Abstract
microRNAs (miRNAs) were discovered nearly two decades ago by researchers who sought to understand how basic developmental mechanisms work in the nematode Caenorhabditis elegans. Since the identification of conserved miRNA families in higher eukaryotes, there has been an explosion of interest into how these tiny RNA molecules function. miRNAs are 20–24 nucleotide non-coding RNA molecules that predominantly regulate transcripts of target genes through translational inhibition. Much recent interest has focused on the influence of miRNAs on homeostatic regulation, and in particular, hypoxic responses. The ability to sense and respond to hypoxia is of fundamental importance to aerobic organisms and dysregulated oxygen homeostasis is a hallmark in the pathophysiology of cancer, neurological dysfunction, myocardial infarction, and lung disease. miRNAs are ideal mediators of hypoxic stress responses as they are able to modify gene expression both rapidly and reversibly. This enables miRNA-mediated gene regulatory circuits to modify metabolic networks with immaculate precision and control. Therefore, one may consider miRNAs as molecular rheostats which effect tuning and switching of regulatory circuits to facilitate survival and adaptation to hypoxic conditions. Such miRNA-mediated regulatory circuits would provide flexible and conditional alternatives to “conventional” transcriptional regulation. Here, I review recent discoveries that have boosted our understanding of miRNA regulation of hypoxia and discuss where future breakthroughs in this area may be made.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
microRNAs
microRNAs (miRNAs) are a class of endogenous 20–24 nucleotide non-protein-coding RNAs that regulate eukaryotic gene expression at the post-transcriptional level [1]. miRNAs are among the more abundant gene regulatory molecules, consisting of ~1% of the predicted genes in animal cells; and it is estimated that more than 30% of all messenger RNA (mRNA) transcripts are regulated by miRNAs [29]. The first miRNAs to be discovered, lin-4 and let-7, were identified in forward genetic screens for mutants with defective developmental timing in Caenorhabditis elegans [46, 56, 60, 78]. These heterochronic genes are required for the correct timing of cell cycle progression and terminal differentiation of the seam cells during worm larval development. In the ensuing years, the diversity of miRNA functions has expanded to many aspects of development and disease such as the control of cell death, neuronal patterning, flower development, metabolism, and oncogenesis [6, 14, 18, 35, 36, 52, 53, 76, 77]. miRNAs predominantly regulate gene expression through repression of target genes by reducing mRNA stability and/or inhibiting translation. The miRNA profile of each cell may be distinct to enable the customization of protein levels depending on the requirements of a cell. In some cases, miRNAs may lower specific proteins to undetectable levels, and in others, miRNAs may adjust the dosage of gene expression depending on their developmental or environmental state. Therefore, miRNAs append an extensive tier of gene regulatory control that integrates transcriptional and other regulatory processes to expand the complexity and adaptability of gene regulation in animals and plants.
Since their discovery, relatively few functions have been attributed to miRNAs. In fact, a survey of miRNA genetic mutants in C. elegans found that the majority of miRNAs are dispensable in normal laboratory conditions [54]. This work suggests that miRNAs have (1) discrete cellular functions, such as for lsy-6 which regulates left/right patterning of the ASE pair of neurons in the C. elegans chemosensory system [35] or (2) act as buffers to provide robustness to genetic networks when subjected to genetic or environment perturbation, such as miR-7 during sensory organ development in Drosophila [51].
miRNA biogenesis
Most miRNAs are transcribed by RNA polymerase II into primary transcripts called primary precursor miRNAs (pri-miRNAs), which are transcribed as autonomous genes or as introns of protein-coding genes (Fig. 1) [49]. pri-miRNAs fold into hairpin structures that are cleaved by the Microprocessor complex, which contains the RNase III enzyme Drosha, into pre-miRNA (miRNA precursor) molecules of ~70 nucleotides that are transported into the cytoplasm by Exportin 5 [47, 48, 80]. For some particular miRNAs that reside within introns, splicing bypasses the Microprocessor requirement and directly produces pre-miRNAs [62]. pre-miRNAs act as substrates for a second RNase III enzyme called Dicer that further processes them into 20–24-bp imperfect miRNA/miRNA* duplexes [33]. Within miRNA duplexes, the strand with the weakest 5′-end base pairing is selected as the mature miRNA and the miRNA* is either degraded or recognizes a different group of target transcripts for regulation of gene expression [39, 64]. The mature miRNA is loaded onto an Argonaute (Ago) protein to form miRNA-induced silencing complexes (miRISCs) [42]. miRISCs are then directed to target mRNAs and induce translational repression or mRNA degradation. Imperfect miRNA::mRNA base pairing favors translational repression, which is the principal mode of regulation in metazoa [7], whereas plant miRNAs predominantly cause cleavage of target transcripts due to near-perfect complementarity [7].
miRNA biogenesis. miRNAs are processed from RNA polymerase II transcripts. Such transcripts may arise from independent genetic loci (canonical pathway) or from introns of protein-coding genes (non-canonical pathway). Canonical miRNA primary precursors (pri-miRNAs) are processed by the RNase III family members Drosha and Dicer. Non-canonical transcripts bypass the Drosha processing step. Following Dicer processing, the mature miRNA strand (red) is incorporated into a miRNA-induced silencing complex (miRISC) whereas the miRNA* strand (black) is degraded. miRISCs are guided to the 3′ UTR of target mRNA transcripts where they induce translational repression or mRNA degradation
miRNAs in metazoa target mRNA transcripts via imperfect base pairing to multiple sites in 3′ untranslated regions (UTRs) [2]. An important determinant for the miRNA::mRNA interface is Watson-Crick base pairing to nucleotides 2–7 of the 5′ end or “seed” region of miRNAs [2]. miRNAs and their target mRNA 3′ UTRs form imperfect hybrids that harbor central bulges between nucleotides 9–12 which enable inhibition of translation. The 3′ ends of miRNAs are thought to be less important than the 5′ end but may contribute to target recognition when seed matches are weak. miRISCs may inhibit translation through a variety of known and unknown mechanisms. miRISC-associated Ago2 can prevent ribosome assembly through binding to the anti-association factor eIF6 [16]. In addition, miRISC can block translation initiation at the mRNA-cap-recognition step by competing with the cytoplasmic cap-binding protein eIF4E [32, 40].
miRNA target identification
One of the major challenges of miRNA research is the identification of miRNA targets. Since miRNAs predominantly regulate gene expression through imperfect matches in their target mRNA 3′ UTR regions [2], computational prediction programs identify hundreds of potential targets, many of which may be artifacts. Therefore, the identification of miRNA targets requires a number of parallel approaches to be undertaken: genetics, proteomics, whole genome sequencing, and bioinformatics. If a miRNA of interest has a deletion mutant available (such as for most C. elegans miRNAs), genetics may be used to identify miRNA targets via suppressor screening approaches, provided that a suppressible phenotype is detected. This is in fact how the let-7 target lin-41 was identified [71]. However, if the miRNA target gene is essential or if the miRNA regulates multiple targets, this may not be a suitable strategy. To circumvent these issues, biochemical and next-generation sequencing technologies are now being used to identify miRNA targets on a genome-wide scale [17, 65, 82]. miRNAs mediate post-transcriptional regulation of gene expression through miRISCs, and the physical interaction between mRNAs and miRISC can be exploited to pull-down miRNA targets [82]. Immunoprecipitation of miRISC-associated proteins will purify ribonucleoprotein complexes that contain miRNAs and their mRNA targets. Immunoprecipitated miRNAs and mRNAs from control and miRNA mutant or a conditioned sample are then analyzed by high-throughput sequencing to identify potential miRNA targets. Recent innovative work has also enabled the identification of miRNA targets by directly measuring changes in cellular protein synthesis using the stable isotope labeling with amino acids in cell culture technique [65]. This technique yields a genome-wide profile of protein synthesis in response to miRNA knockdown or overexpression and enables the measurement of changes in protein synthesis shortly after induction of changes in miRNA expression.
Hypoxia
Sensing and responding to fluctuations in the environment are an essential requirement for all organisms. Mechanisms are required to restore homeostasis or to implement a novel genetic program to enable long-term adaptation to a new environment. Maintaining oxygen homeostasis is a fundamental prerequisite for all aerobic organisms. Consequently, organisms and cells have developed adaptive mechanisms to survive hypoxic insults. The major effector of the hypoxic response in metazoa is the hypoxia-inducible factor (HIF-1α) [66]. This bHLH–PAS transcription factor regulates the expression of target genes to increase anaerobic metabolism within cells and enhance vascularization of hypoxic tissues [34, 59]. HIF-1α mRNA is detected in all human, rat, and mouse organs; however, in normoxic conditions, the half-life of HIF-1α protein is less than 5 min due to its efficient degradation by a conserved proteasomal degradation pathway [21]. This degradation pathway is initiated by hydroxylation of a specific proline residue in the conserved LXXLAP motif of HIF-1α by members of the EGL-9 oxygen-dependent prolyl 4-hydroxylase superfamily [4]. The VHL-1 (von Hippel-Lindau tumor suppressor protein) E3 ubiquitin ligase complex recognizes the hydroxylated proline and targets HIF-1α for ubiquitin-mediated proteasomal degradation [4]. Hypoxic conditions interfere with hydroxylation of HIF-1α, thereby stabilizing the protein. Stabilized HIF-1α subsequently translocates to the nucleus where it forms a heterodimer with the constitutively expressed HIF-1β protein [75]. The HIF-1 complex then functions to transcriptionally regulate the expression of a host of target genes (>100), including growth and survival factors, extracellular matrix proteins, and modifying enzymes, cytoskeletal proteins, proapoptotic proteins, glucose transporters, glycolytic enzymes, and transcription factors [66, 67]. HIF-1 regulates its targets through a conserved cis-regulatory motif called the hypoxia-response element (HRE), which contains the core HIF-1 binding site of 5′-(A/G)CGTG-3′ [68]. In mammalian systems, the battery of genes regulated by HIF-1 differs between cell types, as HIF-1 can act as an activator or repressor in a cell type-specific manner [37]. This suggests that specific cells respond to a build up of nuclear HIF-1α in different ways, presumably dependent on the prior programming fate of the cell. Such programming would be contingent on the presence or absence of HIF-1 co-activators/co-repressors or other potential transcriptional and post-transcriptional regulators of HIF-1α target genes.
Recent studies indicate that in addition to the canonical HIF-1 pathway, further levels of regulation exist to control hypoxic responses. In particular, it was revealed that miRNAs play important roles in hypoxic adaptation [44]. It is proposed that miRNAs may act to regulate the expression of (1) genes normally required in a normoxic environment but must be inhibited in hypoxic conditions and (2) genetic factors that provide alternative metabolic pathways or other advantages in hypoxia. miRNAs may be regulated in an HIF-1-dependent or -independent manner and they may act to repress the expression of HIF-1α, HIF-1β, or a plethora of possible downstream targets to effect hypoxic responses (Fig. 2).
The HIF-1 transcriptional pathway monitors internal oxygen levels and regulates metabolic networks, angiogenesis, neuronal development, cell survival and proliferation and sensory capacity [11, 13, 14, 31, 57, 58]. So, why would cells and tissues also require miRNA-mediated mechanisms to orchestrate HIF-1-dependent and -independent gene regulatory circuits to combat hypoxia? Hypoxic and metabolic stress can be an acute, localized, and transient phenomena and miRNA-mediated regulation has advantages over transcriptional regulation in each of these respects. miRNAs can be generated rapidly due to their small size and non-coding nature, they may be partitioned in cell compartments, and their repressive effect is rapidly reversible which would enable translation of inhibited mRNAs to recommence at the flick of a switch. Therefore, miRNAs may act as rheostats, where they allow cells to quickly and efficiently turn off or dampen protein expression of target genes in hypoxia. Alternatively, miRNAs may initiate new gene expression programs to enable adaptation to long-term hypoxic stress. Previously, the functional roles of miRNAs in hypoxic adaptation were poorly understood; however, recent studies have implicated miRNAs in the control of mitochondrial metabolism, hypoxic responses in tumor cells, and DNA damage responses [13–15, 19, 31, 79]. The importance of better understanding regulatory mechanisms involved in hypoxic responses is crucial, as hypoxia plays a key role in the pathophysiology of cancer, neurological dysfunction, myocardial infarction, and lung disease.
Hypoxia-regulated miRNAs
Control of metabolism
During the past 5 years, various studies have determined miRNA expression profiles, or signatures, from a variety of different organisms, cell types, and disease states in relation to hypoxia [8, 22, 30, 43]. These studies have described more than 90 hypoxia-regulated miRNAs (HRMs); however, many are only regulated in certain cellular contexts. The lack of consistent HRM profiles is likely due to differences in detection methods, the period and severity of oxygen deprivation, and the cellular and organismal context. However, all these studies have identified miR-210 as a HRM that is robustly regulated by hypoxia in all cell types. miR-210 expression is upregulated in a dose-dependent manner when cells are incubated in decreasing levels of oxygen but is not dysregulated when cells are exposed to osmotic stress, low pH, or growth factor deprivation [14, 22]. Genetic and biochemical studies have also now confirmed that miR-210 is directly regulated by HIF-1 via a HRE in the miR-210 promoter [19, 31]. In addition, miR-210 is required for cell survival under a hypoxic microenvironment [14]. How does miR-210 promote hypoxic survival? The identification of miR-210 targets has revealed some interesting insights. Two identified targets of miR-210 are required for the mitochondrial electron transport chain and the tricarboxylic acid cycle to function correctly [14, 15]. The iron–sulfur cluster scaffold homolog and cytochrome c oxidase assembly factor (COX10) proteins are both targeted by miR-210 and their downregulation represses mitochondrial respiration. In normoxia, such repression of mitochondrial respiration would lead to decreased adenotriphosphate (ATP) levels; however, in hypoxia, repression of electron transport balances the reduced oxygen tension and increases ATP levels via enhanced glycolysis (Pasteur effect) [14, 15]. Therefore, upregulation of miR-210 in response to acute hypoxia enables survival and permits cellular adaptation to hypoxia. However, chronic repression of mitochondrial function is linked to various pathological states, including stroke and diabetes mellitus [20, 70]. The effects of miR-210 may therefore be diverse depending on oxygen tension, cell/tissue type, and duration of hypoxic exposure.
Hypoxia is a major hallmark of cancer and work from the Giaccia laboratory has found that hypoxia upregulates the expression of miR-210 in a variety of tumor types [31]. In normoxia, miR-210 is generally expressed at low levels in breast, pancreatic, head and neck, lung, colon, and renal cell lines. After exposure to 24 h of 2% oxygen, miR-210 expression was induced in all cancer cell lines analyzed. Upregulation of miR-210 is controlled by HIF-1 via a HRE in the miR-210 promoter. To identify miR-210 targets, miRISC immunoprecipitation of myc-tagged AGO2 protein was performed in the breast cancer cell line MCF10A in normoxia and hypoxia. AGO2 immunoprecipitation identified 246 genes that were enriched more than twofold in hypoxia. Of these, 50 genes contain miR-210 target sites in their 3′ UTRs and are therefore predicted miR-210 targets. Only one out of the 50 predicted miR-210 targets (EFNA3) was identified previously as hypoxia-inducible. Therefore, it was postulated that HIF-1-induced miR-210 represses these “normoxia-expressed” genes in hypoxia to enable survival and adaptation to hypoxia. The biological requirement for miR-210 was assayed using tumor xenograft growth assays. miR-210 was stably expressed, via retroviral transfection, in the head and neck cancer cell line FaDu and in the pancreatic cell line SU86.86 [31]. Expression of miR-210 did not affect cell growth in normoxia or at 2% oxygen. However, expression of miR-210 dramatically delayed tumor growth when transfected cells were implanted into nude mice. It appears, however, that miR-210 only delays the initiation of tumor growth, as tumors are able to overcome growth inhibition [31]. This is possibly due to utilization of alternative tumorigenic pathways to overcome miR-210’s inhibitory effect.
In the three studies detailed above, miR-210 acts in two distinct ways. First, miR-210 induction in hypoxia represses crucial components that are required for electron transport in mitochondria, leading to increased ATP generation via glycolysis and cell survival. Second, miR-210 represses genes that are expressed under normoxia that are not required for adaptation and survival in hypoxia, which presumably also reduces the energy requirements of the cells.
DNA damage responses
Tumor cells exhibit a high mutational frequency that enables adaptation to adverse tumor microenvironments [5]. Such genetic instability within solid tumors offers a poor prognosis for cancer patients owing to local resistance to anti-cancer therapies and systemic metastasis. The hypoxic tumor microenvironment has been implicated in causing genetic instability through the dysregulation of DNA repair pathways [61]. The roles of miRNAs in hypoxia-induced regulation of DNA repair are beginning to be elucidated. Recent work has found that the expression of miR-210 and miR-373 is induced when HeLa and MCF-7 cells were exposed to 0.01% oxygen for 24 h [19]. miR-210 was induced by ten- to 12-fold and miR-373 by three- to fourfold in both cells lines studied. Both miR-210 and miR-373 contain HREs in their promoter regions suggesting direct regulation by HIF-1, and indeed, hypoxia induction of miR-210 and miR-373 is dependent on HIF-1 function [19]. Bioinformatic analysis identified DNA repair genes that may be potential targets of miR-210 and miR-373. Both miR-210 and miR-373 have predicted binding sites in the 3′ UTR of the RAD52 gene that is a part of the homology-dependent repair pathway [63]. RAD52 promotes annealing of complementary DNA strands and the loading of RAD51 onto DNA to form nucleoprotein filaments [28]. miR-373 also has a predicted binding site in the 3′ UTR of the nucleotide excision repair gene RAD23B which is an important factor required for the recognition of DNA lesions [3]. Both RAD52 and RAD23B are downregulated in hypoxia and forced expression of miR-210 and miR-373 can cause such downregulation via targeting the 3′ UTR of these genes [19]. Such downregulation of DNA repair factors by miRNA-mediated pathways during hypoxia confers a mutator phenotype on cancer cells. In the adverse microenvironment of a tumor, this would provide enhanced adaptive capacity due to augmented mutational frequency and afford advantages to cancer cells over non-transformed cells. In addition, inhibition of DNA repair mechanisms may conserve ATP pools that may be of short supply under metabolic stress.
Angiogenesis
Angiogenesis is a highly coordinated process of tissue remodeling that leads to the formation of new blood vessels [10]. Hypoxic regions during normal development of an embryo and in pathophysiological conditions, such as cancer and ischemic brain injury, modulate the induction of angiogenesis via the regulation of pro- and anti-angiogenic factors [23, 25, 38]. During hypoxia, the transcriptional activity of HIF-1 directly induces the expression of a variety of angiogeneic growth factors, including vascular endothelial growth factor (VEGF), angiopoietin 2, stromal-derived factor 1, and stem cell factor [12, 26, 37, 69]. The cell type-specific expression of these and other factors from a variety of cell types orchestrate the regulation of specific receptors that are expressed on the surface of vascular endothelial cells and smooth muscle cells. Activation of these cells by receptor–ligand interactions promotes angiogenic budding of new capillaries from existing vessels. Angiogenesis is required for tumors to grow beyond a certain size; therefore, a better understanding of how this process is regulated is of therapeutic importance. Recent work has now revealed an additional layer of angiogenic regulation via the action of specific miRNAs.
In human cancer, one of the most frequently mutated genes is p53 [55, 74]. p53 protein normally acts to inhibit cell growth and stimulate apoptosis when induced by cellular stresses. However, perturbation of the p53 pathway has been associated with angiogenesis and tumor growth [72, 81]. Analysis of miRNAs that are dysregulated in p53-induced human colon cancer specimens revealed that miR-107 is a p53 target and that p53 directly regulates miR-107 via its 5′ UTR [79]. Investigations into potential targets of miR-107 identified potential target sites in the HIF-1β 3′ UTR (and not HIF-1α) suggesting a novel means of regulating hypoxic signaling [79]. Indeed, miR-107 overexpression decreases transactivation of a luciferase reporter that contains the HIF-1β 3′ UTR and mutation of the miR-107 target sites abrogates this regulation. Furthermore, endogenous HIF-1β, and not HIF-1α, protein levels are reduced with miR-107 overexpression [79]. HIF-1 transcriptional activation of VEGF is known to induce angiogenesis [26]. But does miR-107 have an impact on VEGF expression and angiogenesis? Overexpression of miR-107 was indeed found to decrease VEGF induction by a hypoxia mimetic suggesting that miR-107 repression of HIF-1β could blunt angiogenesis [79]. This potentiality was validated in vivo through expression of miR-107 via a lentiviral vector system in mice. Tumorigenic HCT116 cells were injected into nude mice and tumors were allowed to develop. After 31 days, overexpression of miR-107 decreased tumor size in addition to reducing the number of blood vessels within the tumor [79]. Therefore, genetic mutations in p53 and subsequent reduction in miR-107 levels induce angiogenesis in hypoxic tumors via HIF-1β derepression and subsequent increased levels of hypoxic signaling. This is a novel means in which p53 can regulate hypoxic signaling where HIF-1β, and not HIF-1α, is the regulatory target.
Another intriguing mode of miRNA regulation of angiogenesis has recently been reported where miR-519c is a hypoxia-independent regulator of HIF-1α [13]. miR-519c was identified as a potential regulator of HIF-1α expression through target prediction. This was corroborated via luciferase-based assays where the efficient downregulation of the HIF-1α 3′ UTR was abolished when the miR-519c target site in the 3′ UTR was mutated [13]. HIF-1α is a known regulator of the angiogenic factors VEGF, bFGF, and IL-8[66], and the authors found that conditioned media of CL1-5 cells transfected with miR-519c exhibited a significant decrease in the levels of these factors [13]. In the converse experiment, inhibition of miR-519c, using antisense oligonucleotides, was sufficient to increase HIF-1α levels and secretion of angiogenic factors from CL1-5 cells. However, unlike HIF-1α, miR-519c expression is not dysregulated in hypoxia [13]. In contrast, miR-519c expression is suppressed in response to hepatocyte growth factor (HGF) in a dose-dependent manner. HGF was already known as a hypoxia-independent regulator of HIF-1α and it appears that the mechanism for such regulation, at least in part, is through HGF suppression of miR-519c, which releases HIF-1α from post-transcriptional repression [13]. HGF expression does not affect the levels of pri-miR-519c or pre-miR-519c but is suggested to act by inhibiting mature miR-519c biogenesis [13]. The in vivo role of miR-519c in HIF-1α-dependent angiogenesis was confirmed in mice in which Matrigel plugs, pre-soaked with conditioned medium, were injected. Using this approach, the angiogenic activity of miR-519c knockdown medium was found to be higher than from control cells and this effect was abolished with dominant-negative HIF-1α. To study the role of miR-519c on tumor angiogenesis, CL1-5/miR-519c overexpressing cells were injected into nude mice. miR-519c overexpression causes a slower growth rate and tumor size when compared to controls and the tumors had significantly fewer blood vessels [13]. These in vivo outcomes suggest that miR-519c may be a potential cancer therapeutic in the future. In addition, this work describes the presence of non-hypoxia-regulated microenvironmental mediators, such as HGF, that provide alternative layers of information used to integrate homeostatic responses.
Future directions
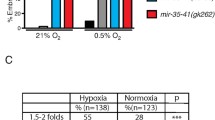
Understanding the roles of miRNAs in the regulation of complex physiological and pathophysiological responses of hypoxic cells is still in its infancy. In the coming years, the study of miRNA-mediated hypoxic responses in animal and plant model systems will help provide a better picture on the importance of post-transcriptional gene regulatory mechanisms in hypoxia. The kinetic properties of miRNAs provide marked advantages over transcriptional regulation for acute and transient hypoxic insults. Their rapid biogenesis and mode of regulation affect gene expression with less delay than nuclear regulatory factors. Such rapidity and efficiency may be crucial to counteract variable fluctuations in the environmental milieu such as oxygen levels. The fact that few functions have been assigned for miRNA mutants that are cultured in standard laboratory conditions suggests that they have discrete functions and perhaps act as responders to fluctuations in environmental stress [45, 54]. Therefore, miRNA profiling after environmental stress and exposure of mutant animals and cells to environmental challenges may reveal novel mechanistic roles. In fact, a survey of miRNA expression levels in C. elegans has identified a number of miRNAs that are dysregulated in 0.5% oxygen, and that mutant animals lacking specific HRMs are sensitive to hypoxia (Pocock, unpublished data). I expect that further work in the nematode, and other models, will elucidate novel miRNA-mediated mechanisms of hypoxic adaptation.
One crucial aspect of miRNA biology that is of particular interest at present is miRNA compartmentalization and transport. Such mechanisms may enable miRNAs to be shuttled intracellularly between cellular compartments (e.g., plasma membrane, mitochondria, endoplasmic reticulum, and synapse) and, in addition, to be carried in a systemic fashion via circulation in the blood and other body fluids [9, 24, 41]. Intracellular localization may provide restricted high concentrations of specific miRNAs, temporal segregation and even control of access to mRNA targets. Sequestration of hypoxia-responsive miRNAs could be especially useful in mitochondria where hypoxic insults may require a rapid and orchestrated response. In addition, the membrane association of miRNAs within and at the periphery of cells may enable the rapid and localized regulation of mRNA translation in response to environment stressors such as hypoxia. One could envision that the internalization of activated plasma membrane receptors may in some cases be used to signal the release of latent miRNAs from subcellular compartments or the plasma membrane itself. Systemically acting miRNAs also open up the possibility of long-range modes of action where miRNAs expressed in one cell type are then packaged and transported to regulate the expression of genes in other cells and tissues. The involvement of endosomal trafficking in miRNA silencing suggests that miRNAs use this route for intracellular and intercellular transmission [27, 50].
Finally, it is crucial to remember that the initial discovery of miRNAs, and many subsequent advances in our knowledge, have hinged on basic research in developmental genetics and the free availability of genome sequences [46, 56, 60, 73, 78]. Thus, continued work on elucidating the biological roles of miRNAs and decoding the mechanisms of miRNA-mediated gene regulation of hypoxia require the utilization of a variety of model systems. This will ensure that we do not overlook important discoveries in the future.
References
Bartel DP (2004) MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Batty D, Rapic'-Otrin V, Levine AS, Wood RD (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol 300:275–290
Bertout JA, Patel SA, Simon MC (2008) The impact of O2 availability on human cancer. Nat Rev Cancer 8:967–975
Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA (2006) Human cancers express a mutator phenotype. Proc Natl Acad Sci USA 103:18238–18242
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148
Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J (2008) Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14:1340–1348
Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L (2010) Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 78:838–848
Carmeliet P (2000) Mechanisms of angiogenesis and arteriogenesis. Nat Med 6:389–395
Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E (1998) Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485–490
Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, Capla JM, Galiano RD, Levine JP, Gurtner GC (2004) Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med 10:858–864
Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, Tan CT, Lin MT, Kuo ML (2010) MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res 70:2675–2685
Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10:273–284
Chen Z, Li Y, Zhang H, Huang P, Luthra R (2010) Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29:4362–4368
Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447:823–828
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460:479–486
Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH (2010) p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17:236–245
Crosby ME, Kulshreshtha R, Ivan M, Glazer PM (2009) MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69:1221–1229
Duchen MR (2004) Roles of mitochondria in health and disease. Diabetes 53(Suppl 1):S96–S102
Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43–54
Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F (2008) MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 283:15878–15883
Ferrara N (2002) VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer 2:795–803
Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 1775:181–232
Folkman J (1971) Tumor angiogenesis: therapeutic implications. N Engl J Med 285:1182–1186
Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 16:4604–4613
Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11:1143–1149
Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52–ssDNA complexes. Nucleic Acids Res 38:2917–2930
Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N (2005) microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 1:e13
Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ (2007) High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer 6:5
Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ (2009) Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 35:856–867
Humphreys DT, Westman BJ, Martin DI, Preiss T (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102:16961–16966
Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838
Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 12:149–162
Johnston RJ, Hobert O (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426:845–849
Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ (2009) The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28:2419–2424
Kelly BD, Hackett SF, Hirota K, Oshima Y, Cai Z, Berg-Dixon S, Rowan A, Yan Z, Campochiaro PA, Semenza GL (2003) Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ Res 93:1074–1081
Kerbel RS (2008) Tumor angiogenesis. N Engl J Med 358:2039–2049
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216
Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z (2007) An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129:1141–1151
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101:2087–2092
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610
Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M (2007) A microRNA signature of hypoxia. Mol Cell Biol 27:1859–1867
Kulshreshtha R, Davuluri RV, Calin GA, Ivan M (2008) A microRNA component of the hypoxic response. Cell Death Differ 15:667–671
Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U (2005) Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121:1097–1108
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21:4663–4670
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim DH, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW (2009) Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11:1150–1156
Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW (2009) A microRNA imparts robustness against environmental fluctuation during development. Cell 137:273–282
Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315:1576–1579
Medina PP, Nolde M, Slack FJ (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467:86–90
Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR (2007) Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3:e215
Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10:431–442
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408:86–89
Pocock R, Hobert O (2008) Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat Neurosci 11:894–900
Pocock R, Hobert O (2010) Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci 13:610–614
Ratcliffe PJ, Pugh CW, Maxwell PH (2000) Targeting tumors through the HIF system. Nat Med 6:1315–1316
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Reynolds TY, Rockwell S, Glazer PM (1996) Genetic instability induced by the tumor microenvironment. Cancer Res 56:5754–5757
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86
San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–257
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63
Semenza GL (2001) Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 7:345–350
Semenza GL (2007) Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J 405:1–9
Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem 271:32529–32537
Simon MP, Tournaire R, Pouyssegur J (2008) The angiopoietin-2 gene of endothelial cells is up-regulated in hypoxia by a HIF binding site located in its first intron and by the central factors GATA-2 and Ets-1. J Cell Physiol 217:809–818
Sims NR, Anderson MF (2002) Mitochondrial contributions to tissue damage in stroke. Neurochem Int 40:511–526
Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5:659–669
Teodoro JG, Parker AE, Zhu X, Green MR (2006) p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science 313:968–971
Then C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018
Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10:789–799
Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA 92:5510–5514
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Welch C, Chen Y, Stallings RL (2007) MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26:5017–5022
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862
Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ (2010) P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA 107:6334–6339
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016
Yu JL, Rak JW, Coomber BL, Hicklin DJ, Kerbel RS (2002) Effect of p53 status on tumor response to antiangiogenic therapy. Science 295:1526–1528
Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR 3rd, Han M (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 28:598–613
Acknowledgements
My work is supported by funding from the Biotech Research and Innovation Centre and by a European Research Council Starting Grant (Grant number 260807). I am thankful to members of my laboratory for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pocock, R. Invited review: decoding the microRNA response to hypoxia. Pflugers Arch - Eur J Physiol 461, 307–315 (2011). https://doi.org/10.1007/s00424-010-0910-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-010-0910-5