Abstract
Changes in root architecture are one of the adaptive strategies used by plants to compensate for local phosphate (Pi) deficiency in soils. Root architecture variables triggered by Pi availability are well documented in Arabidopsis (Arabidopsis thaliana), but the molecular mechanisms behind these adaptive responses remain to be elucidated. By the use of transcriptomic and quantitative RT-PCR analysis, we observed that an AINTEGUMENTA-like gene, named PRD for Phosphate Root Development, was rapidly repressed in roots under low Pi conditions. The physiological function of the PRD gene was analyzed through the null allele mutant prd, which displayed less development of primary and lateral roots under Pi-starvation conditions than wild-type plants. Complementation of the prd mutant with the wild-type gene led to a similar response to Pi starvation as wild-type plants, indicating the complete rescue of the mutant phenotype. These results suggest that PRD gene is involved in the regulation of root architectural responses to Pi starvation by controlling primary and lateral root elongation. This model is in agreement with the tissue-specific pattern of PRD gene expression, which was observed to occur specifically in the apex in both the primary and lateral roots. However, Pi influx, anionic profiles and root expression of genes typically induced by Pi starvation, such as high affinity Pi transporters (PHT1;1 and PHT1;4) and an acid phosphatase (AtACP5), were similar in wild type and prd plants in response to Pi starvation. These results support the hypothesis that the PRD gene is not a checkpoint for Pi-starvation responses, but acts specifically as a regulator of root architectural responses to Pi starvation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphorus is an essential structural constituent of many bio-molecules and plays a pivotal role in energy conservation and metabolic regulation. Paradoxically, phosphorus bioavailability is one of the most challenging problems in crop nutrition and is often a limiting factor even in fertile soils. Although inorganic orthophosphate (Pi), the assimilated form of phosphorus, may be present in sufficient amounts, the high soil binding capacity of Pi leads to very low Pi concentrations available to plants (Hinsinger 2001). As a consequence, under Pi limiting conditions, plants have evolved sophisticated metabolic and developmental strategies to maintain Pi homeostasis and to maximize its acquisition from the rhizosphere. These adaptive responses to increase the availability of internal or external Pi include increased expression of high-affinity Pi transporters, enhanced production of enzymes such as phosphatases and nucleases, and secretion of organic acids (Raghothama 1999; Raghothama and Karthikeyan 2005). Besides allowing roots to explore a larger volume of soil in search of nutrient-rich patches, changes in the root system architecture play a central role in the plant’s adaptive response to Pi starvation. In recent years, this adaptive response has been widely studied and several studies have described the effects of Pi deprivation on root architecture. In Arabidopsis, Pi starvation affects primary root growth and lateral root initiation and elongation (Williamson et al. 2001; Linkohr et al. 2002; López-Bucio et al. 2002, 2005; Al-Ghazy et al. 2003; Nacry et al. 2005). In fine, Pi deprivation alters cell division and elongation at the primary root tip and induces an irreversible shift from an indeterminate to a determinate root growth program in which the quiescent center seems to play a central role (Sánchez-Calderón et al. 2005). However, despite recent extensive studies on global gene expression during Pi deficiency, little is known about the molecular components of the signal transduction pathway that trigger the Pi-starvation responses (Hammond et al. 2003; Uhde-Stone et al. 2003; Wasaki et al. 2003, 2006; Wu et al. 2003; Misson et al. 2005; Muller et al. 2007). To date, only few transcription factors have been shown to be required for Pi-starvation-dependent responses in higher plants. The MYB transcription factor PHR1 was first reported to control a small subset of Pi-starvation responses that including only a few genes whose expression is activated by Pi limitation, such as AtACP5, AtIPS1/3, At4, and RNS1 (Rubio et al. 2001). In addition, phr1 mutants showed reduced anthocyanin accumulation, fresh weight, intracellular Pi, and, to a lesser extent, a reduced root-to-shoot growth ratio in response to Pi starvation, indicating that PHR1 is a positive regulator of Pi-starvation responses. However, other changes in root architecture typically associated with Pi deficiency were unaffected in phr1. Recently, the bHLH transcription factor OsPTF1 was shown to provide tolerance to Pi starvation in rice and to be involved in the efficient utilization of Pi in plants (Yi et al. 2005). The WRKY75 transcription factor has also been characterized as a regulator of Pi stress responses, but its regulatory effect on root architecture was independent of the Pi status of the plant (Devaiah et al. 2007). Moreover, several groups have also isolated mutants impaired in their Pi response including pdr2, which disrupts sensing of low external Pi availability that lead to alterations in root growth (Ticconi et al. 2004), and lpi that fails to arrest primary root growth when grown under low Pi conditions (Sánchez-Calderón et al. 2006). The molecular function of these genes is still unknown and none of these mutations specifically triggered changes in root architectural responses to Pi starvation. By the use of an elegant QTL approach, Reymond et al. (2006) and Svistoonoff et al. (2007) recently identified an LPR gene encoding a multicopper oxidase specifically expressed in the root tip and involved in the sensing of the external Pi concentration.
Thus, the aim of this work was to identify and characterize regulatory genes specifically involved in root architectural responses to Pi deprivation in Arabidopsis. Using transcriptome analysis, we identified a transcription factor named PRD for Phosphate Root Development, belonging to the AINTEGUMENTA-like (AIL) gene family (Nole-Wilson et al. 2005), that was regulated by Pi starvation. In-depth physiological, genetic, histological, and molecular analyses show that this gene regulates root growth responses to Pi deprivation.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana, ecotype Columbia (Col-0) and T-DNA mutant seeds were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al. 2000). Seeds were surface-sterilized in 4% (v/w) Bayrochlor (Bayrol, Mundolsheim, France), 50% (v/v) ethanol for 15 min, and washed three times with 100% ethanol and three times in sterile water. Sterile seeds were sown on 12 × 12 cm Petri dishes (Greiner Bio-One, Frickenhausen, Germany) containing 40 ml of sterile culture medium and sealed with Parafilm. The high Pi culture medium contained 0.5 mM CaSO4, 2 mM KNO3, 0.5 mM MgCl2, 0.05 mM NaFe-EDTA, 1 mM KH2PO4, 2.5 mM Mes, 50 μM H3BO3, 12 μM MnCl2, 1 μM CuCl2, 1 μM ZnCl2, and 30 nM NH4Mo adjusted to pH 5.7 with 1 N KOH and solidified with 0.8% (w/v) Bactoagar (DIFCO, BD Bioscience, Sparks, MD, USA). All chemicals were purchased from Sigma Chemicals (St. Quentin, France). In the low Pi medium, KH2PO4 was replaced by 1 mM KCl and the Pi content was estimated at 3 μM due to the slight Pi content of Bactoagar. After sowing on either high or low Pi medium, Petri dishes were cold treated at 4°C for 24 h in darkness to promote and synchronize germination, subsequently transferred in a vertical position to a growth chamber at a temperature of 21.5°C, and subjugated to a photoperiod of 16 h of light (150 μmol m−2 s−1) from fluorescent and metal halogen lamps.
Morphological analysis
A desktop scanner (resolution: 450 dpi) was used to record images of the root system directly from plants growing in Petri dishes. Images corresponding to different growth times were analyzed using Optimas software version 6.1 (Media Cybernetics, Silver Spring, MD, USA). The length of primary and lateral roots, and the number of lateral roots were determined manually. Data were exported to an Excel work-sheet for final processing. All results were statistically analyzed using Statistica software version 7.1 (Statsoft, Tulka, OK, USA). For the analysis of variance, 2- and 3-factors ANOVA with an LSD post hoc test were used to measure differences (P = 0.05).
Identification of a T-DNA insertion mutant for the PRD gene
The homozygote plants of the T-DNA insertion line (SALK_046920) for the At1g79700 gene were identified by PCR with the following primer sets: 5′-TTCGAATGGGCTTAGAATCG-3′ and 5′-AGGGAACAAAGAAGGTCACG-3′ from the gene and the primer LBb1, from the T-DNA region. Individual homozygous mutants were back-crossed twice with the wild-type Col-0. The genotype of the F2 individuals was checked by PCR using gene-specific primers and T-DNA primers.
Plant transformation
For complementation of the prd mutant, a 5.1-kb fragment, containing the PRD coding region and the 1.34 kb of the 5′ flanking DNA upstream of the starting codon, was amplified by PCR with 5′-AATGGATCCATTCCGGCATGTAAAGATCG-3′ and 5′-GGCAAGCTTAGACGTCACGGATGACGAA-3′ primers and DNA genomic BAC F20B17 as the template. The PCR product was cloned into the binary vector pCAMBIA1300 (Cambia, Canberra, Australia) using BamHI and HindIII restriction sites (underlined in the sequence primers). The construct and empty vector control were used to transform Arabidopsis plants using the floral-dip method (Clough and Bent 1998) and transgenic seedlings were selected by spraying 50 μg l−1 Basta.
RNA extraction and analysis
Frozen roots were ground in liquid nitrogen and total RNA was isolated using TRIzol reagent (Invitrogen, Cergy Pontoise, France) according to the manufacturer’s protocol. The quality and quantity of RNA were measured with OD280/OD260 and gel electrophoresis. RNA extracts were stored at −80°C prior to hybridization onto macroarrays or PCR quantification.
Macroarray analysis
Macroarrays were prepared, hybridized and analyzed as previously described (Al-Ghazi et al. 2003; Tranbarger et al. 2003). Complex probes were prepared from 5 to 10 μg of total RNA, using anchored oligo-dT (VT18) as previously described (Al-Ghazi et al. 2003). Two sets of macroarray hybridizations were performed in two replicate biological experiments and three membranes were used for each hybridization.
Quantitative real-time PCR
Three micrograms of RQ-DNase (Promega, Madison, WI, USA) digested total RNA was used to prepare cDNA by reverse transcription using M-MLV reverse transcriptase (Promega) and oligo-dT (V18) primers, according to the manufacturer’s protocol. Gene expression was determined by quantitative RT-PCR (LightCycler, Roche Diagnostics, Mannheim, Germany) using gene-specific primers (see sequences below) and LightCycler FastStart DNA Master SYBR GREEN I (Roche Diagnostics). Specific primers were designed to generate PCR gene products between 100 and 250 bp. Expression levels of tested genes were normalized to expression levels of the EF1a (At1g07920, At1g07930, At1g07940) or TUBULIN (At5g12250) gene. Gene-specific primer sequences were [5′–3′ and corresponded to forward (F) or reverse (R) gene sequences] for PRD (At1g79700): F, CAAATGCGCTTCGTATCTCC; R, TAAAACAGCGTCTGCGTCTG; for AtACP5 (At3g17790): F, TCATCGATCCTACCTAATTCATCA; R, ACCCCAATCTCCGATGACTA; for PHT1;1 (At5g43350): F, AAAACCAAACATCGCACTCC; R, CCAGGCATCGGAGTTAAGAA; for PHT1;4 (At2g38940): R, TGATAAGCTCGGGAGGAAGA; F, TGGTTGCGGATAAAGGGTAG; for EF1α (At1g07940): F, GTCGATTCTGGAAAGTCGACC; R, AATGTCAATGGTGATACCACGC; for TUBULIN (At5g12250): F, ATAGCTCCCCGAGGTCTCTC; R, TCCATCTCGTCCATTCCTTC.
Whole-mount in situ hybridization
Sense and antisense probes were labeled with UTP-digoxigenin (Roche Diagnostics) during transcription with the T7 MAXIscript kit (Ambion, Applied Biosystems, Courtaboeuf, France). Nucleic probes for PRD and PLT1 (At3g20840) were specially designed to reach a length between 150 and 200 bp. Probes for ribosomal 18S were used as controls. Whole-mount in situ hybridization was performed manually according to Friml et al. (2003) with the following modifications: 8-day-old Arabidopsis seedlings were fixed for 30 min in nylon biopsy bags in a 1:1 (v/v) mixture of fixation solution (4% paraformaldehyde, 15% DMSO, 0.08 M EGTA, 0.1% Tween 20 in PBS pH 7.4) and heptane, with shaking. The fixed seedlings were then incubated in methanol (2 × 5 min), ethanol (3 × 5 min), ethanol/safesolv (Labonord, Templemars, France) mixture (1:1, 30 min), washed with ethanol (2 × 5 min), and rehydrated in ethanol/PBS series (75, 50 and 30%, 10 min each). The rehydrated seedlings were post-fixed in fixation solution (20 min) and washed in PBS (3 × 5 min). To remove proteins from RNA, the samples were treated by proteinase K digestion (40 μg ml−1) for 20 min at 37°C and the reaction was stopped with glycine (0.2% in PBS) for 5 min. The samples were washed in PBST (PBS, 0.1% Tween 20, 2 × 10 min). The treated samples were incubated in 2 ml Eppendorf tubes in a 1:1 mixture (PBST/hybridization mix) for 15 min, pre-hybridized in 400 μl of hybridization mix (50% formamide, 10% dextran sulfate, 5× Denhardt’s solution, 2× SSC and 1 mg ml−1 tRNA) for 2 h at 45°C, and hybridized in 400 μl of hybridization mix with 0.5–1 ng μl−1 denatured riboprobes overnight at 45°C. After hybridization, seedlings were washed for 10, 60, and 20 min in a solution containing 50% formamide, 2× SSC, and 0.1% Tween 20, at 50°C, and then incubated in NTE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 0.5 M NaCl) twice at room temperature. RNaseA digestion (40 μl ml−1 in NTE) was carried out for 30 min at 37°C. The samples were finally washed in 2× SSC-0.1% Tween 20 for 20 min at 50°C and in 0.2× SSC-0.1% Tween 20 for 20 min at 50°C.
For signal detection, samples were equilibrated in PBST for 10 min, blocked for 2 h in 2% BSA in PBST and incubated with gentle agitation in an anti-digoxigenine alkaline phosphatase-conjugated Fab fragment antibody (Roche Diagnostics) 1/1000 in PBST, 2% BSA, overnight at 4°C. After five washes in PBST for 20 min each, the tissues were equilibrated in detection buffer (0.1 M Tris–HCl, pH 8.2, 2 mM Levamisole) for 10 min and in the substrate from Vector Blue (Vector Laboratories, Burlingame, CA, USA) for 2 h. The reaction was stopped by immersion in 0.1 M Tris–HCl, pH 8.2, follow by water. The seedlings were mounted in Mowiol on ESCO slides. The microscope imaging was performed at the Montpellier RIO Imaging Plateform (www.mri.cnrs.fr) with a Leica DM6000 microscope (Leica Microsystems, Rueil-Malmaison, France) and images were processed through Volocity 4.0.1 (Improvision, Lexington, MA, USA). The experiment was repeated four times, and for each experiment, 20 seedlings were used for each treatment.
Pi influx measurements
Pi influx was measured on whole plants grown on vertically oriented agar plates for 12 days. Roots of whole plants were placed for 2 min at 25°C in a 0.05 mM NH4H2PO4 solution adjusted to pH 4.5 with Mes buffer, transferred in an identical solution containing 7.4 kBq ml−1 of [33]Phosphorus radiotracer (GE Healthcare, Aulnay sous Bois, France). After 20 min of incubation, the roots were washed three times for 30 s in ice-cold 0.2 mM CaSO4 solution, harvested, and blotted with absorbent paper. Their soluble content extracted in a 100 mM HCl solution by heating for 30 min at 70°C. The radioactivity of the acidic extract was determined after addition of scintillation liquid (Ultimas Gold, Packard, Rungis, France) using a scintillation counter (Tri-Carb 2101 TR, Packard). Pi influx was measured on plants grown in similar conditions as described earlier for root measurements.
Soluble anionic profiles
Soluble anions were extracted by incubating 20 mg of fresh tissues in 1 ml 70°C water for 30 min, and these water soluble extracts were filtered (0.45 μm, cellulose acetate, Nalgene) to remove particular matter. Pi and major inorganic and organic anions were analyzed by high-performance ion chromatography (LC20, Dionex, Voisins Le Bretonneux, France) using a Dionex IonPac AS11 column and a sodium hydroxide (1–22 mM) linear gradient as described by El Kassis et al. (2007).
Results
The PRD expression is repressed by Pi starvation in Arabidopsis roots
In order to identify transcription factors involved in root architectural responses to Pi-deprivation, a set of 126 putative transcription factor ESTs were arrayed on nylon membranes (Tranbarger et al. 2003). Hybridizations were performed with RNA extracted at 5, 7, 10, and 14 days after germination in both the control and Pi-starved plants. These time points were selected according to the time-course analyses of the root adaptive responses to Pi-deprivation reported by Al-Ghazi et al. (2003). This transcriptomic approach led to the selection of 19 transcription factors up- or down-regulated in response to Pi starvation (data not shown). Among them, one gene (At1g79700) showed a unique expression profile under Pi-starvation conditions characterized by global repression of expression (Fig. 1a). After 5 days of culture on high Pi medium, the root expression of this gene rapidly increased to reach a plateau from day 10. In contrast, on low Pi medium, the expression remain unchanged and led to a maximum fold change (+P/−P) of 1.7 at day 10 after germination. This expression profile was confirmed by quantitative RT-PCR analysis in three independent experiments (Fig. 1b). In parallel, the expression of a high-affinity Pi transporter gene (PHT1;4) was analyzed as a root-specific marker of Pi starvation and, as expected, under Pi-starvation conditions, the expression of this gene increased continuously throughout the growth time course (Fig. 1c).
Kinetic effects of Pi availability on the root expression profiles of PRD and AtPT2 genes in Arabidopsis roots. Wild-type Columbia seedlings were grown for 14 days in the presence of low (3 μM; open circles) or high (1 mM; full circles) concentrations of Pi on vertically orientated agar plates. a Macroarray analysis of PRD expression. b Qantitative RT-PCR analysis of PRD expression. c Qantitative RT-PCR analysis of AtPT2 (At2g38940) expression. Relative expression levels are expressed in arbitrary units. Data correspond to the average values (±SE) of three independent measurements
These data showed that the level of At1g79700 mRNA accumulation in Arabidopsis roots was dependent of the Pi content of the growth medium. This gene, named PRD for Phosphate Root Development, encodes a putative AP2/ERF transcription factor and belongs to the family of AINTEGUMENTA-like (AIL) genes as defined by Nole-Wilson et al. (2005). The AP2 subgroup of this family is comprised of 18 genes including the floral developmental gene AINTEGUMENTA (ANT; Elliott et al. 1996; Klucher et al. 1996) and the root developmental meristem genes PLETHORA1 (PLT1) and PLT2 (Aida et al. 2004).
The PRD gene is involved in the regulation of architectural root responses to Pi starvation
To determine the physiological function of PRD in Arabidopsis, one mutant line that harbors the T-DNA insertion in the PRD gene (Salk_046920) was isolated in the T-DNA insertion collection generated at the Salk Institute (Alonso et al. 2003). The T-DNA insertion was localized in the eighth exon (Fig. 2a). The presence and the location of the T-DNA insertion were verified by junction PCR and sequencing. Nucleotide sequencing of the T-DNA::genomic borders of the homozygous prd mutant revealed that the T-DNA insertion site was 2872-bp downstream of the translation start. Individual homozygous mutants were back-crossed twice with the wild-type Col-0. To ensure that the T-DNA insertion altered PRD gene expression in this mutant, PCR analyses were carried out regardless of Pi growth conditions. No amplification was detected in the prd mutant indicating that it is probably a null allele (Fig. 2b). Under greenhouse conditions with normal fertilization, the aerial parts of prd mutants were phenotypically normal. Their vegetative and floral development and seed production were similar to that of wild-type plants. In contrast, under low-Pi (3 μM) conditions, root growth pattern of prd mutants were found to be distinguishable from the wild-type plants (Fig. 2c).
Isolation of the prd T-DNA insertional mutant. a Intron–exon organization of the Arabidopsis PRD gene (At1g79700) and T-DNA location. Gray shaded boxes indicate untranslated regions, and solid black boxes and the solid line indicate coding regions and the intron, respectively. The position of the T-DNA insertion in the prd allele is indicated by a triangle (not to scale). b PCR analysis of PRD transcript in wild type (Col-0) and mutant alleles. Expression of GAPDH (At3g26650) was analyzed as a control. c Root architecture of prd mutant. Wild type and prd plants were grown for 7 days at high Pi concentration (1 mM) and then transferred for 5 days on media with high (1 mM) or low (3 μM) Pi concentrations on vertically orientated agar plates. Primer locations are indicating in Fig. 2a by asterisks for homozygous mutant line screening and by dark triangles for quantitative RT-PCR analyses and whole-mount RNA in situ hybridization
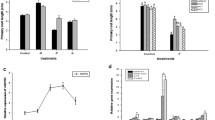
To precisely evaluate the consequences of PRD gene disruption on root architectural responses to Pi starvation, we examined the root growth parameters for mutant plants grown under high- and low-Pi conditions (Fig. 3). Wild type and prd seeds were germinated on medium with high Pi content during 7 days and then transferred to fresh agar plates on media with low- or high-Pi concentrations during 5 days. Temporal analysis of the root system architecture of wild type and prd seedlings was determined 7–12 days after germination (i.e. from day 0 to day 5 after transfer; Supplementary Fig. S1). As confirmed at day 5 after transfer (Fig. 3), when grown on high Pi medium, both prd and wild-type plants showed a similar root phenotype suggesting that, under optimal culture conditions, PRD plays no role in root growth. In contrast, low Pi availability reduced the length of primary roots in wild-type plants (30%), while the reduction was more drastic (45%) in prd plants and their primary root growth was almost arrested (Fig. 3a). Furthermore, low Pi availability promoted lateral root elongation in wild-type plants (37%), while it clearly reduced it in prd mutant plants (22%, Fig. 3b). Regardless of Pi treatment, there were no differences in the lateral root number between wild type and prd mutant plants (Fig. 3c). To assess whether Pi deficiency affected other morphological responses, leaf size was measured on wild type and prd mutant plants grown under high- and low-Pi conditions for 10 days (Table 1). This variable was drastically reduced by almost fivefold under low-Pi conditions for both wild type and prd mutant plants, while no difference was observed between mutant and wild-type plants suggesting a specific role for the PRD gene at the root level.
Effects of Pi availability on root architecture variables. Wild type (black bars) and mutant (white bars) seedlings were grown for 7 days at high Pi concentration (1 mM) and then transferred for 5 days on media with high (1 mM) or low (3 μM) Pi concentrations on vertically orientated agar plates. a Primary root length. b Total lateral root length and c the number of emerged lateral roots were measured at day 12. All results are the average value (±SE) of 16 seedlings. The letters represent statistically homogenous subgroups using the LSD post hoc test at a α = 0.05 significance level
To confirm that the altered root responses to Pi starvation were the result of the PRD gene disruption, complementation analysis of the prd mutant was performed using Agrobacterium tumefaciens-mediated transformation. A 5.2-kb BAC genomic DNA fragment containing the PRD ORF, 1.4-kb upstream and 0.6-kb downstream of the sequence was cloned into the pCAMBIA 1300 binary vector for transformation (Hajdukiewicz et al. 1994). Transgenic lines were able to respond to Pi starvation in a similar way to wild-type plants, indicating the complete rescue of the mutant phenotype (Fig. 4).
Architectural root analyses of complemented prd mutant. Wild type (black bars), prd mutant (white bars) and complemented prd mutant (Cprd, hatched bars) seedlings were grown for 7 days at high Pi concentration (1 mM) and then transferred for 5 days on media with high (1 mM) or low (3 μM) Pi concentrations on vertically orientated agar plates. a Primary root length and b total lateral root length were measured at day 12. All results are the average value (±SE) of 16 seedlings. The letters represent statistically homogenous subgroups using LSD post hoc test at a α = 0.05 significance level
To explore the tissue-specific pattern of expression of the PRD gene in more detail, endogenous PRD transcripts were observed in wild type and prd mutant seedlings using whole-mount in situ hybridization (Fig. 5). This technique allows direct observations of tissues without samples embedding and sectioning, and it is particularly adapted to the localization of gene expression at the root tip level (Friml et al. 2003). The tissue was hybridized with digoxigenin-labeled single-stranded PRD RNA in either the antisense (Fig. 5a) or the sense orientation (Fig. 5d). The sense RNA is a negative control and reveals a weak background, nonspecific staining. The antisense probe specifically hybridizes with the PRD mRNA present in the tissue, thereby revealing its spatial distribution. Several other controls were included in the whole-mount experiment such as the 18S ribosomal RNA antisense (Fig. 5g) and sense probe (Fig. 5h). Thus, only primary and lateral root tips showed a specific signal, strong and detectable in all the cells and in the thickness of root apex, whereas no signal was observed with the sense probe, validating our analysis at the root apex level. The PLT1 probe was used as a positive control because of its well described expression pattern in the root apex using similar techniques (Aida et al. 2004) and it belongs to the same AIL family as the PRD gene (Fig. 7i). The PRD hybridization signal was specifically observed in the root apex, mainly in the central cylinder and the cortex (Fig. 5a). A weak signal was also observed in the epidermal cells. Under similar conditions, PLT1 presented a similar expression pattern but the hybridization signal was strongest in the central cylinder (Fig. 5i). Quantitative RT-PCR showed an accumulation of PRD mRNA in the apex part of the root compared to the whole root confirming the in situ localization of the PRD gene in the root apex (Fig. 5j). Even if the whole-mount in situ hybridization technique was not the adequate technique to quantify gene expression, a slight decrease in the hybridization signal of PRD gene was observed in Pi-starvation plants, especially at the vascular tissue level, compared to those from the control conditions (Fig. 5b). Moreover, the weak and diffuse hybridization signal observed with PRD antisense probe in the prd mutant plants confirmed the tissue-specific pattern of PRD gene expression (Fig. 5c). The weak and diffuse hybridization signal observed in Fig. 5c may reveal weak cross-hybridizations with some of the other genes of the AIL family or the presence of PRD truncated mRNA. All these converging data confirmed that PRD gene expression was tightly linked with meristem activity, in both the primary and lateral roots, and reinforced its functional role in response to Pi privation.
Localization of PRD expression in root apex of Arabidopsis by whole-mount RNA in situ hybridization. PRD antisense probe on primary root of wild-type plants grown in the presence of high (1 mM, a) or low Pi (3 μM, b) Pi concentrations, and on primary root of prd mutant plants grown in the presence of high Pi concentration (c). d PRD sense probe on primary root of wild-type plants grown in the presence of high Pi concentration. e, f PRD antisense probe on lateral roots of wild-type plants grown in the presence of high Pi concentration. g 18S ribosomal RNA antisense probe. h 18S ribosomal RNA sense probe. i PLT1 antisense probe. Seedlings were grown for 8 days in the presence of high or low Pi concentrations. c cortex, e endodermis, ep epidermis, p pericycle, vb vascular bundle. Scale bar = 20 μm. j Quantitative RT-PCR analysis of PRD expression from whole-root (white bars) or dissected root tips (black bars) of wild-type seedlings grown for 12 days at high Pi concentration on vertically orientated agar plates
Together, these results clearly showed that prd mutation reduced growth of both primary and especially lateral roots under Pi starvation. These data suggest that the PRD gene plays a role in the regulation of root architectural responses to Pi starvation through its involvement in root elongation.
Root development defect in prd mutant was not observed under other nutrient deprivations.
To investigate whether PRD was specifically involved in root architectural responses to Pi starvation, we assessed the effects of other nutrient limited conditions including nitrogen (−N), sulfur (−S), and iron (−Fe) on the length of the primary and lateral roots. Nutrient starvation affected either primary root or lateral root growth, or both (Fig. 6). As expected, the Pi-starved medium inhibited primary root growth (Fig. 6a) and stimulated lateral root growth (Fig. 6b) in wild-type plants. Primary root length was mainly affected by Pi- or N-starvation and lateral root length was strongly reduced by Pi-, N-, or S-starvation. However, no significant difference was observed between wild type and prd mutant plants under starvation conditions at the exception of Pi starvation where highly significant differences were measured for both the primary and lateral roots growth. These results suggest that PRD is involved in the regulation of root architecture in responding specifically to Pi-starvation signal.
Effects of nutrient availability on root architecture variables. Wild type (black bars) and prd mutant (white bars) seedlings were grown for 7 days at high Pi concentration (1 mM) and then transferred for 5 days on media with high Pi concentrations (1 mM) or medium lacking a mineral element (Pi, N, S, Fe) on vertically orientated agar plates. a Primary root length and b total lateral root length were measured at day 12. All results are the average value (±SE) of 16 seedlings. Comparison of means (t test) are indicated as non-significant (NS) or very highly significant (***) responses (P < 0.001)
The PRD gene is not a checkpoint for responses to Pi starvation
To check if the loss of PRD affects other characteristic responses to Pi starvation, we conducted several experiments including analysis of gene expression, Pi uptake, and anionic profile.
To study whether the genetic defects in PRD had an effect on the expression of Pi–responsive genes, we used quantitative RT-PCR to analyze the expression level of several Pi-starvation-induced genes such as PHT1;1 and PHT1;4 (two high-affinity Pi transporters), and AtACP5 (an acid phosphatase). As previously described, the steady-state transcript levels of these three genes increased noticeably in wild-type plants subjected to Pi starvation (Fig. 7). Similar increases were also observed in prd mutants that show no significant difference from the wild-type plants grown under Pi deficiency.
Effect of Pi availability on the expression profiles of the Arabidopsis PRD, AtACP5, PHT1;1 and PHT1;4 genes. a Quantitative RT-PCR analyses of PRD, b AtACP5, c PHT1;1, and d PHT1;4 gene expression. Relative levels of expression are expressed in arbitrary units. Wild type (black bars) and prd mutant (white bars) seedlings were grown for 7 days at high Pi concentration (1 mM) and then transferred for 5 days on media with high (1 mM) or low (3 μM) Pi concentrations on vertically orientated agar plates. Root gene expressions were measured at day 12. Data correspond to the average values (±SE) of three independent measurements
With the same rationale, Pi uptake was monitored in wild type and prd mutants using 33P labeling (Supplementary Fig. S2). The results showed that 12 day-old wild-type plants Pi-starved for 5 days increased their Pi uptake rate about fourfold compared to plants supplied with Pi. Phosphate starvation induced a similar increase in Pi uptake by prd mutants.
Finally, anionic profiles of wild type and prd mutant plants were analyzed in roots and leaves after 12 days of Pi treatments (Supplementary Fig. S3). In both the roots and leaves, Pi starvation caused a significant reduction in nitrate and Pi contents, and a significant increase in the accumulation of some organic acids such as malate and citrate. Similar changes were observed in wild type and mutant plants indicating that PRD mutation did not alter anion accumulation in response to Pi starvation, but that the changes in root development in prd mutants were not caused by a general or local decrease in Pi content in the root of this mutant with respect to wild-type plants.
Taken together, these results support the hypothesis that the PRD gene does not function as a checkpoint for Pi-starvation responses, but acts specifically as a regulator of root architectural response to Pi starvation.
Discussion
Since roots often determine the capacity of plants to efficiently explore and exploit the spatially heterogeneous composition of the soil, changes of root architecture are one of the major adaptive responses of plants to Pi starvation. Two of the most consistent root architectural changes that occur during Pi starvation are the reduction in primary root growth and the prolific growth of lateral roots (Williamson et al. 2001; Al-Ghazi et al. 2003; Nacry et al. 2005; Sánchez-Calderón et al. 2006). As expected, we obtained similar results under our conditions with wild-type plants (Figs. 2c, 3). Recently, it was reported that the reduction in primary growth is a consequence of a determinate program induced by low Pi that inhibits cell division in the primary root meristem and promotes differentiation within the root tips (Sánchez-Calderón et al. 2005). However, low Pi conditions did not lead to complete permanent loss of meristem activity. When plants were transferred from low to high Pi medium, primary root growth was rapidly stimulated (Chen et al. 2000). This result suggests a high degree of meristem plasticity in response to external Pi status and an adjustment of root architecture via changes in meristem activity. Moreover, the maintenance of a basal level of meristem function under low Pi conditions could be a basic process to reorient root growth in Arabidopsis and is probably under the control of multiple genes, to increase the flexibility of root plasticity responses.
Despite their important implications, little is known about the molecular events controlling for Pi sensing and the effect of Pi starvation on root system architecture. Several studies have provided new insights into the genetic components involved in root architectural adaptive responses to Pi availability (Ticconi et al. 2004; Sánchez-Calderón et al. 2006; Svistoonoff et al. 2007). The data presented in this work showed that mRNA accumulation of PRD, an AP2/ERF transcription factor, in Arabidopsis roots was regulated by the Pi content of the medium and strongly repressed by Pi deficiency (Figs. 1, 7). The physiological function of the PRD gene was analyzed through the null allele mutant line prd. The effects of Pi availability on architectural root variables were compared between wild type and prd mutant plants (Figs. 2c, 3). Mutants displayed shorter primary roots under Pi-starvation conditions than wild-type plants. Moreover, low Pi availability clearly reduced lateral root elongation in prd mutant plants, whereas it promoted it in wild-type plants. These results suggest that the PRD gene is involved in the regulation of root architectural responses to Pi starvation by controlling primary and lateral root elongation. This is in agreement with the tissue-specific expression pattern of the PRD gene which was specifically observed in the apex in both the primary and lateral roots (Fig. 5). In addition, no difference was observed in the emergence of lateral roots between prd and wild-type plants suggesting that the PRD gene was not directly involved in the initiation and emergence of root primordia, but in the constitutive control of root growth. It should be emphasized that both the prd and wild-type plants showed similar root architectural responses to the lack of other nutrients, including nitrate, sulfur, and iron (Fig. 6), and thus, conferring a specific role for PRD in the regulation of root architecture under Pi-starvation conditions. Other typical Pi-starvation responses such as leaf size reduction (Table 1), induction of high-affinity Pi transporters and acid phosphatase gene expression (Fig. 7), and Pi influx, as well as endogenous Pi content were similar in prd mutant and wild-type plants (Supplementary Figs. S2, S3). Overall, these data suggest that the PRD gene does not function as a checkpoint in the cascade of Pi-starvation responses, but acts specifically as a modulator of root growth in response to Pi deprivation. A complex series of signaling cascades that control plant responses to Pi starvation is emerging to include several transcription factors such as AtWRKY75, a WRKY transcription factor (Devaiah et al. 2007), or PHR1, a myb transcription factor (Rubio et al. 2001), but these genes appear to control a wide range of Pi-starvation responses. To our knowledge, PRD is the first identified molecular element that can act specifically on the root architecture in responses to Pi deprivation and does not seems to be involved in other starvation responses (Fig.7; supplementary data). Our results underline the complexity and the diversity of the signaling cascades involved in the responses to Pi privation. Nevertheless, it is important to notice that public microarray database analysis through Genevestigator (https://www.genevestigator.ethz.ch/) show that At1g79700 gene expression was affected in the shoots by different stress suggesting a more general role of PRD in the adaptive responses of the plant to fluctuant environments.
Interestingly, PRD belongs to the AINTEGUMENTA-like (AIL) gene family (Nole-Wilson et al. 2005). AIL genes are mainly expressed in actively growing and developing tissues and may specify meristematic or division-competent states (Nole-Wilson et al. 2005). Several genes of the AIL family have been functionally characterized. APETALA2 (AP2), one of the founding members of the AP2 family, is a floral homeotic gene that specifies sepal and petal identity (Bowman et al. 1991). AINTEGUMENTA (ANT) is required for integument initiation and promotion of growth within developing floral organs (Elliott et al. 1996; Klucher et al. 1996). BABY BOOM (BBM) has been reported to be involved in a variety of critical plant cellular functions and is preferentially expressed in seeds (Boutilier et al. 2002). BBM is likely to promote cell proliferation and morphogenesis during embryogenesis. Recently, a clade of four PLT homologs was found to be required for specification and maintenance of stem cells within the root apical meristem and was necessary for the root formation (Aida et al. 2004; Galinha et al. 2007). On the other hand, all the AIL genes were expressed in multiple plant tissues, and at higher levels in young tissues (seedlings, roots, inflorescences, seeds, siliques) compared to older tissues (mature rosette leaves, stems). These data confirm that AIL genes are important regulators of floral and root development and are consistent with the possible role of AIL genes in the specification of meristem states. In this context, PRD should be considered as a constitutive element necessary for primary and lateral roots growth. PRD should be considered as a root factor functioning as a fine tune to adjust root growth and architecture in response to low external Pi availability. However, even in prd mutants, we did not observe a completely growth-arrested or truncated root system in response to Pi privation but rather a slowing down of root growth, suggesting that PRD is one of the components of the signaling pathway by which Pi starvation modulates primary and lateral roots growth. The lack of drastic phenotype also raised the issue of the functional redundancy. Based on the Needleman-Wunsch global alignment algorithm to find the optimum alignments, PRD gene presents 72% of similarity with At1g16060, another member of the AIL family (Nole-Wilson et al. 2005). These high sequence similarities may imply a partial functional redundancy. Future studies will define at a cellular level the role of PRD gene in cell division and/or cell elongation processes and will focus on dissecting the interacting components involved in the regulation of root architecture.
Abbreviations
- BSA:
-
Bovine serum albumin
- EST:
-
Expressed sequence tag
- PBS:
-
Phosphate buffer saline
- QTL:
-
Quantitative trait locus
- RT-PCR:
-
Real-time PCR
- SSC:
-
Sodium sodium citrate
References
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120
Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TJ, Nacry P, Rossignol M, Tardieu F, Doumas P (2003) Temporal response of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signalling. Plant Cell Environ 26:1053–1066
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, Custers JB, Van Lookeren Campagne MM (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Bowman JL, Drews GN, Meyerowitz EM (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3:749–758
Chen DL, Delatorre CA, Bakker A, Abel S (2000) Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211:13–22
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
El Kassis E, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC (2007) Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiol 143:1231–1241
Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8:155–168
Friml J, Benkova E, Mayer U, Palme K, Muster G (2003) Automated whole mount localisation techniques for plant seedlings. Plant J 34:115–124
Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swamp R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132:578–596
Hinsinger P (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237:173–195
Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8:137–153
Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29:751–760
López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129:244–256
López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L (2005) An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137:681–691
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC (2005) Genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102:11934–11939
Muller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143:156–171
Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138:2061–2074
Nole-Wilson S, Tranby T, Krizek BA (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57:613–628
Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol 50:665–693
Raghothama KG, Karthikeyan AS (2005) Phosphate acquisition. Plant Soil 274:37–49
Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29:115–125
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46:174–184
Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140:879–889
Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124:1477–1480
Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39:792–796
Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37:801–814
Tranbarger TJ, Al-Ghazi Y, Muller B, Doumas P, Touraine B (2003) Transcription factor gene with expression correlated to nitrate-related root plasticity of Arabidopsis thaliana. Plant Cell Environ 26:459–469
Uhde-Stone C, Zinn KE, Ramírez-Yánez M, Li A, Vance CP, Allan DL (2003) Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to P deficiency. Plant Physiol 131:1064–1079
Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Safkata K, Sasaki T, Kishimoto N, Kikuchi S, Yamagishi M, Osaki M (2003) Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 26:1515–1523
Wasaki J, Shimano T, Onishi K, Yonetami R, Yazaki J, Fujii F, Shimbo K, Ishikawa M, Shimatani Z, Nagata Y, Hashimoto A, Ohta T, Sato Y, Miyamoto C, Honda S, Kojima K, Sasaki T, Kishimoto N, Kikuchi S, Osaki M (2006) Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J Exp Bot 57:2049–2059
Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126:875–882
Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132:1260–1271
Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138:2087–2096
Acknowledgments
This work was supported in part by the INRA ECOGENE programme. JJCC and JR received a research fellowship from Consejeria de Educacion y Ciencia (Junta de Andalucía, Spain).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Camacho-Cristóbal, J.J., Rexach, J., Conéjéro, G. et al. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta 228, 511–522 (2008). https://doi.org/10.1007/s00425-008-0754-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0754-9