Abstract
The moss Physcomitrella patens is so far the only plant species in which it is possible for nuclear genes to be modified by homologous recombination at a reasonably efficiency. Here we describe the use of homologous recombination for another moss, Ceratodon purpureus. Our approach is based on the repair of the ptr116 mutant allele. In this mutant, codon 31 of the heme oxygenase gene CpHO1 is mutated to a stop codon. Heme oxygenase is necessary for the conversion of heme to biliverdin, the precursor of the phytochrome chromophore. Thus, in ptr116 the phytochrome-mediated responses of phototropism, chlorophyll accumulation and branching are lost. Protoplast transformation with DNA encoding the wild-type protein resulted in a rescue of 0.8% of regenerated protoplasts. In about half of the analyzed lines, formation of CpHO1 concatemers was observed at the CpHO1 locus, whereas in the other half, the mutant CpHO1 gene was replaced by a single DNA copy. This gene repair led to the exchange of single bases, and thus provides the first demonstration of efficient site-directed mutagenesis in a plant nuclear genome. Our studies also revealed an effective mechanism for gene inactivation in Ceratodon. When wild-type protoplasts were transformed with intact or modified CpHO1 genes, approximately 40% of regenerated protoplasts showed the ptr phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene targeting (GT) through homologous recombination (HR) is a powerful method by which to assign gene functions in an organism. Whereas in prokaryotes, yeast and lately even in mice GT has become a routine technique, HR frequencies in seed plants are low, requiring sophisticated screening strategies for efficient GT (Hanin et al. 2001; Terada et al. 2002; Puchta 2002). For example, in an experiment where positive and negative selection markers were used to target the rice waxy gene, the ratio of homologous to illegitimate recombination events was estimated as approximately 6.5×10−4 (Terada et al. 2002). The leafy moss Physcomitrella patens is as yet the only plant species for which efficient GT of nuclear genes has been established (Schaefer 2002; Puchta 2002). Consequently, Physcomitrella has become a valuable model system for plant molecular biologists over recent years. In the present paper we describe efficient GT for another moss, Ceratodon purpureus.
This species has been used as model system for studies on gravitropism (Lamparter et al. 1998a) and phototropism (Hartmann et al. 1983; Lamparter et al. 1997, 1998b; Esch and Lamparter 1998; Brücker et al. 2000). The genome of Ceratodon has about 240 Mbp (Lamparter et al. 1998b) and is thus significantly smaller than that of Physcomitrella, which has about 500 Mbp (Reski 1999; Rensing et al. 2002). Presumably, Physcomitrella has undergone a recent genome duplication and thus contains more redundant genes than Ceratodon.
For GT studies with Ceratodon, we took advantage of the phenotype of class-1 ptr (phototropism) mutants. Wild-type protonemata show a positive phototropic response, which is controlled by the red-light photoreceptor phytochrome (Hartmann et al. 1983). Other effects such as chlorophyll synthesis (Lamparter et al. 1997) and side-branch initiation (Kagawa et al. 1997) are also mediated by phytochrome. In class-1 mutants, all phytochrome responses are dysfunctional because the synthesis of the phytochrome chromophore is blocked. Phototropism, branching and chlorophyll accumulation can be rescued by biliverdin or phycocyanobilin (PCB) (Kagawa et al. 1997; Lamparter et al. 1997; Brücker et al. 2000). Biliverdin is a precursor of phytochromobilin (PφB), which is the natural phytochrome chromophore of Ceratodon. Based on these rescue studies it is likely that in the class-1 ptr mutants the heme oxygenase (HO) enzyme, which catalyses the conversion of heme into biliverdin, is defective. Indeed, microinjection of expression plasmids with the HO genes HO-1 from rat and AtHO1 from Arabidopsis rescued phototropism and red-light induced chlorophyll accumulation (Brücker et al. 2000). For the present approach we aimed at altering the Ceratodon HO gene by means of HR. For this purpose, an HO gene homolog from the wild type and the class-1 mutant ptr116 was cloned and analyzed, and used for two different transformation studies. HR was performed without the help of a resistance cassette and resulted in an exchange of single nucleotides.
Materials and methods
Strains and culture conditions
The Ceratodon purpureus wild-type wt4 (Hartmann et al. 1983) and the class-1 mutants ptr115, ptr116, ptrp4 and ptr120 (Esch and Lamparter 1998) were used. For propagation and physiological assays, the moss filaments were grown on medium 1b (Hartmann et al. 1983) containing 1.2% agar, at 20°C on vertically oriented Petri dishes. For chromophore feeding experiments, dark-adapted moss filaments were transferred to 1b medium containing 1 μM PCB.
Sequencing of Ceratodon heme oxygenase CpHO1 and construction of plasmids
The following oligonucleotides were used for analytical and preparative PCR:
-
f98, GARGARATGMGITTYGTIGCNATG;
-
f106, GCCTCCTTCTCCCCCTCCTTCG;
-
f108, CACATCGTCACCATCAATCG;
-
f109, GCCTGATGGAATCACATACG;
-
f116, TTGGACTGCACACCCCATACC;
-
f117, CGAGTAGGCTTCACGTGAAC;
-
f118, GGTTCTGGATGATGGCTATGG;
-
f119b, TTATGCTGAGACAATCAGGCG;
-
f122, GCCATGGCTATGGCGGTAGGGGAGC;
-
f123b, CCCCCATGGCTGCTGAGACAATCAGGCGC;
-
f126, GCCATGGCCAGCTCGGTTGCCCC;
-
f155, CCCATTCTGAATCACACTTTTC;
-
f156, CCCAGTAGAGCGCCGACGTCGGCCCAT;
-
g5, CTCGTCAGGCTTAGATGTGC;
-
g9, CAATTGCTGTGAGCAAGGGCGAGGAG;
-
g10, CAATTGCGCTTTACTTGTACAGCTCG;
-
g13, GTAATAATA-ACAGAGTTTGAATCATC;
-
g14, CCCAACCCACTGCCACTG;
-
g18, AAGATGCGTTGT-AGTTTACCTC;
-
mz28, TATCCCGACTCTCTTCTCAA.
All cloning steps were carried out according to standard methods (Sambrook and Russell 2001). Genomic DNA of Ceratodon was purified as described by Rogers and Bendich (1985); cDNA was obtained according to Lessard et al. (1997). For PCR amplification of a Ceratodon HO gene fragment, a degenerate primer f98 was designed based on the conserved peptide sequence EEMRFVAM. The primer was used for 3′-RACE (rapid amplification of cDNA ends) with Ceratodon cDNA. The sequence of the 5′ end was obtained by 5′-RACE (SMART; Clontech, Palo Alto, CA, USA) with the primer f106. The cDNA sequence has 1,722 bp. Primers f117 and f116 were used to PCR-amplify the genomic DNA of CpHO1 (Fig. 1). The 2,522-bp PCR product was cloned into pT-Adv (Clontech), to obtain pG17.
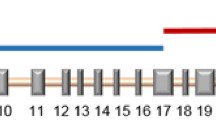
Structure and sequence analysis of the Ceratodon purpureus CpHO1 gene. a Representation of the CpHO1 gene displaying exons, introns and UTRs. The positions of PCR primers g14, g18, f106, f109, f116, f117 (black arrowheads), restriction sites (H=HpaI, M=MfeI, N=NheI), the position of the CpHO1 mutation in ptr116 and the probe (red bar) are indicated. b Enlarged detail of a showing the nucleotide and amino acid sequences of the wild type, ptr116 and the construct used for knock-in transformations. Nucleotides that differ from the wild-type sequence are printed in red letters. The stop codon of ptr116 is given by an asterisk in the protein sequence. c DNA sequences of the same region as in b from the knock-in lines G4 and C1. Red arrows indicate nucleotides that are expected after a gene replacement; the black arrow shows a nucleotide that was not changed
Moss expression plasmids encoding full-length CpHO1 and truncated CpHO1, missing the predicted chloroplast transit peptide (Emanuelsson et al. 1999), were constructed by cloning the CpHO1 cDNA sequence into the expression vector pBAS (Brücker et al. 2000). The resulting plasmids are termed pBASCpHO1 and pBASΔCpHO1, respectively. For the construction of pBASCpHO1, the CpHO1 cDNA was PCR-amplified with primers f118 and f119b, blunted and ligated into the SmaI site of pBAS. The pBASΔCpHO1 plasmid was constructed similarly, using the primer f126 instead of f118. Expression plasmids for green fluorescent protein (GFP) fusion proteins were constructed starting with the vector pBASGFP (Brücker et al. 2000). Full-length and truncated CpHO1 genes were PCR-amplified with the primer pairs f122/f123b and f126/f123b, respectively. Both constructs were cloned into the NcoI site of the GFP expression vector pBASGFP to produce pBASCpHO1GFP and pBASΔCpHO1GFP, respectively.
Four constructs pG18as, pG18s, pG19 and pG22 were made for gene-inactivation studies. Construction of these vectors started with the plasmid pG17. A hygromycin cassette containing an hpt gene driven by the rice actin 1 promoter (McElroy et al. 1990) was cloned into a unique, blunted MfeI site, which lies within the first exon of the CpHO1 gene (Fig. 1a). Blunt-end cloning produced two different plasmids in which the hygromycin cassette interrupts the CpHO1 gene either in sense (pG18s) or antisense (pG18as) direction. To obtain pG19, pG17 was digested with MfeI, blunted and re-ligated. In this way, a frame shift is introduced into the CpHO1 gene such that codon 158 is changed to a stop codon. In pG22, a GFP gene (Brücker et al. 2000) was cloned into the MfeI site of the first exon of the CpHO1 gene in sense orientation using the primers g9 and g10.
For gene repair of the ptr116 mutant, the f117/f116 PCR product of genomic CpHO1 from pG17 was cloned into pBluescript II SK− (Stratagene, La Jolla, CA, USA) to obtain pG20. Silent mutations around the ptr116 mutant site (Fig. 1b) were introduced by “inverse” PCR around the plasmid using the primers f155 and f156. The PCR product was re-circularized to obtain pG23 and correct cloning was confirmed by DNA sequencing.
Microinjection
Microinjection and analysis of the phototropic response were performed as described by Brücker et al. (2000). Before microinjection, the moss filaments were grown in darkness for 6 days to obtain vertically aligned filaments with low chlorophyll content. After injection, the filaments were directly placed in unilateral red light and analyzed 24 h later. The concentration of the injected plasmids was 1.5 μg μl−1. Fluorescence of chlorophyll and GFP was observed with a Zeiss Axioplan 100 fluorescence microscope (filter set BP 450–490; FT 510; LP 520).
Protoplast isolation, transformation and regeneration
Protoplasts were isolated and regenerated as described by Lamparter et al. (1998b). Polyethylene glycol (PEG)-mediated protoplast transformation was performed as described for Physcomitrella (Schaefer and Zrÿd 1997). The DNAs of pG17, pG18as, pG18s, pG19, pBASCpHO1GFP and pBASΔCpHO1GFP were used to transform wild-type protoplasts, and pG23 DNA was used to transform protoplasts of the mutant ptr116. Plasmid DNA used for moss transformations was purified using the Endofree Plasmid Maxi-Kit (Qiagen, Hilden, Germany). Before transformation, the DNA was linearized by an HpaI/NheI double digest (see Fig. 1 for the position of the cleavage sites) and purified by phenol/chloroform extraction. Thus, the excised fragments cover 1,977 bp of the genomic CpHO1 sequence.
After transformation, around 50,000 protoplasts per Petri dish were dispersed on recovery medium (Schaefer and Zrÿd 1997). Initially, protoplasts were kept for 1 day in darkness; thereafter, they were transferred to a 16/8 h light/dark culture chamber. After 2 days, the regenerated protoplasts were transferred to 1b medium and kept in continuous unilateral red light (10 μmol photons m−2 s−1) on vertical Petri dishes. When the wild type was transformed with pG18s, pG18as or pG19, a high number of aphototropic cultures were observed after several days in unilateral red light. For a qualitative phototropic analysis, cultures were kept for 7 days in the dark and subsequently irradiated for 24 h with unilateral red light.
Molecular analysis of the transgenes
Genomic DNA of isolated lines was extracted from young protonemal tissue after 3 weeks growth under standard conditions. For PCR analysis, DNA was purified from 30 to 50 mg protonemata using the Plant DNA Extraction Kit (Qiagen). For Southern blot analysis, DNA was purified from 5 to 10 g moss tissue (Rogers and Bendich 1985). The Southern probe template was a 446-bp DNA segment of the CpHO1 open reading frame (ORF), which lies inside the region used for the construction of the knock-in construct (Fig. 1a). The probe was digoxigenin (DIG) labeled by a PCR-based technique (DIG DNA labeling kit, Roche, Mannheim, Germany) using the primers f108 and f155. Hybridization was carried out for 12 h with DIG Easy Hyb at 42°C and then washed under high stringency at 68°C in 0.2×SSC/0.1% SDS. Thereafter, DIG was detected with the chemiluminescent substrate CDP-Star (Roche). The blots were exposed for 5–60 min to Kodak X-OMAT UV film.
Phylogenetic analysis of heme oxygenases
Amino acid sequences were aligned with ClustalX (Thompson et al. 1997) using default parameters. The tree was constructed with the PHYLIP program package (Felsenstein 2004) using the PROTDIST and FITCH algorithms. For bootstrapping, 100 datasets of sequences were generated; bootstrap values were usually above 90%.
Results
Aphototropic Ceratodon class-1 mutants are defective in the CpHO1 gene To obtain the sequence for Ceratodon HO CpHO1, a degenerated primer f98 was designed that allowed the 3′ part of the cDNA to be amplified by PCR. The missing cDNA sequence was obtained by 5′-RACE. PCR products from genomic DNA were sequenced to obtain the entire coding region of CpHO1. The gene encodes a protein with 305 amino acids including a predicted N-terminal chloroplast target (Emanuelsson et al. 1999) of 80 amino acids. There are 544 bases 5′-UTR and 263 bases 3′-UTR on the cDNA sequence, and the genomic sequence contains 3 introns (Fig. 1a). The overall similarity to Arabidopsis HO AtHO1 (Muramoto et al. 1999) at the protein level is 80%. Phylogenetic analyses were performed with other known HO protein sequences. As expected, CpHO1 is closely related to an HO homolog of the moss Physcomitrella. All land-plant HOs are found together in one group, which is clearly separate from HOs of other organisms (Fig. 2). Other groups are formed by cyanobacteria, proteobacteria, animals and algae. In agreement with another publication (Terry et al. 2002), the plant branch point is rather distant from the cyanobacterial branch point, but closer to the group of proteobacteria.
Phylogenetic tree based on HO protein sequences. Land-plant species are given in green, mammalian sequences in red, cyanobacteria in blue and proteobacteria in grey. The NCBI–gi numbers of the protein sequences are as follows: N7120HO (Nostoc sp. PCC 7120): gi6175555; AgrtuHO (Agrobacterium tumefaciens): gi15157689; ArathHO1 (Arabidopsis thaliana): gi26983816; ArathHO2: gi25370566; ArathHO3: gi14485563; ArathHO4: gi14485565; BraspHO (Bradyrhizobium sp. ORS278): gi11993215; CerpuHO1 (Ceratodon purpureus): gi26189984; CordiHO (Corynebacterium diphtheriae): gi4586514; GlymaHO1 (Glycine max): gi14485567; GlymaHO3: gi14485569; GuithHO (Guillardia theta): gi11467702; LycesHO1 (Lycopersicon esculentum): gi14485575; LycesHO2: gi14485577; N7120HO1 (Nostoc sp. PCC 7120): gi17230617; N7120HO2: gi17229389; NeiflHO (Neisseria flava): gi8163800; NospuHO (Nostoc punctiforme): gi4105611; OrysaHO (Oryza sativa): gi29824473; OrysaHO2: gi14029014; PhypaHO (Physcomitrella patens): gi26190434; PintaHO (Pinus taeda): gi14485579; PorpuHO (Porphyra purpurea): gi11465737; PromaHO2 (Prochlorococcus marinus str. MIT 9313): gi33635536; PromaHO6 (Prochlorococcus marinus subsp. marinus str. CCMP1375) gi33241199; PsesyBphO (Pseudomonas syringae): gi28852345; PsesyHO: gi28851731; RhileHO (Rhizobium leguminosarum): gi16304840; RhospHO (Rhodobacter sphaeroides): gi22960642; RhoviHO (Rhodella violacea): gi2323282; S6803HO1 (Synechocystis sp. PCC 6803): gi16330811; S6803HO2: gi16329456; S7942HO (Synechococcus sp. PCC 7942): gi4105613; SorbiHO1 (Sorghum bicolor): gi14485571; SorbiHO2: gi14485573
To see whether the phenotype of aphototropic class-1 mutants results from a CpHO1 defect, we sequenced the gene of the mutant ptr116. In this line, the codon 31 of CpHO1 has mutated to a stop codon (Fig. 1b). In another class-1 mutant, ptrP14, the CpHO1 gene could not be amplified by PCR, and digested genomic DNA gave no signal on Southern blots (data not shown). We thus assume that the entire gene was lost in ptrP14. These data imply that the phenotype of class-1 mutants results from a defect in the CpHO1 locus and that CpHO1 is the dominating HO in Ceratodon filaments.
Two expression plasmids, pBASCpHO1 and pBASΔCpHO1, were constructed to test for functional complementation. The pBASCpHO1 plasmid encodes the full-length protein, whereas pBASΔCpHO1 encodes a truncated HO lacking the 80 N-terminal amino acids of the putative chloroplast target. The plasmids were microinjected into dark-adapted tip cells of the class-1 mutants ptr115, ptr116, ptr120 (Table 1 and Fig. 3a,b for ptr115). Between 24 and 41% of the injected filaments showed a normal wild-type phototropic response under unilateral red light. The phototropic filaments also had an elevated chlorophyll level, as seen by fluorescence microscopy. These results are comparable to results from previous microinjection experiments in which HO genes of rat and Arabidopsis were used (Brücker et al. 2000), and show that CpHO1 is functional in the Ceratodon cells. The putative chloroplast target was not required for restoration of phototropism and chlorophyll synthesis, but the curvature obtained with pBASΔCpHO1 was slightly lower than the curvature of wild-type cells or cells that had been injected with the full-length version (Table 1). Another difference between the two constructs was found for the onset of chlorophyll synthesis. After injection of pBASCpHO1, the increase in chlorophyll preceded the change of growth direction (Fig. 4b), whereas increased chlorophyll levels were only found in the curved tip cell of the filament after pBASΔCpHO1 injection (Fig. 4a). Both observations suggest that the truncated protein is less active with respect to biliverdin production.
Phenotypical analysis of transformed moss filaments. a,b Microinjection of CpHO1 genes into tip cells of Ceratodon protonemata. Bright-field view (a) and chlorophyll autofluorescence (b) after injection of pBASCpHO1 into the aphototropic mutant ptr115. After the injection, filaments were irradiated for 48 h with red light from the right (except in e). The tip cell of the right filament did not survive the injection. c,d GFP fluorescence and chlorophyll auto fluorescence after injection of pBASCpHO1GFP (c) and pBAS.CpHO1GFP (d) into wild-type tip cells. e Regenerated wild-type filaments after transformation with pG18as. Filaments were irradiated for 3 days with red light from behind the Petri dish. Note the high number of colonies with pale filaments growing negatively gravitropically. f,g Regenerated ptr116 filaments after transformation with pG23. Filaments were either irradiated for 7 days (f) or 4 weeks (g) with red light. The vertical lines are unmodified, pale ptr116 filaments. h–j Phototropic response of wild-type (h) ptr116 (i) and the knock-in line G4 (j) after 24 h red light from the right. Bars = 50 μm (a,b), 10 μm (c,d), 500 μm (e), 200 μm (f), 1,500 μm (g), 300 μm (h–j)
Microinjection of HO expression plasmids in tip cells of class-1 ptr mutants of Ceratodon. a pBASΔCpHO1 was injected into ptr115; fluorescence image. Note that the chlorophyll increase (red fluorescence) in the apex of the tip cell succeeds the phototropic response. b,c Injection of pBASΔCpHO1GFP into ptr120. Bright-field (b) and fluorescence (c) images. Note that GFP accumulates in the cytoplasm and the nucleus. d Injection of pBASCpHO1GFP into ptr120 tip cells; analysis as in c. GFP is detectable only in the chloroplasts. Filaments were assayed after 24 h (a), 48 h (b,c) and 36 h (d) red light irradiation from the right. Bars = 50 μm (a–c), 25 μm (d)
To test for intracellular localization, we constructed the plasmids pBASCpHO1GFP and pBASΔCpHO1GFP encoding for full-length and truncated HO–GFP fusion proteins, respectively. Both plasmids were injected into dark-adapted wild-type cells. Full-length CpHO1–GFP fusion was only detected in the chloroplasts: blue-light excitation induces yellow/green GFP fluorescence and red chlorophyll fluorescence. Therefore, the chloroplasts of injected cells appear orange (Fig. 3c), whereas chloroplasts of non-injected cells appear red. The truncated CpHO1–GFP fusion was not translocated to the chloroplasts but found in the cytosol and in the nucleus (Fig. 3d). A similar intracellular distribution was found with GFP alone (Brücker et al. 2000). When CpHO1–GFP fusion constructs were injected into ptr115, ptr116 or ptr120 tip cells, a similar rescue of phototropism was obtained as with the GFP-free constructs (Fig. 4b–d).
In the following step, we tested whether the phenotype of the class-1 mutants can be induced by transforming wild-type cells with an interrupted or modified CpHO1 gene. This strategy was considered to test for GT by HR. Linearized DNA of the plasmids pG18s, pG18as and pG19 was used for protoplast transformation. For linearization, the plasmids were double-digested with HpaI and NheI, which cleave upstream of the start codon and within the third intron of the CpHO1 gene, respectively (Fig. 1a). In pG18s and pG18as, a hygromycin cassette, which is oriented in sense or antisense direction, respectively, interrupts the CpHO1 gene, whereas in pG19 the codon 158 is mutated to a stop codon. After transformation, regenerated protoplasts were screened for HO-deficiency in continuous unilateral red light. Under these conditions, wild-type filaments grow towards the light source and appear green, whereas class-1 ptr mutants grow upwards and appear pale. With all three constructs, an unexpectedly high rate of approximately 40% of regenerated protoplasts had the ptr phenotype (Fig. 3e for pG18as and Table 2). The phenotype was stable during a period of more than 12 months. The high frequency of aphototropic filaments is clearly a result of the modified HO gene. In control experiments, aphototropic filaments were never obtained when regenerating Ceratodon protoplasts were directly assayed for phototropism or when the GFP plasmid was used for transformation (Table 2).
Although the phenotype of the aphototropic lines could result from GT by HR, the very high frequency of these filaments speaks against this possibility. In GT experiments with the moss Physcomitrella, the frequency of homologous recombinants was between 0.01 and 0.6% of regenerated filaments (Schaefer 2001). We screened 57 aphototropic lines from the pG18as transformation and 10 lines from the pG18s transformation by PCR, using the primers g14 and g18. Both primers bind to regions on the CpHO1 gene that lie outside the DNA sequence used for the construction of the transformation vectors (Fig. 1a). With the wild type, a 2,522-bp PCR product is expected, whereas an HR event should result in a 5.2-kb product. The PCR products of all lines had a size of approximately 2.5 kb and were indistinguishable from the product obtained from the wild type (data not shown). Thus, in none of the 67 lines did the phenotype result from an HR event.
We performed further transformation experiments with linearized pG17, which contains the intact CpHO1 gene, and with non-digested expression plasmids pBASCpHO1GFP and pBASΔCpHO1GFP, which had been used for the above microinjection experiments. High rates of around 40% and 20% aphototropic filaments were obtained with the linearized pG17 and the circular expression constructs, respectively (Table 2). These experiments show that the inhibitory effect is also found with intact CpHO1 genes, and that it is neither dependent on the DNA form (linear vs. circular) nor on the presence or position of a bona fide promoter element within the transformed DNA.
Phototropism of HO-deficient mutants can be rescued with PCB (Lamparter et al. 1997). We tested three arbitrarily selected aphototropic lines from each transformation experiment for PCB rescue. After 24 h in red light, filaments of all lines showed a clear phototropic response, similar to the response of the wild type or the ptr116 mutant on PCB. This indicates that the phenotype of these lines was indeed caused by chromophore deficiency.
Transforming protoplasts of the class-1 mutant ptr116 with modified wild-type CpHO1
In an independent, “knock-in” approach we aimed to repair the phenotype of the mutant ptr116 by transformation with a functional CpHO1 gene. For that purpose, ptr116 protoplasts were transformed with the linearized plasmid pG23. The CpHO1 gene of this plasmid is derived from the wild type and thus contains the correct CAG codon 31, whereas the ptr116 gene has a TAG stop codon at this position. Additionally pG23 contains two silent mutations in codons 34 and 36 (Fig. 1b), which results in an alteration of the restriction pattern. In this way, lines that originate from a gene-replacement event will be distinguishable from ptr116 and from the wild type by restriction digestion of PCR products or by Southern blotting.
In an experiment with approximately 600,000 protoplasts, approximately 12,000 were regenerated after the transformation procedure, 93 (0.8%) of which showed the wild-type phototropic phenotype. The altered phenotype became easily visible after 7 days in unilateral red light by the phototropic growth direction and the greenish color (Fig. 3f). Prolonged growth increased the difference between the rescued lines and the bulk of aphototropic filaments (Fig. 3g). For physiological and molecular analyses, 10 lines were arbitrarily selected. Phototropism was analyzed with filaments that had been grown in darkness for 7 days. The curvature of all lines after a 48 h treatment with unilateral red light was indistinguishable from that of the wild type (Fig. 3h–j). To see whether the phenotype of the rescued lines results from a gene-replacement event, we amplified the CpHO1 sequence with the g14/g18 primer pair by PCR. The g14/g18 product was only obtained with five of the ten analyzed lines (Table 3), although other primer combinations with one primer outside and the other inside the transformed sequence gave products with all analyzed lines (data not shown). The missing g14/g18 product might be explained by an insertion of concatemers, as frequently observed in Physcomitrella GT experiments (Strepp et al. 1998; Hiwatashi et al. 2001; Schaefer 2002).
We checked for concatemer formation by PCR using the primer pair f106/f109. These primers point in opposing directions on the CpHO1 gene and will only produce PCR products with two or multiple adjacent CpHO1 copies. DNA from eight out of ten lines gave one or more f106/f109 products of variable size (Table 3, Fig. 5). Based on the PCR results we assume that three different recombination events have occurred in the rescued lines: in one group (A1, A3, B1, H1, I1), HR resulted in the insertion of concatemers at the CpHO1 locus. In the second group (A2, F1, H4), the CpHO1 locus was repaired by insertion of a single copy, but concatemers were probably introduced at other sites of the genome or inherited extrachromosomally. The third kind of recombination was obtained for C1 and G4: in these lines, the CpHO1 locus was replaced by a single insertion and no additional copies are present in the genome.
PCR analysis with the primers f106/f109 of Ceratodon wild type (wt4), ptr116 and the knock-in lines A1, A2, A3, B1, C1, F1, C1, G4, H1 and H4. This PCR shows that all knock-in lines, except C1 and G4, contain concatemers of the pG23 construct, which were either introduced into the genome or persist as extrachromosomal structures. The signal below 1 kb is unspecific. We have also obtained this band under various PCR conditions solely with primer f106 in the wild type and the other analyzed lines (data not shown). M Markers
The g14/g18 PCR products of the five rescued lines, ptr116 and wild type were tested for the presence or absence of the HinfI, ApaI and NarI restriction sites. These digests allow CpHO1 sequences of the wild type, ptr116 and the pG23 plasmid used for transformation to be distinguished (Fig. 1a). It should be noted that the g14/g18 PCR product contains seven cleavage sites for the four-base cutter HinfI. As expected, the wild-type PCR product was cleaved at all HinfI sites and by NarI, but not by ApaI, and the ptr116 product was digested by NarI and ApaI, but not at the specific HinfI site mutated in this line. If the HO gene of ptr116 is repaired by the pG23 sequence, the PCR product should contain this HinfI site and should be cleaved by ApaI, but not by NarI. This pattern was obtained in four out of the five rescued lines. In the fifth line, C1, the specific ApaI site was absent (Table 3, Fig. 6).
Restriction analysis of the 2,522-bp g14/g18 PCR product from Ceratodon wild type wt4, ptr116 and knock-in lines A2, C1, F1, G4 and H4. A1, A3, B1, F1 and H1 gave no g14/g18 PCR product. All analyzed knock-in lines, except for the ApaI digest of C1 (indicated by an asterisk on the gel image), show the restriction pattern of pG23, which is expected after gene replacement by homologous recombination. M Markers
The restriction pattern was confirmed for the two lines C1 and G4 by sequencing of the CpHO1 locus. For that purpose, g14/g18 PCR products were ligated into a cloning vector. As expected, the sequence from G4 was identical to the knock-in construct (Fig. 1c), confirming that the CpHO1 gene of ptr116 had been repaired by HR. The sequence from C1 shows that the HO gene of this line also codes for a wild-type protein but that the ApaI site is indeed missing (Fig. 1c). This result implies that during the repair of the CpHO1 gene of C1, multiple crossovers occurred.
Southern blot analysis was performed with genomic DNA from the wild type, ptr116 and the knock-in lines A2 and G4 after double digestion with ApaI/EcoRV and ApaI/HindIII. With the wild type and with ptr116, both enzyme combinations gave a single band around 5.5 kb. The DNA from G4 gave a 500-bp band after the ApaI/HindIII digest, and a signal above 2 kb after the ApaI/EcoRV digest (Fig. 7). This result also shows that the specific ApaI cleavage site was introduced into G4 by HR. The same bands were also found with DNA from A2, which indicates that the same rearrangement has occurred at the CpHO1 locus. However, DNA from A2 gave an additional, rather strong signal in the range above 5 kb. Thus, the A2 genome contains additional copies of the introduced DNA, as already suggested above by PCR results with the f106/f109 primers.
Southern blot of ApaI/HindIII-digested (a) and ApaI/EcoRV-digested (b) Ceratodon genomic DNA. After site-directed mutagenesis of the ptr locus by HR a smaller signal is expected in targeted lines than in the wild type because of the newly generated ApaI site (Fig. 1b). The positions of the HindIII and EcoRV restriction sites are not known. M Markers
Discussion
In the present study we cloned an HO gene from Ceratodon, showed that overexpression of the corresponding cDNA rescues the aphototropic ptr mutant phenotype after microinjection (Fig. 3a,b), and used this strong phenotype in loss- and gain-of-function experiments. All land-plant HOs form one group of rather closely related isofunctional homologs, which is separate from HOs of other species. As for other land-plant HOs (Terry et al. 2002), the CpHO1 protein contains an N-terminal chloroplast target sequence and is transported to the plastid (Muramoto et al. 1999). The intracellular localization suggests that plant HOs are derived from the cyanobacterial endosymbiont that gave rise to plant plastids, and that the gene was transferred from the plastid to the nucleus and subsequently eukaryotized. However, phylogenetic analysis performed by us (Fig. 2) and others (Terry et al. 2002) supports the theory that plant HOs have been inherited from an ancient proteobacterium, probably via the mitochondrial endosymbiont. Our finding that HO in Ceratodon can function outside the plastid shows that during early plant evolution HOs might have been located in the cytosol or mitochondrion as well. Specific or non-specific transport mechanisms must exist that shuttle heme and biliverdin through the plastid membranes. The ferredoxin cofactor that is required for HO activity (Terry et al. 2002) must also be present in the cytosol. Since the cytosolic version of CpHO1 was less active with respect to the rescue of phytochrome function, the advantage of a plastidic version seems clear. This advantage might have produced evolutionary pressure towards plastid localization.
Two different strategies for investigating gene function were tested in this work. Both were aimed to establish HR for Ceratodon. In the first strategy, which is based on a loss-of-function screen, wild-type cells were transformed with an interrupted, mutated or unmodified CpHO1 gene. In all cases, the rate of regenerated protoplasts with an HO-minus phenotype was unexpectedly high. In all 67 aphototropic lines that were tested by PCR, a wild-type signal was obtained. Thus, in the tested lines, the effect did not result from an HR event. The high frequency of aphototropic lines also speaks against this possibility. The inhibitory effect might be explained by DNA silencing or an indirect RNAi effect, caused by RNA transcribed from the introduced DNA template. Gene inactivation by RNAi has become a general technique, which has also been extended to the moss Physcomitrella (Bezanilla et al. 2003). Six of our transformation constructs contained the strong rice actin promoter (McElroy et al. 1990) as a part of the hygromycin cassette or expression plasmid. The other two constructs carried no defined promoter sequences. Because the rate of inactivation was comparable in all cases, we assume that the inhibitory effect is not based on an RNAi effect, although we recognize that the presence of covert promoter sequences in the homologous fragments might lead to an RNAi effect. Since the inhibition was clearly mediated by the introduced homologous DNA, our results demonstrate that specific gene silencing is highly effective in Ceratodon. Thus, rapid loss-of-function tests can be performed with any gene, especially if the phenotype can be observed at the cellular level after protoplast regeneration.
The second strategy was based on a gain-of-function screen by repair of the defective HO gene of the ptr116 mutant. In this approach, the frequency of regenerated protoplasts with an HO-positive phenotype was low, compared to the high rate of HO-minus lines in the above experiments. PCR-based analyses showed that in ten out of ten tested lines in which the PTR phenotype had been rescued, the HO locus of ptr116 was altered by HR. In five lines, the HO gene of ptr116 was replaced by a single copy of the introduced DNA, which could be PCR-amplified by the external primer pair g14/g18. However, PCR with the primers f106/f109 in the other five lines implied that in these lines the endogenous gene was replaced by concatemers formed from the introduced DNA (Strepp et al. 1998; Hiwatashi et al. 2001; Schaefer 2002). Since the same primer combination also produced PCR fragments from three of the five lines of the first group, we conclude that these lines also contain concatemers of introduced DNA, either incorporated into the genome by illegitimate recombination, or inherited as extrachromosomal DNA (Ashton et al. 2000). Gene replacement was confirmed for the first five lines by restriction digestion of the PCR fragments, and for two of these lines by Southern blotting. The restriction pattern of one line, C1, was unexpected but could be confirmed by sequencing of the CpHO1 gene of that line; the gene sequence of another line, G4, was also confirmed by sequencing (Fig. 1c). The C1 line is particularly interesting as it implies that multiple crossovers between homologous sequences occur. We note, however, that in the homologous region of pG23, three mismatches are present (Fig. 1b). HR events like this might provide a valuable insight into the underlying mechanisms.
Since for our knock-in experiment, it was not necessary to clone a resistance cassette into the introduced DNA, the gene repair might be termed as site-directed mutagenesis of ptr116 (ho1). Our knock-in experiments thus provide an example of efficient site-directed mutagenesis in a plant nuclear genome. In seed plants, site-specific point mutation based on the action of chimeric RNA/DNA oligonucleotides has been described in tobacco (Nicotiana tabacum; Beetham et al. 1999; Kochevenko and Willmitzer 2003) and maize (Zea mays; Zhu et al. 1999). However, after a pre-selection, the overall frequency of site-specific targeting in these studies was around 10−4, and thus much lower than in our approach. Moreover, the specificity of the targeting in these studies was not as precise as had been hoped. The chimeric oligonucleotides induced mutations not only at the specific mismatch but also elsewhere within the DNA stretch complementary to the target. Based on our results, pooled genomic DNA from numerous transformed Ceratodon lines could be used as starting material for PCR-based screenings for site-directed mutagenesis. With such an approach it should be possible to identify clones with any single nucleotide gene modification within a realistic time frame. Moreover, as our experiments imply a high rate of HR in Ceratodon, comparable to that in Physcomitrella, this is likely to be a general characteristic of mosses.
Abbreviations
- GFP :
-
Green fluorescent protein
- GT :
-
Gene targeting
- HO :
-
Heme oxygenase
- HR :
-
Homologous recombination
- PCB :
-
Phycocyanobilin
- ptr :
-
Phototropism
References
Ashton NW, Champagne CEM, Weiler T, Verkoczy LK (2000) The bryophyte Physcomitrella patens replicates extrachromosomal transgenic elements. New Phytol 146:391–402
Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD (1999) A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc Natl Acad Sci USA 96:8774–8778
Bezanilla M, Pan A, Quatrano RS (2003) RNA interference in the moss Physcomitrella patens. Plant Physiol 133:470–474
Brücker G, Zeidler M, Kohchi T, Hartmann E, Lamparter T (2000) Microinjection of heme oxygenase genes rescues phytochrome-chromophore-deficient mutants of the moss Ceratodon purpureus. Planta 210:529–535
Emanuelsson O, Nielsen H, von Heije G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Esch H, Lamparter T (1998) Light regulation of phytochrome content in wild-type and aphototropic mutants of the moss Ceratodon purpureus. Photochem Photobiol 67:450–455
Felsenstein J (2004) PHYLIP: phylogeny inference package 3.6 (computer program distributed by the author). Department of Genetics, University of Washington, Seattle
Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J (2001) Gene targeting in Arabidopsis. Plant J 28:671–677
Hartmann E, Klingenberg B, Bauer L (1983) Phytochrome mediated phototropism in protonemata of the moss Ceratodon purpureus BRID. Photochem Photobiol 38:599–603
Hiwatashi Y, Nishiyama T, Fujita T, Hasebe M (2001) Establishment of gene-trap and enhancer-trap systems in the moss Physcomitrella patens. Plant J 28:105–116
Kagawa T, Lamparter T, Hartmann E, Wada M (1997) Phytochrome-mediated branch formation in protonemata of the moss Ceratodon purpureus. J Plant Res 110:363–370
Kochevenko A, Willmitzer L (2003) Chimeric RNA/DNA oligonucleotide-based site-specific modification of the tobacco acetolactate synthase gene. Plant Physiol 132:174–184
Lamparter T, Esch H, Cove D, Hartmann E (1997) Phytochrome control of phototropism and chlorophyll accumulation in the apical cells of protonemal filaments of wildtype and an aphototropic mutant of the moss Ceratodon purpureus. Plant Cell Physiol 38:51–58
Lamparter T, Hughes J, Hartmann E (1998a) Blue light- and genetically-reversed gravitropic response in protonemata of the moss Ceratodon purpureus. Planta 206:95–102
Lamparter T, Brücker G, Esch H, Hughes J, Meister A, Hartmann E (1998b) Somatic hybridisation with aphototropic mutants of the moss Ceratodon purpureus: genome size, phytochrome photoreversibility, tip-cell phototropism and chlorophyll regulation. J Plant Physiol 153:394–400
Lessard P, Decroocq V, Thomas M (1997) Isolation and analysis of mRNA from plant cells: cloning of cDNAs. In: Clark MS (ed) Plant molecular biology: a laboratory manual, 1st edn. Springer, Berlin Heidelberg New York, pp 154–201
McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11:335–348
Puchta H (2002) Gene replacement by homologous recombination in plants. Plant Mol Biol 48:173–182
Rensing SA, Rombauts S, Van de PY, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7:535–538
Reski R (1999) Molecular genetics of Physcomitrella. Planta 208:301–309
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schaefer DG (2001) Gene targeting in Physcomitrella patens. Curr Opin Plant Biol 4:143–150
Schaefer DG (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53:477–501
Schaefer DG, Zrÿd J-P (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11:1195–1206
Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95:4368–4373
Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20:1030–1034
Terry MJ, Linley PJ, Kohchi T (2002) Making light of it: the role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem Soc Trans 30:604–609
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Zhu T, Peterson DJ, Tagliani L, St. Clair G, Baszynski C, Bowen B (1999) Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci USA 96:8768–8773
Acknowledgements
G. Brücker and F. Mittmann contributed equally to this work. We thank Dr. Jon Hughes (JLU Giessen) for helpful discussions and critical reading of the manuscript and Viola Eckl for technical assistance. The DNA sequence of CpHO1 is deposited in GenBank under accession number AJ489940. This work was partly supported by the Deutsche Forschungsgemeinschaft (La799/7-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brücker, G., Mittmann, F., Hartmann, E. et al. Targeted site-directed mutagenesis of a heme oxygenase locus by gene replacement in the moss Ceratodon purpureus. Planta 220, 864–874 (2005). https://doi.org/10.1007/s00425-004-1411-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1411-6