Abstract
Arabidopsis END1-LIKE (AtEND1) was identified as a homolog of the barley endosperm-specific gene END1 and provides a model for the study of this class of genes and their products. The END1 is expressed in the endosperm transfer cells (ETC) of grasses. The ETC are responsible for transfer of nutrients from maternal tissues to the developing endosperm. Identification of several ETC-specific genes encoding lipid transfer proteins (LTP), including the END1, provided excellent markers for identification of ETC during seed development. To understand how AtEND1 forms complexes with lipid molecules, a three-dimensional (3D) molecular model was generated and reconciled with AtEND1 function. The spatial and temporal expression patterns of AtEND1 were examined in transgenic Arabidopsis plants transformed with an AtEND1 promoter-GUS fusion construct. The AtEND1 promoter was found to be seed and pollen specific. In contrast to ETC-specific expression of homologous genes in wheat and barley, expression of AtEND1 is less specific. It was observed in ovules and a few gametophytic tissues. A series of AtEND1 promoter deletions fused to coding sequence (CDS) of the uidA were transformed in Arabidopsis and the promoter region responsible for AtEND1 expression was identified. A 163 bp fragment of the promoter was found to be sufficient for both spatial and temporal patterns of expression reflecting that of AtEND1. Our data suggest that AtEND1 could be used as a marker gene for gametophytic tissues and developing endosperm. The role of the gene is unclear but it may be involved in fertilization and/or endosperm cellularization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arabidopsis has provided an excellent model for the study of various aspects of seed development. Patterns of development of Arabidopsis and cereal endosperm share many features, particularly during the early stages (Olsen 2004). In both types of endosperm, the early stage of endosperm development is defined by free-nuclear divisions without cell wall deposition, which results in the formation of the multinucleate syncytium. Development of the endosperm coenocyte and cytokinesis in Arabidopsis occurs along the anterior–posterior axis (micropylar-chalazal axis). Cellularization of the free nuclei surrounded by the integument occurs in a wave-like pattern. It is initiated in the micropylar zone (anterior), moves rapidly through the central cell zone and finally reaches the chalazal zone (posterior) (Brown et al. 1999). These three distinct developmental domains in the Arabidopsis coenocyte were designated as the embryo surrounding region (ESR) or micropylar endosperm (MCE), the peripheral endosperm (PEN) around the central vacuole, and the chalazal endosperm (CZE) (Brown et al. 1999; Boisnard-Lorig et al. 2001; Sørensen et al. 2002). During cellularization, the embryo sac gradually enlarges forming an inverted “U-shape”. All phases of the establishment of polarized nucleic cytoplasmic domains (NCDs) and formation of tube-like structures (alveoli) can be observed at this stage of seed development (Brown et al. 1999).
Endosperm cellularization in Arabidopsis seeds begins at late globular to early heart stage (3 DAP) of embryo development. The typical feature of this stage is co-existence of a cellular MCE with a gradient stage of alveolation process in the PEN and endosperm nodules in the CZE of the embryo sac. A large coenocytic cyst is positioned in the corner of the chalazal region above the nucellar proliferating tissue (Brown et al. 1999; Boisnard-Lorig et al. 2001). At late torpedo stage of embryo development (about 6 DAP), the completion of cellularization in most endosperm domains except the cyst, marks the end of cytokinesis in the developing Arabidopsis endosperm (Brown et al. 1999; Boisnard-Lorig et al. 2001; Otegui et al. 2001; Guitton et al. 2004; Lid et al. 2004). The peripheral, cell-originated epidermal layer in eudicots is the counterpart of the aleurone layer of cereals, as both have the same cell fates (Chamberlin et al. 1994; Keith et al. 1994; Brown et al. 1999). Similar to basal endosperm transfer cells, the endosperm cyst in Arabidopsis is thought to play a role in transfer of assimilates and solutes to endosperm via its connection to the funiculus-chalazal pad. In contrast to cereals, which have a large endosperm, the two upturned cotyledons remain the main storage organ in mature Arabidopsis seeds, for consumption during seed germination.
Endosperm transfer cells (ETC) are specialized cells in cereal grains, which facilitate transport of nutrients and solutes from maternal tissue to developing endosperm (Becraft and Gutierrez-Marcos 2012). A distinct feature of ETC is the presence of cell wall ingrowths, which enlarge cellular membrane surface and consequently increase influx of assimilates and solutes to developing grains. The morphology and ultrastructure of ETC have been extensively studied in grasses (Thiel et al. 2012b; Wang et al. 2012; Monjardino et al. 2013) but little is known about the presence or possible role of this structure in eudicots. Initiation and cellularization of Arabidopsis endosperm are complex processes, which require involvement of large numbers of genes. So far, marker genes for Arabidopsis seed tissues responsible for transfer of nutrients from maternal tissues to seed have not been identified. To find genes that are responsible for nutrient uptake and seed protection from pathogens, research has focused on the isolation and characterization of cereal genes that are gametophyte and/or endosperm specific, and particularly those which are expressed in ETC. Among such genes is barley END1 (Acc. No. Z69631), encoding a lipid transfer protein (LTP), which is expressed in the coenocyte above the nucellar projection during the free-nuclear division stage of grain development. At the end of cellularization (8–10 DAP), END1 transcripts are accumulated in ETC and the adjacent starchy endosperm (Doan et al. 1996). END1 is commonly used as an endosperm marker gene for seed development studies. Several other LTP genes with patterns of expression similar to END1 have been reported (reviewed in Li et al. 2013). Among them is a wheat homolog of END1, designated as TaPR60 (Acc. No. EU64062) (Kovalchuk et al. 2009).
Here we investigate the possibility of using the model plant Arabidopsis to study the role of ETC-specific genes and proteins. The first task was to identify cells with ETC function in Arabidopsis seed using the promoter of the Arabidopsis homolog of barley END1, designated as AtEND1. The spatial and temporal expression of the AtEND1 was studied using real-time PCR and promoter-GUS fusion constructs in transgenic Arabidopsis. A 163 bp-long fragment of the promoter was identified to contain all functional elements necessary for tissue-specific AtEND1 expression. Three-dimensional (3D) models of AtEND1 was created and reconciled with biological function. In addition, structural analyses of several END1 proteins are presented. The results suggest that Arabidopsis can be used to study the role of ETC-related genes but the results need to be interpreted with some caution since the expression patterns of the genes differ and the developmental role of END1 genes in eudicots and monocots may also differ.
Materials and methods
Plant material and growth conditions
The donor plants used in this research were Arabidopsis (Arabidopsis thaliana L. Heynh) ecotypes Columbia-0 (Col-0) and Columbia-4 (Col-4) (CSIRO, Plant Industry, Waite Campus, Adelaide, Australia). Arabidopsis plants were grown in 12 cm pots in Arabidopsis soil mix containing white sand, peat and perlite at a ratio of 1:1:1, supplemented with 1 g/L FeSO4, 4 g/L pH amendment (2 g dolomite; 1 g gypsum; 1 g lime) and 3 g/L slow-release multi-nutrient fertilizer (Osmocote Plus; Nutri-Tech Solutions, Yandina, Australia). Initially, plants were grown at 22–24 °C with an 8 h photoperiod. Plants were watered every second day. Upon the appearance of well-developed rosette leaves, healthy plants were transferred to long-day growth conditions (22 °C, 16 h photoperiod) for induction of reproductive growth. Primary bolts were trimmed off at 6–7 days prior to transformation to encourage lateral bolts to emerge. Plants were transformed when bolts were 2–10 cm long.
Generation of promoter-GUS fusion constructs and transformation of Arabidopsis
Barley END1 cDNA sequence (GenBank accession Z69631) (Doan et al. 1996) was retrieved from the NCBI database and homology searches were performed using tBLASTx with the translated amino acid sequence of END1 against the translated EST databases from higher plants. Two genes with high sequence identity to barley END1 were identified and designated as AtEND1 (At1g32280) and AtEND2 (At5g56480). A 1,258 bp-long nucleotide sequence upstream of the translational start codon of AtEND1, which included promoter and 5′-UTR, was amplified by PCR using genomic DNA of Arabidopsis thaliana ecotype Col-0 as template. BamHI and HindIII restriction sites were introduced in the forward and reverse primers, respectively. The resulting full-length AtEND1 promoter-GUS fusion construct was designated as pBIAtEND1. Five AtEND1 promoter deletions were amplified by PCR using a series of forward primers with a BamHI adaptor, and the same reverse primer which was used for cloning of the full-length AtEND1 promoter (Suppl. Table S1). The PCR amplifications resulted in 1,062 bp (D1), 877 bp (D2), 699 bp (D3), 479 bp (D4), and 316 bp (D5) promoter fragments. Each fragment was sub-cloned into pGEM T-Easy vector (Promega) and sequenced. The inserts were cut from pGEM T-Easy with BamHI and HindIII and ligated into the binary vector pBI101 (Clontech). The transcriptional GUS fusion constructs were designated pBIAtEND1-D1, -D2, -D3, -D4, and -D5, respectively. All constructs were verified by sequencing and transformed into Agrobacterium strain LBA4404 by electroporation. Positive Agrobacterium clones were selected on 50 µg/mL kanamycin and 25 µg/mL rifampicin and plasmids were isolated using an Agrobacterium plasmid miniprep method described by Matthews et al. (2001). The presence of integrated inserts was verified using restriction analysis with BamHI and HindIII. The confirmed clones were used for transformation of Arabidopsis by a modified floral dipping method (Clough and Bent 1998).
Selection of plant transformants was carried out on agar plates containing half-strength MS basal medium (Sigma) supplemented with 1 % sucrose (w/v), 50 µg/mL kanamycin and 150 mg/mL Timentin (ticarcillin sodium and potassium clavulanate; GlaxoSmithKline, Australia). PCR analysis was used to confirm the integration of neomycin phosphotransferase (NPTII) and uidA genes into the plant genome. Transgene copy numbers in T1 plants were determined by Southern blot hybridization analysis using a 709-bp fragment from GUS coding sequence as a probe.
Real-time quantitative RT-PCR (qRT-PCR)
Arabidopsis WT Col-0 tissue types were collected at various stages of development from at least 12 individual plants with four biological replicates. The following tissues were selected to test expression levels of AtEND1 and AtEND2 genes: rosette leaf, cauline leaf, stem, apical meristem, bulk of flower buds, flowers at anthesis, root of 3-week-old seedling from liquid culture, above-ground part of seedling (3-week-old from liquid culture), green siliques at 0–5 days post anthesis.
TRIzol-like reagent was used to isolate plant total RNA. Prior to cDNA synthesis, RNA samples were purified with RNeasy Mini Kit (Qiagen) and on-column DNase I treatment (Qiagen) to remove genomic DNA according to the manufacturer’s instructions. First-strand cDNA was synthesized using SuperScript III Reverse Transcriptase and Oligo(dT)12–18 Primer (Invitrogen). cDNA template quality was assessed by PCR using primer pairs derived from the control genes for cyclophilin (AtCylophilin), actin (AtActin), tubulin (AtTublin), and glyceraldehyde 3-phosphate dehydrogenase (AtGAPdH). The expression levels of AtEND1 and AtEND2 genes were normalized against control genes as described by Burton et al. (2004). Primers used for qRT-PCR are given in Suppl. Table S1.
Histochemical GUS analysis of transgenic plants
Whole-mount GUS staining (Grossniklaus and Schneitz 1998) was performed to detect the spatial and temporal expression patterns of GUS expression driven by the AtEND1 promoter in transgenic Arabidopsis plants. The ovules and developing seeds from Arabidopsis were cleared with Hoyer’s “Light” medium (Stangeland and Salehian 2002). Whole-mount GUS-stained tissues were dissected and observed under a Zeiss Stemi dissecting microscope using bright field illumination. Ovules and developing seeds of Arabidopsis were observed under a Zeiss Axioskop microscope or a laser micro-dissecting microscope (Leica) with differential interference contrast (DIC) optics, and images were captured with a digital camera. Images were processed using the Photoshop Elements 2.0 program.
Construction of three-dimensional (3D) models of AtEND1 and AtEND2 from Arabidopsis by homology modeling
Construction of 3D models by homology modeling relies on spatial restraints of a suitable structural template through the Modeler algorithm (Šali and Blundell 1993). An Arabidopsis LTP defense protein, Protein Data Bank (PDB) accession number 2rkn, chain A from A. thaliana, was identified as a suitable template for the AtEND1 and AtEND2 proteins via 3D-PSSM (Kelley et al. 2000), LOMETS (Wu and Zhang 2007), MUSTER (Wu and Zhang 2008) and the Structure Prediction Meta-server (Ginalski et al. 2003). The AtEND1 and AtEND2 sequences were aligned by PROMALS3D (Pei et al. 2008) and analyzed by Hydrophobic Cluster Analysis (HCA) (Callebaut et al. 1997). The aligned sequences were used as input parameters to generate 3D models of AtEND1 and AtEND2 (both 80 amino acid residues), using Modeler 9v7 (Šali and Blundell 1993), running the Fedora 12 operating system on a Linux station. The final 3D molecular models of the AtEND proteins were selected from a pool of 40 models. The models with the lowest value of the Modeler 9v7 objective function and the most favorable Discrete Optimized Protein energy scoring parameters were chosen for optimization with a Tripos force field within the Sybyl 8.0 suite of programs (Tripos International). A Ramachandran plot of the AtEND1 and AtEND2 optimized models indicated that 100 % of residues were in the most favored, additionally allowed and generously allowed regions, when excluding glycine and proline residues. The overall G-factors (estimates of stereochemical parameters) evaluated by PROCHECK (Laskowski et al. 1993), were 0.06, −0.22, and −0.22 for 2rkn:A [LTP defense protein with Protein Data Bank (PDB) accession number 2rkn, chain A from A. thaliana], AtEND1 and AtEND2, respectively. The Z-score values deduced from Prosa2003 (Sippl 1993), reflecting combined statistical potential energy, were −7.7, −5.0, and −4.9 for 2rkn:A, AtEND1, and AtEND2, respectively. The RMSD values, between 2rkn:A and AtEND1 and AtEND2, determined with a ‘super’ algorithm in PyMol (http://www.pymol.org) were 0.32 and 0.36 Å in Cα positions. The buriedness of the HvNIP2;1 channel was calculated by PocketPicker (Weisel et al. 2007). Molecular graphics were generated with PyMol.
Results
Identification of END1 homologs from Arabidopsis
Database searches identified four proteins from Arabidopsis, which share high sequence similarities with END1 from barley. All four proteins contain a conserved domain PF00234 (Pfam) commonly found in seed storage proteins, plant LTPs and trypsin-α-amylase inhibitors (Skriver et al. 1992; Marchler-Bauer et al. 2005). The two genes with highest identity to barley END1 have Accession numbers: At1g32280 (designated as AtEND1) and At5g56480 (designated as AtEND2). AtEND1 (NM_102961) is an annotated gene. A number of ESTs were found for AtEND2 (NM_125031) in the TAIR database. Both genes contain a single intron situated in identical positions near to the 3′ end of the coding regions. The products of these genes, AtEND1 and AtEND2, are small cysteine-rich proteins which contain a putative domain often seen in lipid transfer proteins, and have structural similarity to barley END1 protein (Fig. 1). AtEND1 comprises 113 amino acid residues and shares 75.9 % amino acid sequence identity with AtEND2.
Multiple sequence alignment of six END1-like sequences from different plant species using ProMals3D (Pei et al. 2008). The predicted secondary structures are shown in magenta (α-helices) and black lines (loops). The conservation of residues on a scale of 9–5 is given at the top of the diagram. Absolutely conserved residues are highlighted in green boxes. Accession numbers for HvEND1, OsPR602, TaPR60 AtEND1, AtEND2, and BnEND1 are Z69631, CA767165, FJ459807, NM_102961, NM_125031, and AY208878, respectively
A gene encoding a seed-specific nsLTP protein from Brassica napus, Bn15D18B (AY208878), was retrieved from the NCBI database based on homology to AtEND1. Transcript profiling databases indicated that transcripts of this gene are abundant in canola seed during early stages of seed development. Several ESTs from other cereals with high similarity to barley END1 were also identified in the TIGR EST databases. These ESTs were predominantly found in cDNA libraries prepared from reproductive tissues, including flower, spike, developing caryopsis, and a mix of all developmental stages from ovules at meiosis to endosperm at seed maturation. The C-terminal sequences of these proteins are nearly identical, while other regions are also highly conserved, particularly the positions of cysteine residues (Fig. 1). A signature for all non-specific LTPs bears a conserved C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C motif, also known as an eight cysteine motif (8CM). This motif is present in most LTPs (José-Estanyol et al. 2004).
Structural alignments of the 2rkn:A and AtEND sequences and molecular modeling
The most suitable template for AtEND1 and AtEND2 was found to be the LTP defense protein 2rkn:A from Arabidopsis. This analysis was guided by PsiPred (McGuffin et al. 2000), SAM-T08 (Karplus 2009), STRIDE (Frishman and Argos 1995), DSSP (Kabsch and Sander 1983), PROMALS3D (Pei et al. 2008) and Robetta (Kim et al. 2004) searches. The LTP 2rkn:A defense protein occurs as a monomer and was crystallized in complex with two monoacylated phospholipid molecules [(7R)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacosan-1-aminium 4-oxide)], bound side by side in the internal cavity, and in the presence of Zn2+ ions. The sequence 2rkn:A was aligned with those of AtEND1 and AtEND2, taking care to ensure that α-helices remained undisturbed. The positional sequence identity and similarity values (Epprofile algorithm) between AtEND1 and AtEND2 were 94 and 80 %, respectively, and the positional sequence identities between 2rkn:A and AtEND1 or AtEND2 were 25 and 28 %, respectively. The latter sequence identities fall in the ‘twilight zone’, emphasizing that molecular modeling carried a certain degree of complexity. However, the presence of the eight-cysteine motif (8CM) in the sequence alignment indicated that the template sequence was suitable for molecular modeling (Fig. 2a). The positions of secondary structural elements were further examined with bi-dimensional HCA plots (shown for 2rkn:A and AtEND1 in Fig. 2a). It was of note that the first 23 residues in both AtEND proteins exhibited characteristics of a hydrophobic signal peptide as predicted by a SignalP, using neural networks and hidden Markov models trained on eukaryotes. Pairwise alignments between the template and target sequences 2rkn:A and AtEND1 and AtEND2 indicated that there was one two-residue deletion in both alignments (data not shown). In summary, the models generated by comparative modeling and evaluated by PROCHECK (Laskowski et al. 1993) and Prosa2003 (Sippl 1993) were of satisfying quality.
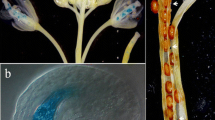
Molecular model of AtEND1. a Hydrophobic cluster analysis of AtEND1 and 2rkn:A. The positions of an NH2-terminal hydrophobic signal peptide in AtEND1 (vertical arrow), four paired, conserved cysteines (arrowheads) and α-helices (lines) are marked. Proline residues are shown as red stars, glycine residues as black diamonds, serine residues are empty squares, and threonine residues are shown as squares containing a black dot in the centre. Negatively charged residues are colored in red and positively charged residues are in blue. Other residues are shown by their single amino acid letter codes. Numbers of amino acid residues are read from the top to the bottom of the plots (in duplicate) in a left to right direction. b Model of AtEND1 and 3D structure of 2rkn:A, illustrating protein folds. The dispositions of two lipid molecules are shown in cpk sticks in internalized protein cavities, delineated by white to blue circles (PocketPicker analysis) (Weisel et al. 2007) and the positions of four invariant disulfide bridges (yellow) are also indicated. The models are shown in rainbow colors, with blue and red indicating NH2- and COOH-termini, respectively. The small right- and left-hand side arrows indicate NH2- and COOH- termini, respectively. The light blue arrows point to a deletion or insertion and to NH2-terminal α-helices, respectively. c Molecular surface representations of AtEND1 and 2rkn:A (gray) with two lipid molecules (cpk sticks) are enclosed in their internal cavities
The sequence identity between the two AtEND proteins was 80 %. Consequently, the folds and dispositions of key residues were very similar in both models. For simplicity, we present only the model of AtEND1 in this work (Fig. 2). It is evident from Fig. 2b, c that AtEND1 and 2rkn:A contain a series of α-helices that fold into a global scaffold bundle of the ‘all alpha protein’ class, according to SCOP protein classification (Pasquato et al. 2005). The overall folds of AtEND1 are stabilized by four di-sulfide bridges (Fig. 2b). These most likely play decisive roles in protein structural integrity, whereby the COOH-terminal loop is wrapped around the bundle. The characteristic feature of both AtEND1 and 2rkn:A is the presence of an internalized central cavity with a hydrophobic interior that could potentially enclose lipidic molecules (Fig. 2c). The volumes of these cavities were estimated to be 1,122 and 1,069 cubic Å for AtEND1 and 2rkn:A, respectively, using CASTp (Dundas et al. 2006). Two lipidic molecules in extended conformations were modeled in the central cavity of AtEND1, which is lined with a series of hydrophobic residues. These residues make close contacts with both lipid molecules (Fig. 2c). The major structural difference between the AtEND1 model and the 2rkn:A template is a much more open entrance to the central cavity of AtEND1 compared to that of 2rkn:A (Fig. 2c). This larger opening results from a shorter NH2-terminal helix (small, vertical blue arrows in Fig. 2b). The other structural difference reflects the presence of a 2-residue deletion in AtEND1 that leads to replacement of a short α-helix in 2rkn:A by a loop structural motif in AtEND1 (light blue vertical arrows in Fig. 2b).
qRT-PCR analysis of AtEND1 and AtEND2 expression in Arabidopsis
Investigation of expression patterns of the AtEND gene family members was initially performed using the AtGenExpress Visualization Tool (AVT) (http://jsp.weigelworld.org/expviz/expviz.jsp) from TAIR (The Arabidopsis Information Resource). It was found that AtEND2 (Array element #248019_At) was predominantly expressed in mature pollen (6-week-old plants) and in developing seeds (8-week-old plants) from stage 8 (walking stick to early curled-cotyledons embryos) to stage 10 (green cotyledons of embryos). AtEND2 was also expressed at a lower level in rosette leaves. Unfortunately, information about AtEND1expression was not available from either the EST database or from multi-array experiments (TAIR).
Quantitative RT-PCR analysis was performed to identify expression patterns of AtEND1, and compare expression levels of AtEND1 and AtEND2 in a variety of Arabidopsis tissues (Fig. 3). Transcripts of AtEND1 were found to be abundant in flowers before pollination and their number increased 1.5-fold in young green siliques, while the number of transcripts was relatively low in other tested tissue types. In contrast, transcripts of AtEND2 accumulated mainly in flowers before pollination and their number reduced to 50 % in green siliques, where levels of AtEND2 expression were lower than in other tested tissues. Although both genes demonstrated high transcript levels in flowers and developing siliques, the expression level of AtEND1 was nearly threefold higher than that of AtEND2. Peak transcript levels for AtEND1 were observed in green siliques, while the maximum levels for AtEND2 were found in flower buds.
Quantitative RT-PCR analysis of AtEND1 and AtEND2 expression in different tissues of WT Col-0 Arabidopsis plants. The level of AtEND1 transcripts was very high in flowers and 0–1.5 cm long green siliques, while it was relatively low in other tested tissues. The expression levels of AtEND2 were relatively high in flowers, and lower in green siliques. The expression of AtEND1 was stronger than expression of AtEND2 in all tested tissues
Spatial and temporal GUS activity under the control of the AtEND1 promoter in Arabidopsis
Eight independent transgenic T1 lines were selected based on resistance to kanamycin and the presence of transgene was confirmed by PCR and Southern blot hybridization (data not shown). Various tissues were analyzed throughout plant development using a whole-mount GUS assay. All eight T1 lines demonstrated similar spatial and temporal GUS expression patterns, with slight variations in strength of GUS activity. Multiple copies of the transgene were detected in the three lines with strong GUS activity using Southern blot hybridization (data not shown). Lines 1, 3, and 7 possessed three, five, and five copies of the transgene, respectively, while the remainder lines possessed single copy of uidA. GUS activities were analyzed in all eight T1 lines and their corresponding T2 progenies. No GUS activity was detected in either WT Arabidopsis plants or null-allele segregants, which were used as negative controls.
Promoter activity of AtEND1 was not detected in plant tissues until shortly before anthesis. During the vegetative growth phase, GUS was not found in the rosette or cauline leaves (Fig. 4I-a, b), or stems (not shown). During reproductive growth, promoter activity was initially detected at anthesis in mature pollen grains (Fig. 4I-c, f, h) and ovules (Fig. 4I-c). It was especially strong in the non-fused central cell nuclei (Fig. 4II-a). A high level of GUS activity was found in developing seed (Fig. 4I-d, e) and was initially detected in the embryo sac during fertilization and free-nuclear stage of endosperm development (Fig. 4II). After fertilization, very strong GUS activity was observed in fertilized egg cells and fertilized central nuclei (Fig. 4I-b). Promoter activity was detected in dividing endosperm nuclei and zygotes (Fig. 4II-b, c), and retained in developing embryo and free nucleate endosperm during the first 2 DAP (Fig. 4II-d–f). At 48 h after pollination (Stages 2, the globular stage of the embryo development) GUS activity was detected within the embryo sac; it was particularly strong in the three mitotic domains: MCE, PEN, and CZE of the developing endosperm with the strongest expression in the anterior and posterior poles, and along the adaxial arch of the embryo sac (Fig. 4II-f). At this stage of seed development, endosperm in the MCE zone was already cellularized while nuclei could still be seen in the PEN (Fig. 4II-f). During the globular stage of embryo development, strong GUS activity was detected in endosperm nodules (Fig. 4II-c, f). Interestingly, during the first 48 h after pollination, in lines with strong GUS expression, GUS activity was also found in the funiculi and the adjacent maternal tissues below the chalazal pad (Fig. 4II-c, f). This expression pattern was not observed in lines with moderate GUS expression. At about 3 DAP, between the late globular and early heart stage of embryo development, GUS activity diminished in the embryo sac but was very strong in CZE zone (Fig. 4II-g). At the end of endosperm cellularization (about 5–6 DAP), GUS activity was observed only in the posterior pole (cyst) (Fig. 4II-h–j). No GUS activity was detected elsewhere in the embryo sac at this or later stages including cotyledon phase (Fig. 4II-l), except in one of the strongest expressing lines, where promoter activity in the uncellularized cyst could still be detected at the torpedo embryo stage (Fig. 4II-k). For comparison, seeds of control plants at different developmental stages are shown in Fig. 4II-m–v.
GUS activity under the control of the 1,258 bp-long promoter of AtEND1, in transgenic Arabidopsis plants transformed with pBIAtEND1 construct. GUS expression was detected in ovules (I-c) and mature pollen (I-f, h) at anthesis and later in seeds at early stages of development (I-d, e). No GUS activity was observed in rosette leaf (I-a), cauline leaf (I-b), sepal, petal, filament, gynoecium or pedicel (I-c) during vegetative or reproductive stages of plant development. Expression of GUS was not found in pollen of control plants (I-g). Ovules (I-c), developing seeds (I-d) and mature pollen grains (I-f) are indicated by arrows. GUS activity under the control of AtEND1 promoter was detected in the embryo sac of ovules (II-a) at anthesis, in fertilized embryo sac (II-b) and in the entire developing seeds during the first 2 DAP (Stages 1–3, globular embryo, II-c–f). GUS activity reduced in seed from 3 DAP (heart embryo, II-g) but was retained in the endosperm cyst until about 5–6 DAP (Stages 4–6, heart-to-torpedo embryo, II-h–j). GUS activity was very weak at late torpedo to walking-stick embryo (Stages 7–8, II-k) but undetectable during cotyledon stage (Stage 10, II-l) of embryo development. Seeds from control plants did not show GUS activity at any stage of development (II-m–v). an anther, cot cotyledon, cz chalazal pole, CZE chalazal endosperm, DAP days after pollination, fu funiculus, em embryo, mp micropylar pole, MCE micropylar endosperm, PEN peripheral endosperm, ov ovule. Bars 2 mm (I-a, b), 200 µm (I-c, d, i), 300 µm (I-e, f, h), 500 µm (I-g). Bars in II = 200 μm
The observed spatial and temporal GUS expression patterns of T1 transgenic Arabidopsis plants were also observed in T2 and T3 progenies of all independent T1 lines. Some of the pollen grains and ovules, as well as developing seeds, did not show GUS activity in the T2 lines due to segregation of the transgene.
In silico analysis of the AtEND1 promoter
Analysis of the AtEND1 promoter sequence was conducted using PLACE software (http://www.dna.affrc.go.jp/PLACE/) (Ogawa et al. 1998; Higo et al. 1999). Several cis-elements, which have been previously characterized in higher plants were identified. Promoter elements related to specific expression in pollen and seeds are shown in Fig. 5. The promoter region contained four AAAG motifs specific to members of the Dof family of transcription factors (TFs), which are found only in higher plants (Yanagisawa 1997). The AAAG motif forms the core of the Dof recognition sequence while the flanking sequences show broader diversity (Yanagisawa 2000; 2001; 2004). Dof binding sites are found in the promoters of many genes encoding seed storage proteins, such as α-, γ-gliadins, and LMW glutenins (Yanagisawa and Schmidt 1999; Yanagisawa 2000). A Prolamin box (P Box) and Opaque-2 box (O2 Box) were also identified in the AtEND1 promoter (Fig. 5). These two motifs were initially identified in the promoter of the maize gene encoding a seed storage protein (zein) and it was demonstrated that they are important for endosperm-specific expression (Singh 1998). Three pollen-specific TF binding sites (core sequence AGAAA) (Bate and Twell 1998; Filichkin et al. 2004) were also predicted in the promoter sequence of AtEND1 (Fig. 5).
Computational prediction of transcription factor binding sites in the promoter region of AtEND1. The initial amino acid residues for AtEND1 are indicated in red at the end of the upstream sequences. The 5′ end of each promoter deletion is indicated by a solid red arrow and the starting nucleotide is in large bold font. Conserved sequences of transcription factor binding sites are shown different colors: blue binding sties for Dof factors; purple Opaque-2 binding site; orange pollen-specific TF binding site; green Prolamin box
Identification of promoter regions responsible for tissue-specific activation of AtEND1
As revealed by patterns of GUS activity controlled by the AtEND1 promoter, AtEND1 expression in Arabidopsis is gametophyte and seed specific. To identify the minimal length of the regulatory 5′ flanking sequence which retains spatial and temporal patterns of AtEND1 promoter activity and to reveal possible cis-elements conferring male and/or female gametophyte- and/or seed-specific expression, five AtEND1 promoter deletions fused to the GUS gene, designated D1–D5, were generated and stably transformed in Arabidopsis (Fig. 6I). Transgenic lines were selected for all five promoter deletion constructs. Numbers of T1 transgenic lines for D1–D5 constructs were 3, 9, 13, 7, and 5, respectively. Flowers and seeds of all T1 individuals were analyzed for GUS activity at various stages of development. GUS activity was not detected in ovules and developing seed from control plants (Fig. 6II-a–g).
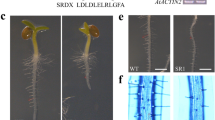
GUS activity under the control of a series of AtEND1 truncated promoters. I Schematic representation of promoter deletions with indicated length of fragments and presence of GUS activity. II Patterns of GUS activity controlled by promoter deletions. Micrographs a–g show an ovule (a) and developing seeds of control plants from early globular to cotyledonal embryo development after GUS staining (b–g). The activity of AtEND1 promoter was observed in ovules and mature pollen grains at anthesis (h from D2; k and l from D3; m from D4). GUS activity was detected in only the embryo sac after pollination (p and q from D3; n and o from D2) and during the globular stage of embryo development (r from D2, s from D3 and t from D4). Promoter activity could only be detected in the endosperm cyst during heart embryo stage (u, v) and was undetectable at later stages until the end of seed maturation (w, walking-stick stage and x, cotyledon stage). Bars 200 μm (a–g, n–x), 1.5 mm (h, k), 0.5 mm (i, j), 100 μm (m), 50 μm (l)
GUS activity under the control of the D1, D2, D3, and D4 promoter deletions showed very similar spatial and temporal patterns to that of the full-length promoter in Arabidopsis. The active time window was consistent from anthesis to cellular endosperm formation (Fig. 6II-h–v). GUS activity was initially detected in mature pollen grains (Fig. 6II-h, II-k–m) (n = 215) and ovules (Fig. 6II-h, i, k, n–p) (n = 378) before pollination. After fertilization, GUS expression persisted in the embryo sac until 5–6 DAP and in the chalazal zone of the cellular endosperm (Fig. 6II-j, II-q–v) (n = 318). At about 1 day before pollination, GUS activity was detected in the embryo sac before the central cell nuclei had fused (D2: Fig. 6II-n and D3: Fig. 6II-o), and shortly after, in fused polar nuclei and the egg apparatus (D4: Fig. 6II-p). Consistent with the activity of the full-length promoter, continuous expression was detected in the zygote and dividing nuclei of the endosperm and embryo from fertilization to 72 h after pollination in the three nucleic cytoplasmic domains (NCDs) of the developing endosperm (D2: Fig. 6II-r, D3: Fig. 6II-s and D4: Fig. 6II-t). Cyst-specific expression at 3 DAP in the cellular endosperm was observed (n = 298) in all GUS positive promoter deletion lines (Fig. 6II-u, v). The mature pollen expression of GUS before and shortly after pollination (D2: Fig. 6II-h, D3: Fig. 6II-k and l and D4: Fig. 6II-m) resembled the pattern obtained from the full-length promoter. No GUS activity was found in any other tissues or cell types during vegetative growth and reproductive development.
GUS activity was not detected in any lines transformed with the D5 promoter deletion (data not shown), although the transgenic status of these lines was confirmed by PCR analysis using uidA-specific primers (not shown).
Discussion
AtEND1 and AtEND2 are homologs of barley END1, TaPR60, and OsPR602. Several genes encoding small cysteine-rich protein family members, which are specifically expressed in the developing grain, were identified in rice (Li et al. 2008) and Arabidopsis EST databases. They all originate from cDNA libraries prepared from developing caryopses. Two of these genes, designated as AtEND1 and AtEND2, encode nsLTPs and belong to the type VI subfamily of nsLTPs from Arabidopsis (Molina and García-Olmedo 1997). The product of OsPR602 (Li et al. 2008), CA767165, also belongs to the Type VI subfamily of nsLTPs (José-Estanyol et al. 2004). It is known that nsLTPs are abundant in the seeds of higher plants and are capable of binding fatty acids and mobilizing phospholipids between membranes (Arondel et al. 2000). The 8CM motif form a backbone structure (C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C) which is common to most nsLTPs. These cysteine residues form four disulfide bonds and a hydrophobic cavity, which binds lipids or other hydrophobic molecules in vitro. Using 3D modeling, it was predicted that the eight cysteine residues in AtEND1 paired to form four di-sulfide bonds (C29–C69, C40–C58, C59–C94, and C71–C104), which are crucial for formation of the hydrophobic pocket to bind lipids (Frishman and Argos 1995; Heinig and Frishman 2004). Unprocessed nsLTPs possess an NH2-terminal signal peptide and are thought to be involved in secretion of cutin and wax to the cell wall (Arondel et al. 2000). Further roles have been proposed for LTPs, in membrane biosynthesis and as antimicrobial agents. The latter has been demonstrated for several LTPs (Arondel et al. 2000; de Ruijter et al. 2003). However, the precise biological functions of nsLTPs are unclear.
Structural models of the AtEND proteins were constructed using spatial restraints from the LTP defense protein 2rkn:A from Arabidopsis. The 2rkn:A structure was identified as the optimal template by several prediction servers. The LTP defense protein represents a canonical ‘all alpha protein’ fold and belongs to the ‘protease inhibitor/seed storage/LTP’ family of the Pfam protein classification system (Finn et al. 2010) and to the ‘proteinase/alpha-amylase inhibitors’ family of the SCOP classification (Pasquato et al. 2005).
Based on molecular modeling, we predict that both AtEND1 and AtEND2 bind lipid molecules. AtEND proteins could also associate with plant plasma membranes through bound lipids or via receptors. Minor shape variability could exist between individual AtEND proteins and these disparities could lead to differences in lipid-binding efficiencies or in their surface electrostatic properties. Further, the 2rkn:A, AtEND1 and AtEND2 sequences contain a ‘proline’ motif (PxxPxxP in 2rkn:A and PxxPxP in AtEND proteins) that is specific for a certain type of lipid-binding protein (Lascombe et al. 2008). These proline-rich regions could be involved in protein–protein interactions. Another possibility is that AtEND proteins could interact with other interacting partners and become components of larger protein complexes involved in cell signaling. Such hypotheses could be tested by generating variant molecules of Arabidopsis AtEND proteins through site-directed mutagenesis and plant transformation. In particular, replacements of highly conserved disulfide bridges, hydrophobic residues in the cavities and the “proline” motifs could be investigated. One approach could include the substitution of cysteine residues involved in the formation of disulfide bonds with alanine via site-directed mutagenesis, for example, C29A or C69A, to disrupt the secondary structure and potentially block AtEND functionality. However, it remains to be seen if these variations would lead to structural re-arrangements or abolishment of lipid binding.
It is postulated that AtEND1 is involved in lipid binding and lipid transport during free-nuclear division and/or phragmoplast formation during endosperm cellularization. One report has shown that a nsLTP from Arabidopsis is a cell wall protein, predominantly localized to epidermal cells as revealed by immunolocalization (Bouyer et al. 2011). It could be useful to investigate sub-cellular location of the product of AtEND1 in Arabidopsis. Functional analysis of AtEND1 could be achieved by analyzing mutants, for example, T-DNA insertion lines, for this locus and determining if viable seed can form or if endosperm development is disrupted. Alternatively, gene expression could be knocked down using RNA interference with an RNAi cassette under the control of the AtEND1 promoter. However, in this case, it would be important to avoid knocking down both AtEND1 and AtEND2 and perhaps other genes encoding members of nsLTPs.
AtEND1 is predominantly expressed in seed during early endosperm development
Expression of AtEND1 and AtEND2 was initially studied using qRT-PCR analysis and transgenic Arabidopsis plants transformed with the AtEND1 promoter-GUS fusion construct. The qRT-PCR data showed that the highest transcript level was observed in flower buds at anthesis. This result correlated with the expression pattern for AtEND2 (At5g56480) derived from the AtGenExpress Visualization Tool (ATH5). It was demonstrated that AtEND2 (probe set 248019_at) was expressed in mature pollen and developing seed at development stages 8–10, which correspond to the upturned to green cotyledon stages of embryo development (http://jsp.weigelworld.org/expviz/expviz.jsp?experiment=development&normalization=normalized&probesetcsv=At5g56480&action=Run).
Spatial and temporal patterns of GUS activity under the control of AtEND1 promoter during seed development correlated with our qRT-PCR data. The expression of GUS was very strong in egg apparatus, nuclei of central cell and antipodal cells (Fig. 4II-a), mature pollen grains (Fig. 4I-c, f, h) and developing seeds (Fig. 4I-d, e, II-b–g). As it was shown by qRT-PCR, transcript level of AtEND1 was found high in flower buds containing anthers and ovules as well as in the less than 1.5 cm long green siliques containing developing seeds up to heart stage of embryo development (Fig. 3). A much stronger GUS activity has been detected in the embryo sac where endosperm nuclei are undergoing rapid divisions accompanied by cell wall depositions. Particularly strong GUS activity has been observed in dividing nuclei, endosperm nodules and developing embryos at globular stage of embryo development (Fig. 4II-c–f). These data suggest that AtEND1 is very likely to be involved in lipid transfer during endosperm cellularization. No GUS activity has been detected in tissues, where transcript numbers were lower than 2,000 copies per microliter of the cDNA.
No transgenic lines transformed with the AtEND2 promoter-GUS construct were generated or analyzed in this study. Recently, Royo et al. (2014) demonstrated GUS expression patterns driven by promoters of the group VI nsLTP genes in transgenic Arabidopsis plants. It was shown that in reproductive tissues, AtEND1 (At1g32280) and AtEND2 (At5g56480) promoters had very similar patterns of GUS activity. Royo et al. (2014) observed that AtEND2 promoter was active in ovules before and at anthesis and could be detected in funiculi of the young developing seeds. Expression pattern of the AtEND1 (At1g32280) described by these authors was similar to what we observed in our AtEND1 promoter-GUS transgenic lines. However, due to lack of details of the GUS images presented in their paper, a suggestion by the authors (2014) that AtEND1 and AtEND2 have non-overlapping yet distinctive spatial and temporal patterns of promoter activities seems to be inconclusive.
In contrast to END1 from barley, which was expressed in the endosperm coenocyte and ETC starting from 4 DAP (Doan et al. 1996), the mRNA levels of AtEND1 were already high in flowers at anthesis. The level of AtEND1 transcripts increased in green siliques up to the heart embryo stage during endosperm cellularization (Fig. 3). At the heart-to-torpedo embryo stage, GUS activity driven by the AtEND1 promoter was detected only in the cyst (Fig. 4II-h–j). After late torpedo stage of embryo development, AtEND1 was not expressed in the developing seed (Fig. 4II-l). Taken together, AtEND1 could have a role in lipid transfer in fertilization, nuclear divisions in the coenocyte and endosperm cellularization.
The strength of GUS activity is dependent on the number of copies and positions of transgene insertions in the target genome. In this study, we did not find direct correlation between the strength of GUS activity and the transgene copy numbers in the transgenic lines. Similar results were obtained earlier for transgenic rice and barley plants (Li et al. 2008; Kovalchuk et al. 2010). According to our observations, transgene copy number does not often affect the level of GUS activity and very rarely changes the spatial or temporal patterns of the reporter expression. In our analyses of GUS reporter lines, we always compare a large number of independent transgenic lines. If no difference in patterns of expression is found in preliminary analyses, we usually select two to three lines with the strongest GUS expression for a more thorough analysis. We have never observed influence of promoter-uidA construct insertions on plant phenotypes, although we realized that higher chances of phenotypic change could occur to plants with high copy number of transgene than those with single or low copies. We have not quantified GUS activity dependence on positional effects of transgene insertions.
Endosperm cyst may play a role similar to the ETC in cereals
Recent studies of seed transfer cells have been extensively with cereals, including maize (Monjardino et al. 2013), barley (Thiel et al. 2008, 2009, 2012a, b) and sorghum (Wang et al. 2012). Although these studies focused on the morphology and ultrastructure of wall ingrowths in the transfer cell, some novel findings have come to light. Using 454 sequencing, membrane bound receptors have been identified to be expressed in ETC during differentiation of the ETC region. Transcript profiling revealed that specific phosphorelays were activated during ETC cellularization and differentiation (Thiel et al. 2012b). It has been demonstrated that cell wall deposition in endosperm requires biosynthesis of phospholipids, pectins, and ethylene for the formation of membrane microdomains, which are potentially involved in ETC development (Thiel et al. 2008).
The endosperm cyst in the chalazal zone of Brassicaceae seeds is thought to play a role akin to transfer cells in cereals. These cells are considered to mobilize nutrients from the maternal chalazal pad (Thompson et al. 2001). However, the multinucleate endosperm at this posterior pole never undergoes cellularization during seed development (Brown et al. 1999; Boisnard-Lorig et al. 2001; Otegui et al. 2001; Guitton et al. 2004; Lid et al. 2004). Therefore, it is possible that a different mechanism regulates solute transport in Arabidopsis, and that the cyst and CZE may not be involved in this process. Moreover, cellular endosperm in Arabidopsis is non-persistent as it is consumed by the developing embryo during seed development. Consequently, at the end of embryo development, endosperm is presented by two thin cell layers. No cell or tissue types of Arabidopsis seed were yet reported, which serve a role similar to that of ETC in cereals. Nutrient uptake by the chalazal endosperm from funiculi via integuments of developing seeds was not so far clearly demonstrated for either Arabidopsis or other Brassicaceae species.
AtEND1 is a marker gene for gametophyte and early seed development
GUS expression analysis showed that the 1,258 bp-long 5′ flanking sequence upstream of the translational start site of AtEND1 could guide gene expression in mature pollen and the embryo sac at anthesis. During fertilization, AtEND1:GUS was expressed in the egg apparatus, polar nuclei and the central cell nucleus (Fig. 5). After fertilization, promoter activity was observed in the zygote and developing embryo until the late-heart stage (Fig. 5). In the triploid endosperm, AtEND1 was found in the dividing primary endosperm nucleus and later in the syncytium throughout the free-nuclear and cellular stages. Only cells at the posterior pole showed GUS activity under the control of the AtEND1 promoter in the cellular endosperm at the heart stage of embryo development. No expression beyond late-heart stage in the embryo sac was found. The data suggest that the full-length AtEND1 promoter is active in both gametophytes and in developing seeds of Arabidopsis. The homozygous AtEND1:GUS lines are of interest for genetic analysis of genes involved in seed initiation and endosperm development at early stages.
The spatial and temporal expression pattern of AtEND1 in Arabidopsis is very similar to that of the wild-type FIS genes in developing seed (Luo et al. 2000). The initial expression of MEA::GUS and FIS2::GUS was detected in the synergids, egg cell and central cell nucleus. Later, it was found in the free-nuclear endosperm, and only the endosperm cyst nuclei retained GUS activity when cellularization terminated (Luo et al. 2000). FIE::GUS is also transiently expressed in the microspores before pollination which is similar to that in AtEND1:GUS lines. The conserved expression patterns observed for AtEND1 and FIS genes suggest that, like FIS2, AtEND1 may play a role in fertilization and endosperm development.
The 479 bp-long 5′ flanking sequence of AtEND1 is sufficient for tissue-specific gene expression
Analysis of GUS expression in transgenic plants transformed with a series of promoter deletions revealed that four promoter sequences, D1 to D4, showed the same temporal and spatial patterns as the full-length promoter of AtEND1. A 479 bp-long fragment upstream of the translation start site (D4) was sufficient for directing the gametophyte and seed expression during free-nuclear and cellularization phases of endosperm development. No GUS activity was detected in transgenic Arabidopsis plants under the control of the D5 (316 bp) truncated promoter. Thus, a 163-bp fragment (the red box in Fig. 6I) appears to contain all necessary cis-elements responsible for the specificity of AtEND1 expression in Arabidopsis. Promoter element analysis using PLACE (Prestridge 1991; Higo et al. 1999) predicted three binding sites for Dof factors and one binding site for a pollen-specific factor within the 163 bp promoter fragment. This supports our hypothesis that the 163 bp fragment contains sufficient information for the specific temporal and spatial pattern of AtEND1 expression. It also implies that other predicted potential cis-elements beyond this crucial region have no real function in transcriptional regulation. The 163 bp-long promoter fragment was used in a yeast one hybrid screen of an Arabidopsis cDNA library prepared from flowers and developing siliquas (data not shown). Unfortunately, no transcription factors were identified in the screen. The 163 bp-long promoter segment should be further dissected and analyzed to identify novel or confirm predicted cis-elements responsible for pollen- and/or cyst-specific expression.
Our results suggest that AtEND1 is a gametophyte and early seed gene which may be involved in fertilization and/or nuclear endosperm formation and cellularization during early stages of Arabidopsis seed development. A 163 bp fragment (−479 to −316) of the AtEND1 promoter contains elements sufficient for the correct spatial and temporal expression of AtEND1. Although there are clear differences in the expression patterns of the END1 genes in cereals and Arabidopsis, there is also sufficient overlap to suggest that the proteins are likely to show similar functions and Arabidopsis may provide a suitable model for exploring the role of the ETC in supporting the developing endosperm of cereals. Homozygous transgenic Arabidopsis lines generated in this research may be used to study the influence of over-expression or mutations in other genes on fertilization and endosperm cellularization.
Author contribution
ML conducted analyses of transgenic Arabidopsis plants including growth, molecular and microscopic evaluations, designed and validated qRT-PCR assays, contributed to preparation of cDNA from the wild-type Arabidopsis tissue series, interpreted experimental data and prepared the manuscript. SL identified the AtEND1 and AtEND2 and the promoter of AtEND1, contributed to experimental design and revised the manuscript. MH conducted three-dimensional protein modeling of AtEND1 and revised the manuscript. NS conducted qRT-PCR experiments and data processing. MP conducted Arabidopsis transformation and selected transformants. AMK supervised the project, contributed to interpretation of the results and critically read the manuscript. PL supervised the project and critically read the manuscript.
Abbreviations
- CZE:
-
Chalazal endosperm
- DAP:
-
Days after pollination
- END1:
-
Endosperm1
- EST:
-
Expressed sequence tags
- ETC:
-
Endosperm transfer cells
- GUS:
-
uidA/5-bromo-4-chloro-3-indolyl-β-d-glucuronidase
- LTP:
-
Lipid transfer protein
- MCE:
-
Micropylar endosperm
- nsLTP:
-
Nonspecific lipid transfer protein
- PEN:
-
Peripheral endosperm
References
Arondel V, Vergnolle C, Cantrel C, Kader JC (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci 157:1–12
Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37:859–869
Becraft PW, Gutierrez-Marcos J (2012) Endosperm development: dynamic processes and cellular innovations underlying sibling altruism. Wiley Interdiscip Rev Dev Biol 1:579–593
Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm Is divided in mitotic domains. Plant Cell 13:495–509
Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, Schnittger A (2011) Polycomb repressive complex 2 controls the mmbryo-to-seedling phase transition. PLoS Genet 7:e1002014
Brown RC, Lemmon BE, Nguyen H, Olsen OA (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12:32–42
Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134:224–236
Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon JP (1997) Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci 53:621–645
Chamberlin MA, Horner HT, Palmer RG (1994) Early endosperm, embryo, and ovule development in (Glycine max L.) Merr. Int J Plant Sci 155:421–436
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
de Ruijter NCA, Verhees J, Leeuwen W, Krol AR (2003) Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol 5:103–115
Doan DNP, Linnestad C, Olsen OA (1996) Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Mol Biol 31:877–886
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:W116–W118
Filichkin SA, Leonard JM, Monteros A, Liu PP, Nonogaki H (2004) A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol 134:1080–1087
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer ELL, Eddy SR, Bateman A (2010) The Pfam protein families database. Nucleic Acids Res 38:D211–D222
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23:566–579
Ginalski K, Elofsson A, Fischer D, Rychlewski L (2003) 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19:1015–1018
Grossniklaus U, Schneitz K (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9:227–238
Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of fertilisation independent seed Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131:2971–2981
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500–W502
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
José-Estanyol M, Gomis-Rüth FX, Puigdomènech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42:355–365
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Karplus K (2009) SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res 37:W492–W497
Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6:589–600
Kelley L, MacCallum R, Sternberg M (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299:499–520
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:W526–W531
Kovalchuk N, Smith J, Pallotta M, Singh R, Ismagul A, Eliby S, Bazanova N, Milligan A, Hrmova M, Langridge P, Lopato S (2009) Characterization of the wheat endosperm transfer cell-specific protein TaPR60. Plant Mol Biol 71:81–98
Kovalchuk N, Li M, Wittek F, Reid N, Singh R, Shirley N, Ismagul A, Eliby S, Johnson A, Milligan AS, Hrmova M, Langridge P, Lopato S (2010) Defensin promoters as potential tools for engineering disease resistance in cereal grains. Plant Biotechnol J 8:47–64
Lascombe M-B, Bakan B, Buhot N, Marion D, Blein J-P, Larue V, Lamb C, Prangé T (2008) The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci 17:1522–1530
Laskowski R, MacArthur M, Moss D, Thornton J (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:191–283
Li M, Singh R, Bazanova N, Milligan AS, Shirley N, Langridge P, Lopato S (2008) Spatial and temporal expression of endosperm transfer cell-specific promoters in transgenic rice and barley. Plant Biotechnol J 6:465–476
Li M, Lopato S, Kovalchuk N, Langridge P (2013) Functional genomics of seed development in cereals. In: Gupta PK, Varshney RK (eds) Cereal genomics II. Springer, Netherlands, pp 215–245
Lid SE, Al RH, Krekling T, Meeley RB, Ranch J, Opsahl-Ferstad HG, Olsen OA (2004) The maize disorganized aleurone layer 1 and 2 (dil1, dil2) mutants lack control of the mitotic division plane in the aleurone layer of developing endosperm. Planta 218:370–378
Luo M, Bilodeau P, Dennis E, Peacock W, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97:10637–10642
Marchler-Bauer A, Anderson J, Cherukuri P, DeWeese-Scott C, Geer L, Gwadz M, He S, Hurwitz D, Jackson J, Ke Z, Lanczycki C, Liebert C, Liu C, Lu F, Marchler G, Mullokandov M, Shoemaker B, Simonyan V, Song J, Thiessen P, Yamashita R, Yin J, Zhang D, Bryant S (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Res 33:D192–D196
Matthews PR, Wang M-B, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, Jacobsen JV (2001) Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Mol Breed 7:195–202
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Molina A, García-Olmedo F (1997) Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J 12:669–675
Monjardino P, Rocha S, Tavares A, Fernandes R, Sampaio P, Salema R, Câmara Machado A (2013) Development of flange and reticulate wall ingrowths in maize (Zea mays L.) endosperm transfer cells. Protoplasma 250:495–503
Ogawa M, Hiraoka Y, Taniguchi K, Aiso S (1998) Cloning and expression of a human/mouse Polycomb group gene, ENX-2/Enx-2. Biochem Biophys Acta 1395:151–158
Olsen OA (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16:S214–S227
Otegui MS, Mastronarde DN, Kang B-H, Bednarek SY, Staehelin LA (2001) Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13:2033–2051
Pasquato L, Pengo P, Scrimin P (2005) Nanozymes: functional nanoparticle-based catalysts. Supramol Chem 17:163–171
Pei J, Kim B-H, Grishin NV (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300
Prestridge D (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7:203–206
Royo J, Gómez E, Sellam O, Gerentes D, Paul W, Hueros G (2014) Two maize END-1 orthologs, BETL9 and BETL9like, are transcribed in a non-overlapping spatial pattern on the outer surface of the developing endosperm. Front Plant Sci 5:180
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Singh KB (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol 118:1111–1120
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17:355–362
Skriver K, Leah R, Muller-Uri F, Olsen F, Mundy J (1992) Structure and expression of the barley lipid transfer protein gene Ltp1. Plant Mol Biol 18:585–589
Sørensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jürgens G, Somerville C, Lepiniec L, Berger F (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129:5567–5576
Stangeland B, Salehian Z (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Biol Rep 20:107–114
Thiel J, Weier D, Sreenivasulu N, Strickert M, Weichert N, Melzer M, Czauderna T, Wobus U, Weber H, Weschke W (2008) Different hormonal regulation of cellular differentiation and nunction in nucellar projection and endosperm transfer cells: a microdissection-based transcriptome study of young barley grains. Plant Physiol 148:1436–1452
Thiel J, Müller M, Weschke W, Weber H (2009) Amino acid metabolism at the maternal–filial boundary of young barley seeds: a microdissection-based study. Planta 230:205–213
Thiel J, Hollmann J, Rutten T, Weber H, Scholz U, Weschke W (2012a) 454 Transcriptome sequencing suggests a role for two-component signalling in cellularization and differentiation of barley endosperm transfer cells. PLoS One 7:e41867
Thiel J, Riewe D, Rutten T, Melzer M, Friedel S, Bollenbeck F, Weschke W, Weber H (2012b) Differentiation of endosperm transfer cells of barley: a comprehensive analysis at the micro-scale. Plant J 71:639–655
Thompson RD, Hueros G, Becker HA, Maitz M (2001) Development and functions of seed transfer cells. Plant Sci 160:775
Wang H-H, Wang Z, Wang F, Gu YJ, Liu Z (2012) Development of basal endosperm transfer cells in Sorghum bicolor (L.) Moench and its relationship with caryopsis growth. Protoplasma 249:309–321
Weisel M, Proschak E, Schneider G (2007) PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. doi:10.1186/1752-153X-1-7
Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35:3375–3382
Wu S, Zhang Y (2008) MUSTER: improving protein sequence profile–profile alignments by using multiple sources of structure information. Proteins 72:547–556
Yanagisawa S (1997) Dof DNA-binding domains of plant transcription factors contribute to multiple protein–protein interactions. Eur J Biochem 250:403–410
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21:281–288
Yanagisawa S (2001) The transcriptional activation domain of the plant-specific Dof1 factor functions in plant, animal, and yeast cells. Plant Cell Physiol 42:813–822
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45:386–391
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17:209–214
Acknowledgments
This research was funded by the Grain Research and Development Corporation (GRDC) and an International Postgraduate Research Scholarship (IPRS). The authors acknowledge the Australian Centre for Plant Functional Genomics (ACPFG) and Common Wealth Scientific and Industrial Organisation (CSIRO), Plant Industry (PI) at Waite Campus for provision of research facilities. We thank Dr Matthew Tucker and Ms Susan Johnson at CSIRO Plant Industry, Waite Campus and Dr Gwen Mayo for assistance with microscopy, Natalia Bazanova for technical support and Dr Julie Hayes for critically reading and editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, M., Lopato, S., Hrmova, M. et al. Expression patterns and protein structure of a lipid transfer protein END1 from Arabidopsis. Planta 240, 1319–1334 (2014). https://doi.org/10.1007/s00425-014-2155-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2155-6