miscanthus sinensis. PLoS One 7:e33821. https://doi.org/10.1371/journal.pone.0033821
Mallet J (2007) Hybrid speciation. Nature 446:279–283. https://doi.org/10.1038/nature05706
Moon YH, Cha YL, Choi YH, Yoon YM, Koo BC, Ahn JW, An GH, Kim JK, Park KG (2013) Diversity in ploidy levels and nuclear DNA amounts in Korean Miscanthus species. Euphytica 193:317–326. https://doi.org/10.1007/s10681-013-0910-6
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nie G, Zhang XQ, Huang LK, Xu WZ, Wang JP, Zhang YW, Ma X, Yan YH, Yan HD (2014) Genetic variability and population structure of the potential bioenergy crop Miscanthus sinensis (Poaceae) in southwest China based on SRAP markers. Molecules 19:12881–12897. https://doi.org/10.3390/molecules190812881
Peakall R, Smouse PE (2012) GenAlEx 65: genetic analysis in excel population genetic software for teaching and research - an update. Bioinformatics 28:2537–2539
Qin Y, Kabirm A, Wangh W, Leey H, Hongs H, Kimj Y, Yook MJ, Kim DS, Kim C, Kwon H, Kim W (2013) Assessment of genetic diversity and relationships based on RAPD and AFLP analyses in Miscanthus genera landraces. Can J Plant Sci 93:171–182. https://doi.org/10.4141/cjps2012-166
Rayburn AL, Crawford J, Rayburn CM, Juvik JA (2009) Genome size of three Miscanthus species. Plant Mol Biol Report 27:184–188. https://doi.org/10.1007/s11105-008-0070-3
Renny-Byfield S, Ainouche M, Leitch IJ, Lim KY, Comber SCL, Leitch AR (2010) Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin. Ann Bot 105:527–533. https://doi.org/10.1093/aob/mcq008
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, 2.1 edn. Applied Biostatistics, New York
Sang T, Zhu W (2010) China’s bioenergy potential. GCB Bioenergy 3:79–90. https://doi.org/10.1111/j.1757-1707.2010.01064.x
Shimono Y, Kurokawa S, Nishida T, Ikeda H, Futagami N (2013) Phylogeography based on intraspecific sequence variation in chloroplast DNA of Miscanthus sinensis (Poaceae) a native pioneer grass in Japan. Botany 91:449–456. https://doi.org/10.1139/cjb-2012-0212
Slavov G, Robson P, Jensen E, Hodgson E, Farrar K, Allison G, Hawkins S, Thomas-Jones S, Ma XF, Huang L, Swaller T, Flavell R, Clifton-Brown J, Donnison I (2013) Contrasting geographic patterns of genetic variation for molecular markers vs phenotypic traits in the energy grass Miscanthus sinensis. GCB Bioenergy 5:562–571. https://doi.org/10.1111/gcbb.12025
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea Mays L): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173. https://doi.org/10.1007/s001220050544
Sun Q, Lin Q, Yi Z, Yang Z, Zhou F (2010) A taxonomic revision of Miscanthus sl (Poaceae) from China. Bot J Linn Soc 164:178–220. https://doi.org/10.1111/j.1095-8339.2010.01082.x
Swaminathan K, Chae WB, Mitros T, Varala K, Xie L, Barling A, Glowacka K, Hall M, Jezowski S, Ming R, Hudson M, Juvik JA, Rokhsar DS, Moose SP (2012) A framework genetic map for Miscanthus sinensis from RNAseq-based markers shows recent tetraploidy. BMC Genomics 13:142–158. https://doi.org/10.1186/1471-2164-13-142
Swigonova Z (2004) On the tetraploid origin of the maize genome. Comp Funct Genomics 5:281–284. https://doi.org/10.1002/cfg.395
Tang J, Daroch M, Kilian A, Jeżowski S, Pogrzeba M, Mos M (2015) DArT-based characterisation of genetic diversity in a Miscanthus collection from poland. Planta 242:985–996. https://doi.org/10.1007/s00425-015-2335-z
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:295–420
Xu WZ, Zhang XQ, Huang LK, Nie G, Wang JP (2013) Higher genetic diversity and gene flow in wild populations of Miscanthus sinensis in southwest China. Biochem Syst Ecol 48:174–181. https://doi.org/10.1016/j.bse.2012.11.024
Yan J, Chen W, Luo F, Ma H, Meng A, Li X, Zhu M, Li S, Zhou M, Zhu W, Han B, Song G, Li J, Sang T (2012) Variability and adaptability of Miscanthus species evaluated for energy crop domestication. GCB Bioenergy 4:49–60. https://doi.org/10.1111/j.1757-1707.2011.01108.x
Yook MJ, Lim SH, Song JS, Kim JW, Zhang CJ, Lee EJ, Ibaragi Y, Lee GJ, Nah G, Kim DS (2014) Assessment of genetic diversity of korean Miscanthus using morphological traits and SSR markers. Biomass Bioenergy 66:81–92. https://doi.org/10.1016/j.biombioe.2014.01.025
Zhang QX, Shen YK, Shao RX, Fang J, He YQ, Ren JX, Zheng BS, Chen GJ (2013) Genetic diversity of natural Miscanthus sinensis populations in China revealed by ISSR markers. Biochem Syst Ecol 48:248–256. https://doi.org/10.1016/j.bse.2012.12.024
Zhao H, Wang B, He J, Yang J, Pan L, Sun D, Peng J (2013) Genetic diversity and population structure of Miscanthus sinensis germplasm in China. PLoS One 8:e75672. https://doi.org/10.1371/journal.pone.0075672
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (31271352 and 31071471) and the Department of Science and Technology of Shandong province (2013GNC11102). We thank Dr. Ji-Ping Zhao (USA) and Dr. Yanyan Wang (Associate Professor in University of Illinois, College of Medicine at Urbana-Champaign) for their English improvement and constructive comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(XLSX 40 kb)
Rights and permissions
About this article
Cite this article
Ge, C., Liu, X., Liu, S. et al. Miscanthus sp.: Genetic Diversity and Phylogeny in China. Plant Mol Biol Rep 35, 600–610 (2017). https://doi.org/10.1007/s11105-017-1048-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-017-1048-9