Abstract
The maize B-chromosome consists mainly of heterochromatin and is considered to be genetically inert. However, the B-chromosome contains euchromatin that carries control elements that direct its behaviors during cell division, such as nondisjunction during the second pollen mitosis. To determine the transcriptional activity of the B-chromosome, complementary DNA-amplified fragment length polymorphism analysis was applied to five inbred maize lines with and without B-chromosomes. Six putative B-chromosome-related transcripts were identified, four of which were cloned and characterized via Southern hybridization, fluorescence in situ hybridization, and sequence comparison to further confirm their B-chromosome origin. All the analyzed B-chromosome-related transcript sequences were repetitive and showed homology to A-chromosomes. Quantitative real-time reverse transcriptase-polymerase chain reaction revealed that the B-chromosome-specific transcribed sequences B3547-179 and B3849-212 were transcribed in a B-chromosome-dosage-dependent manner. Expression of B3849-189 and B3849-147 was not specific to the B-chromosome; however, the former showed a transcriptional pattern with B-chromosome dosage compensation, and the latter displayed down-regulation of transcription due to higher B-chromosome numbers. Using four B-10L translocations, B3849-212 was mapped to the B-chromosome region that contains the nondisjunction control elements of the B-chromosome. Taken together, our results suggested that the maize B-chromosome harbors few transcriptionally active sequences and might influence the transcription of A-chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
B-chromosomes are extra or nonessential chromosomes containing large selfish DNA elements that have been identified in a variety of fungi, animal, and plant species (Jones and Rees 1982). They do not pair or recombine with any of the normal chromosome complement (A-chromosomes) during meiosis, and they accumulate in a non-Mendelian manner. In general, B-chromosomes are considered genetically inert, as they do not carry any genes that are indispensable for plant development (Jones et al. 2008). However, B-chromosomes are not entirely without genetic activity. For example, the maize (Zea mays) B-chromosome can improve the recombination frequencies of A-chromosomes (Rhoades 1968), elicit breakages in A-chromosomes during paternal meiosis (Rhoades and Dempsey 1972), and cause leaf striping (Staub 1987). Similarly, the rye (Secale cereale) B-chromosome can change the distribution of chiasmata in A-chromosomes (Jones and Rees 1967). In addition, the B-chromosome of Drosophila albomicans significantly affects fertility (Zhou et al. 2012), while in cichlid fish, the B-chromosome functionally affects female sex determination (Yoshida et al. 2011), and in Nectria haematococca, the B-chromosome plays an important role in pathogenicity and antibiotic resistance (Coleman et al. 2009). Through fluorescence in situ hybridization (FISH), ribosomal RNA genes were found to be present on the B-chromosomes of Brachycome dichromosomatica (Donald et al. 1995), Crepis capillaris (Maluszynska and Schweizer 1989; Leach et al. 2005), and Plantago lagopus (Dhar et al. 2002).
Molecular approaches have revealed the transcriptional activity of B-chromosomes in various animal and plant species. In the grasshopper Eyprepocnemis plorans, ribosomal RNA genes located on the B-chromosome were shown to be functional and to be transcribed to form the nucleolus attached to the B-chromosome (Ruiz-Estévez et al. 2012). In Drosophila albomicans, an actively transcribed unit of the B-chromosome was identified, which showed sequence homology with the subcentromeric region of both the ancient X and neo-X chromosomes (Zhou et al. 2012). In the Siberian roe deer, Capreolus pygargus, three A-chromosome-located genes were found on the B-chromosome, one of which was transcribed in fibroblast culture (Trifonov et al. 2013).
In plant species, there is an extensive literature on the transcription of rye B-chromosomes. Two rye B-chromosome-specific repetitive sequence families, E3900 and D1100, display transcriptional activities with different tissue-type dependencies (Carchilan et al. 2007). Additionally, highly conserved E3900-related sequences have been identified in rye A-chromosomes, and the transcription of these sequences was shown to be up-regulated in meiocytes exclusively in plants carrying B-chromosomes (Pereira et al. 2009). Through comparative complementary DNA-amplified fragment length polymorphism (cDNA-AFLP) analysis, rye B-chromosomes have been shown to be weakly transcribed and to potentially alter the transcriptional activity of A-chromosome sequences (Carchilan et al. 2009). A recent next-generation sequencing analysis of sorted rye A- and B-chromosomes showed that rye B-chromosomes are rich in gene-derived sequences (Martis et al. 2012). Approximately 15 % of the selected gene-like fragments of rye B-chromosomes displayed transcriptional activity in a tissue-type and genotype-specific manner, and these B-chromosome-located sequences were shown to cause in trans down- or up-regulation of A-chromosome-encoding genic fragments (Banaei-Moghaddam et al. 2013). Furthermore, B-chromosome-enriched repeats with transcriptional activity were found to accumulate in the nondisjunction control region of rye B-chromosomes (Klemme et al. 2013).
The maize B-chromosome has evolved several mechanisms to ensure its survival, and at least three properties allow the B-chromosome to maintain and expand its numbers, including nondisjunction during the second pollen mitosis (Longley 1927; Roman 1947), preferential fertilization of the egg by sperm carrying B-chromosomes (Roman 1948), and prevention of univalent losses through meiosis (Carlson 1986). Regions of the B-chromosome that condition these accumulation mechanisms have been identified. The nondisjunction mechanism requires trans-acting factors located in the proximal and distal euchromatic regions of the B-chromosome long arm (Ward 1973; Lin 1978). In addition, the centromeric knob and the short arm of the B-chromosome are associated with nondisjunction in a cis-acting manner (Carlson 1973; Lin 1979). The third heterochromatic region on the B-chromosome long arm is required to prevent univalent B-chromosome loss during meiosis (Carlson and Roseman 1992). Although the elements required for B-chromosome survival have been mapped, no genes have been identified and characterized in the B-chromosome of maize.
A large number of studies have been conducted on the DNA composition of the maize B-chromosome (Chilton and McCarthy 1973; Alfenito and Birchler 1993; Stark et al. 1996; Cheng and Lin 2003; Lamb et al. 2005; Lo et al. 2009a; Peng and Cheng 2011). In contrast, only one investigation has focused on the transcriptional activity of the maize B-chromosome (Lamb et al. 2007). Here, we used the cDNA-AFLP technique to isolate B-chromosome-related transcripts and to test whether the maize B-chromosome contains transcriptionally active sequences and whether the presence of the B-chromosome can affect the transcription of maize A-chromosome-located gene sequences.
Materials and methods
Plant materials
Maize inbred lines carrying B-chromosomes were developed from a B-chromosome-containing L289 plant (developed by J. B. Beckett) through continuous backcrossing to four different inbred lines, A619, A632, Mo17, and W23, for at least ten generations. Plants from the five inbred lines without a B-chromosome (+0B) or with one B-chromosome (+1B) were used to isolate total RNA for cDNA-AFLP analysis. Specimens from line L289 carrying zero, one, or nine B-chromosomes (L289 + 0B, L289 + 1B, and L289 + 9B, respectively) were subjected to Southern hybridization and reverse transcriptase-PCR (RT-PCR) analysis. cDNAs from L289+0B, L289+1B, and L289+3B were used for quantitative real-time RT-PCR (qRT-PCR) analysis. In addition, genomic DNA from L289+ B, L289+1B, L289+6B, W23+0B, W23+1B, and four B-10L translocations was employed in genomic PCR analyses, and root-tips from L289 + 1B were employed to prepare mitotic chromosome spreads for FISH. The chromosome composition of each plant was confirmed through chromosome counting.
The four B-10L translocations (TB-10L26, TB-10L34, TB-10L36, and TB-10L38) in the W23 inbred background (Lin 1972) were used to generate translocation heterozygous, tertiary trisomic, and hypoploid plants, as described by Cheng (2010). Each translocation has one breakpoint at different positions along the B-chromosome long arm and a second breakpoint on the long arm of chromosome 10 (10L) (Supplementary Fig. 1a). As shown in Supplementary Fig. 1b, a B-10L translocation consists of two chromosomes, 10-B and B-10. The 10-B bears the short arm, the centromere, and the proximal portion of 10L connected to the distal portion of the B-chromosome long arm, while the B-10 contains the short arm and the centromere of the B-chromosome, in addition to the proximal portion of the B-chromosome long arm attached to the terminal portion of 10L. A translocation heterozygote contains 10, 10-B, and B-10; a tertiary trisomy includes 10, 10, and B-10; and a hypoploid carries 10 and 10-B.
RNA isolation and cDNA-AFLP analysis
The cDNA-AFLP analysis was performed following the protocol described by Vuylsteke et al. (2007), with some modifications. Total RNA was isolated from the third leaf of plants at the four-leaf stage following the instructions provided in the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). A minimum of 2 μg of total RNA was used to synthesize cDNA fragments using random sequence hexamer primers with a cDNA synthesis system kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The resulting double-stranded cDNA was then digested with EcoRI and MseI, and the digestion products were ligated with EcoRI- and MseI-adapters to generate PCR templates. Pre-amplification was performed using an EcoRI + A primer in combination with an MseI + C primer. The resulting products were used as templates for selective amplification with selective primers carrying three-base extensions (Supplementary Table 1). The PCR products were resolved via electrophoresis in a 4 % denaturing polyacrylamide gel, then silver stained and scanned. Fragments of interest were excised from the gel and reamplified using the same primer combinations and PCR conditions as in the selective amplification. The PCR products were subsequently cloned into a T-vector plasmid (Yeastern Biotech, Taipei, Taiwan) for further characterization.
Genomic DNA isolation, Southern hybridization, and FISH
The procedure for maize genomic DNA isolation was previously described by Lin and Chou (1997), and Southern hybridization analysis was conducted using the method outlined by Lin et al. (1997). Mitotic metaphase chromosome spreads were prepared according to published protocols (Cheng and Lin 2004). FISH was performed according to Cheng (2010). The fluorescence signals were captured using a cooled charge-coupled device camera (DP73, Olympus) on an Olympus BX51 fluorescence microscope and processed using Photoshop software (Adobe, San Jose, CA, USA).
Genomic PCR, RT-PCR, and qRT-PCR
A total of 80 ng of genomic DNA or cDNA was used in each amplification reaction. Primers were designed for candidate B-related transcript sequences (Supplementary Table 2). The PCR conditions were as follows: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, various annealing temperatures for 30 s (the annealing temperatures are listed in Supplementary Table 2), and 72 °C for 45 s, with a final extension at 72 °C for 5 min. The PCR products were separated on 2.5 % agarose gels and subsequently cloned into a T-vector plasmid (Yeastern Biotech). The clones resulting from each PCR amplification were sequenced, followed by sequence alignment. The maize actin gene was used as an internal control for RT-PCR analysis, according to Li et al. (2010).
For qRT-PCR, cDNAs of L289 + 0B, L289 + 1B, and L289 + 3B were used, and the SYBRGreen RT-PCR kit (Applied Biosystems, Foster City, CA, USA) was employed. To correct for differences in the amount of starting material, the maize actin gene was used as a reference gene. All PCR experiments were carried out in an ABI 7700 thermocycler. The data were analyzed with SDS software, version 1.9.1 (Applied Biosystems). The cycle thresholds (C T) were determined for the samples and actin. The comparative expression level (CEL) is defined as \( {2}^{-\varDelta \varDelta {C}_{\mathrm{T}}} \), which is derived from the equation Δ C T = C T (sample) − C T (actin). The expression of B-related transcripts in L289 + 1B was chosen as the baseline signal. The equation ΔΔ C T = Δ C T (sample) − Δ C T (baseline) was used to determine the CEL. The obtained data are expressed as the mean of three independent experiments (n = 3), each assayed in triplicate, and are presented as the mean ± standard deviation. Student’s t-test was performed to determine statistical significance.
Sequence analysis
Sequence comparisons were performed using the Basic Local Alignment Search Tool (NCBI, http://www.ncbi.nlm.gov/BLAST/) and MaizeGDB BLAST (http://blast.maizegdb.org/home.php?a=BLAST_UI). Sequence alignments were conducted with Vector NTI software (version 10.3.0). The sequences of B-related transcripts obtained in this study were deposited in GenBank under accession numbers JY274297 to JY274300.
Results
The B-chromosome affects the transcription of the maize genome
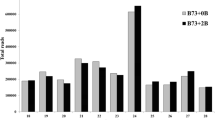
To investigate the transcriptional activity of the maize B-chromosome, cDNA-AFLP analyses were performed on five maize inbred lines (A619, A632, L289, Mo17, and W23) with zero B-chromosomes (+0B) or one B-chromosome (+1B). To minimize transcriptional variability among individuals, cDNA samples from at least six plants from each inbred line, grown under identical conditions, were pooled for the cDNA-AFLP analysis. Using 28 primer combinations, approximately 1,230 clearly visible bands per inbred line were amplified (listed in Supplementary Table 3). A total of 141 additional bands (11.46 % of the average number of bands per inbred line) were observed in +1B materials from at least one inbred line. Only one of these +1B bands (0.08 % of the average number of bands per inbred line) exhibited an identical size and intensity in all inbred lines (Fig. 1a). Together with five +1B bands (0.41 % of the average number of bands per inbred line) present in four of the inbred lines, the six +1B bands B3348-142, B3547-179, B3849-147, B3849-189, B3849-212, and B4049-180 were selected as putative +1B-related transcript candidates (Fig. 1b). The other +1B bands, which were detected in less than four inbred lines, were not analyzed further. As B-chromosomes might also affect the transcription of A-chromosomes, the additional bands observed in +0B materials were also determined. A total of 105 additional bands were observed in +0B materials (8.54 % of the average number of bands per inbred line) from either one or two inbred lines (Supplementary Table 3). Only two (A3749-329 and A3749-261) of these +0B bands (0.16 % of the average number of bands per inbred line) were found in two of the inbred lines, which were considered putative +0B-related transcript candidates. The results of the cDNA-AFLP analysis suggest that the B-chromosome likely affects the transcriptional activity of the maize genome.
Amplification of cDNA-AFLP fragments from maize inbred lines with and without the B-chromosome. a The cDNA-AFLP pattern was obtained from five maize inbred lines (A619, A632, L289, Mo17, and W23) carrying zero (0B) or one (1B) B-chromosome by the primer combination E35M47. b Six additional bands were present in +1B materials of at least four inbred lines. Arrows indicate positions of bands specific to plants with the B-chromosome. The band unique to +0B materials is indicated by an arrowhead
Sequence analysis of putative B-related transcript candidates
Four out of the six +1B bands that were present in +1B materials from at least four inbred lines and the two +0B bands that were observed in two inbred lines were successfully isolated from L289 lines and cloned for sequence analysis (Table 1). The four +1B-related transcripts cloned from L289+1B (B3547-179, B3849-147, B3849-189, and B3849-212) averaged 182 bp (147–212 bp) in length and showed an average 44 % (32–50 %) G/C content. The entire sequence of B3849-189 shared 94 % identity with the StarkB element of the maize B-chromosome (pBK118-8 LL repeat; Lo et al. 2009b), while that of B3849-212 shared 100 % identity with the B-chromosome-specific AFLP fragment in maize (p3849; Peng et al. 2005). The sequences of B3849-147 displayed homology to a maize mRNA sequence of unknown function, with 96 % identity, and the partial sequences of B3547-179 showed homology to the mRNA sequence of the oxygen-independent coproporphyrinogen III oxidase-like protein of maize, with 96 % identity. The two +0B-related transcripts (A3749-329 and A3749-261) were cloned from L289+0B. The entire A3749-329 sequence shared 99 % identity with a maize mRNA sequence, and the partial A3749-261 sequences were identical to chloroplast sequences from maize.
Sequence composition of B-related transcripts
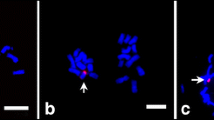
To understand the sequence composition of B-related transcripts, sequence comparisons, Southern hybridization, and FISH analyses were performed. According to sequence comparisons with the maize B73 inbred line genome (B73 RefGen_V2 in the MaizeGDB database) (Supplementary Fig. 2), B3849-147 showed homology to 1,028 sequences in the B73 reference genome with at least 80 % identity. Additionally, B3849-189 and B3849-212 presented homology to 209 and 138 sequences, respectively, whereas B3547-179 exhibited homology to only five sequences. Based on Southern hybridization assays, all four +1B-related transcripts showed multiple hybridization signals, among which B3547-179 displayed the weakest signals and B3849-147 the strongest (Supplementary Fig. 3). The hybridization patterns of these transcripts were nearly identical among the genomic DNA of L289 + 0B, L289 + 1B and L289 + 9B. However, hybridization signals presenting B-specificity or B-dosage dependency, showing stronger hybridization in L289 + 9B than in L289 +1B, were observed for all four transcripts (Supplementary Fig. 3), indicating that the sequences of the four +1B-related transcripts are also found in the B-chromosome. Southern hybridization of the two +0B-related transcripts showed multiple hybridization signals that were identical among all of the L289 lines (data not shown). FISH analysis further revealed the chromosomal distribution of transcriptionally active B-related sequences. Of the six B-related transcripts, only B3849-147 and B3849-189 showed detectable FISH signals that were distributed across all maize B- and A-chromosomes (Fig. 2). The absence of FISH signals for the other four B-related sequences may have been due to their short lengths, low copy numbers, or widely distributed locations on the chromosomes, such that the resulting FISH signals could not be captured by the fluorescence equipment. Thus, the obtained data indicate that the four +1B-related transcripts are present on the B-chromosome and show a similar sequence organization on the maize B- and A-chromosomes.
Fluorescence in situ hybridization of B-related transcripts. Maize mitotic metaphase cells containing one B-chromosome (arrowed) were hybridized with the B-specific centromere repeat ZmBs, and with the +1B-related transcripts B3849-147 (a) and B3849-189 (b), respectively. The red signal displays the hybridization of ZmBs, and the green signal shows the hybridization of +1B-related transcripts. Chromosomes are counterstained with DAPI (in blue). Bars represent 10 μm
Expression of B-related transcripts
To evaluate the transcriptional activity of the B-related transcripts, RT-PCR was performed on cDNA from L289+0B, L289+1B, and L289 + 9B individuals using primers complementary to regions showing the least homology among the four +1B-related transcripts and their top ten homologous sequences from the maize B73 genome database (Supplementary Table 2; Supplementary Fig. 4). RT-PCR products with the expected sizes were observed for all of the +1B-related transcripts (Fig. 3a). RT-PCR analyses using B3547-179- and B3849-212-specific primers yielded products only from the cDNA templates from L289 lines carrying B-chromosomes. In qRT-PCR analyses conducted in L289+0B, L289+1B, and L289+3B, the expression of the two +1B-related transcripts was not detected in L289 + 0B; these transcripts showed B-dosage dependence, with higher transcription being observed in L289 + 3B than in L289 + 1B (Fig. 3c). The RT-PCR patterns obtained for B3849-147 and B3849-189 were similar among the three L289 lines (Fig. 3a). qRT-PCR analysis revealed that the expression of B3849-189 displayed the same level among the three L289 lines, whereas that of B3849-147 was slightly reduced in L289 + 3B (Fig. 3c). The two +0B-related transcripts also showed similar RT-PCR patterns among the three L289 lines (Fig. 3b). The nearly equal amounts of PCR products observed suggested similar levels of A-chromosome transcription, regardless of the presence of B-chromosomes. Because multiple PCR products of unexpected size were generated for both +0B-related transcripts, determining the precise expression levels of these transcripts among the three L289 lines via qRT-PCR was not possible.
Transcriptional activity of B-related transcripts. a, b RT-PCR analyses of six B-related transcripts (B3547-179, B3849-147, B3849-189, B3849-212, A3749-329, and A3749-261) on L289+0B (0B), L289+1B (1B), and L289+9B (9B). Primer combinations correspondent to each B-related transcript were listed in Supplementary Table 2. To confirm that equal amount of cDNA was present, the maize actin gene was used as an internal control. Molecular weights of expected products are shown on the right and unexpected products are marked by arrows. c qRT-PCR of four +1B-related transcripts (B3547-179, B3849-147, B3849-189, and B3849-212) in L289+0B, L289+1B, and L289+3B. In all experiments three individual plants of each genotype were used. CELs were calculated by using the \( {2}^{-\varDelta \varDelta {C}_{\mathrm{T}}} \) method. Experiments were performed in triplicate and data were expressed as mean ± standard deviation. Values were normalized to L289+1B RNA levels. (*) and (**) indicate significant difference at the 0.05 level and 0.01 level, respectively compared with L289+1B RNA levels using Student’s t-test
Mapping of +1B-related transcripts on the B-chromosome based on B-10L translocations
Genomic PCR analysis of L289 + 0B, L289 + 1B, L289 + 6B, W23 + 0B, and W23 + 1B was applied to detect whether the +1B-related transcripts were encoded by the maize B-chromosome. A B3547-179 PCR product of the expected size was only observed in L289 lines carrying B-chromosomes, whereas this product was present in W23 lines both with and without a B-chromosome (Fig. 4a). Additionally, a product of a smaller size was consistent among the L289 and W23 lines, regardless of the presence of B-chromosomes. A unique B3849-212 product showing B-dosage dependency was found in all three L289 lines, whereas this product was specific to the W23 line bearing the B-chromosome (Fig. 4b). However, identical PCR products were observed for B3849-147 and B3849-189 in all L289 and W23 lines (data not shown). These results suggested that the four +1B-related transcripts might be encoded by both B- and A-chromosomes.
Genomic PCR analysis and mapping of two +1B-related transcripts. Genomic DNA of L289+0B, L289+1B, L289+6B, W23+0B, W23+1B, and four B-10L translocations (TB-10L34, TB-10L38, TB-10L26, and TB-10L36) was amplified by B3547-179 (a) and B3849-212 (b) specific RT-PCR primers. P cloned plasmid as positive control, N water as negative control, H translocation heterozygote, T tertiary trisomy, O hypoploid. PCR products of a were electrophoresed in 6 % polyacrylamide gel, and that of b were separated in 2.5 % agarose gel. c Map position of B3849-212 (black line) in relation to breakpoints (arrows) of the four B-10L translocations. BS B-chromosome short arm, CK B-centromere and centromeric knob, PE proximal euchromatin, DH1–DH4 four distal heterochromatins, DE distal euchromatin
To physically map the locations of +1B-related transcripts on the B-chromosome, four B-10L translocations (TB-10L34, TB-10L38, TB-10L26, and TB-10L36) in the W23 background were used. As shown in Fig. 4c, the breakpoints of the four B-10 L translocations are dispersed along the B-chromosome long arm as follows: TB-10L34 is located close to the junction of the proximal euchromatin and distal heterochromatic region 1; TB-10L38 is located between distal heterochromatic regions 1 and 2; TB-10L26 is located near the junction of distal heterochromatic regions 2 and 3; and TB-10L36 is located between distal heterochromatic regions 3 and 4. The principle used to carry out this mapping was as follows: when a B-specific PCR product was amplified from the tertiary trisomic DNA of a B-10L translocation, it was mapped to the portion of the B-chromosome born on the B-10 chromosome. In contrast, when a B-specific PCR product was amplified from the hypoploid DNA of a B-10L translocation, it was mapped to the B-chromosome section carried by the 10-B chromosome (see Supplementary Fig. 1b). However, due to the absence of B-specific PCR products from W23 + 0B and W23 + 1B, mapping the B-chromosome positions of B3547-179, B3849-147, and B3849-189 was not possible based on the set of B-10L translocations (Fig. 4a). In contrast, PCR products corresponding to B3849-212 were prominent in W23 + 1B, but absent in W23 + 0B (Fig. 4b). The B3849-212 PCR products were present in all tertiary trisomies but absent in all hypoploids, suggesting that B3849-212 is located in the B-chromosome region proximal to the breakpoint of TB-10L34, which contains the short arm, the centromeric knob, and the proximal euchromatin of the B-chromosome (Fig. 4b and c).
Sequence analysis of genomic PCR and RT-PCR products
To investigate sequence variations in the genomic PCR and RT-PCR products of the four +1B-related transcripts, we cloned and sequenced the PCR products from each L289 line included in the sequence alignment. As shown in Fig. 5, the genomic PCR products of B3547-179 presented two sequences of different lengths (115 and 137 bp). The 115-bp sequence was amplified from L289 + 0B DNA, suggesting an A-chromosome-derived origin, while the 137-bp sequence was only amplified from L289 DNA from individuals harboring B-chromosomes, indicating a B-chromosome-derived origin. On the other hand, the RT-PCR products of B3547-179 were all 137 bp in length. The B3849-212 sequences were homogeneous in the genomic PCR and RT-PCR products among all L289 lines (Supplementary Fig. 5a). The remaining two +1B-related transcripts, B3849-147 and B3849-189, showed a higher degree of sequence variation in their genomic PCR products, regardless of the presence of B-chromosomes. However, the sequences of the RT-PCR products for B3849-147 and B3849-189 obtained from L289 + 0B were homogeneous, while those from L289 lines carrying B-chromosomes presented a greater amount of sequence variation (Supplementary Fig. 5b and c).
Alignment of genomic and transcribed sequences of B3547-179. PCR products amplified from genomic DNA (L289 + 0B, L289 + 1B, and L289 + 6B) and cDNA (L289 + 0B, L289 + 1B, and L289 + 9B) by B3547-179 specific RT-PCR primer were cloned, sequenced, and aligned with the corresponding B3547-179 sequences. The A-chromosome-derived 115-bp and B-chromosome-derived 137-bp sequences are indicated. Conserved nucleotides are shaded with gray and black
Discussion
The maize B-chromosome is not essential for plant development, but phenotypic effects can still be detected as its numbers increase (Randolph 1941). In the current study, we employed the additional cDNA-AFLP bands present in maize inbred lines lacking the B-chromosome to assess whether the maize B-chromosome can alter the transcriptional activity of A-chromosome sequences. A total of 105 + 0B additional bands (8.54 %) were observed among inbred lines without a B-chromosome (Supplementary Table 3), suggesting that a negative effect on the expression of A-chromosome-located gene sequences occurred in the presence of the B-chromosome. Similarly, by using cDNA-AFLP to analyze three different near-isogenic rye 0B/+B lines, 132 (5.8 %) +0B extra bands were found among plants without B-chromosomes (Carchilan et al. 2009). The data from maize and rye supported the hypothesis that B-chromosomes are capable of affecting the transcriptional activity of the normal genome.
The presence of the four +1B-related transcript sequences on the maize B-chromosome was verified by the observation of B-specific and B-dosage-dependent hybridization signals in Southern hybridization analyses (Supplementary Fig. 3). Moreover, the FISH signals of B3849-147 and B3849-189 observed on the B-chromosome (Fig. 2) as well as the B-specific or B-dosage-dependent genomic PCR products of B3547-179 and B3849-212 found in plants carrying B-chromosomes (Fig. 4a and b) provided further evidence that the four +1B-related transcript sequences are located on the B-chromosome. However, homologs of these sequences were also detected on A-chromosomes, which raises the question of whether the four +1B-related transcripts are expressed from the B-chromosome or from A-chromosome-located homologs in the presence of the B-chromosome.
Sequence analysis of the genomic PCR and RT-PCR products of B3547-179 showed that an A-chromosome-derived 115-bp sequence and a B-chromosome-derived 137-bp sequence were present in the genomic DNA, whereas only the B-chromosome-derived 137-bp sequence was transcribed (Fig. 5), indicating that the 137-bp B3547-179 sequence is specifically expressed from the B-chromosome. B3849-212 was also found to be expressed only in plants with B-chromosomes (Fig. 3a and c), although the B3849-212 sequences detected in genomic PCR products were homogeneous among the L289 lines, regardless of the presence of B-chromosomes, closely resembling the obtained RT-PCR products (Supplementary Fig. 5a). Therefore, B3849-212-like sequences located on A-chromosomes are also transcriptionally active.
While B3849-147 and B3849-189 were found to be expressed in plants both with and without B-chromosomes (Fig. 3a and c), their RT-PCR products were homogeneous in the L289+0B line but showed a higher degree of variation in L289+Bs lines (Supplementary Fig. 5b and c). In rye B-chromosomes, the polymorphisms between B- and A-chromosome-located paralogous sequences are primarily due to single nucleotide polymorphisms (SNPs), deletions, and insertions (Banaei-Moghaddam et al. 2013), and the SNP frequency in the genic sequences of rye B-chromosomes is higher compared with their A-chromosome-located homologs (Martis et al. 2012). Accordingly, B3849-147- and B3849-189-like sequences located on both types of chromosomes are transcriptionally active.
Genotype-dependent transcriptional behavior was observed among +1B-related cDNA-AFLP fragments. As shown in Fig. 1b, five of the +1B cDNA-AFLP fragments were produced in only four of the five maize inbred lines, with B3348-142 being absent in A632+1B; B3849-147 being absent in W23+1B; and B3849-189, B3849-212, and B4049-180 missing from Mo17+1B. A similar genotype-dependet expression pattern was observed in the expression of a B-chromosome-specific transcribed sequence and B-located gene-like fragments on rye B-chromosomes (Carchilan et al. 2009; Banaei-Moghaddam et al. 2013). qRT-PCR analysis revealed that the expression of B3547-179 and B3849-212 was specific to L289 inbred lines carrying B-chromosomes, with the expression level in L289 + 3B being significantly higher than in L289+1B, indicating B-dosage-dependent expression (Fig. 3c). In maize, it has been suggested that changing the dosage of the A-chromosome segment can alter the expression of genes, generally in a negative manner (Birchler et al. 1979; Birchler et al. 2001). The expression of B3849-189 showed B-chromosome dosage compensation, in which the total transcriptional activity did not increase with increasing B-chromosome numbers (Fig. 3c), as observed for the sex chromosomes of different animal and plant species (Birchler et al. 2001; Muyle et al. 2012). Interestingly, the expression level of B3849-147 was identical between L289 + 0B and L289 + 1B but was slightly decreased in L289 + 3B (Fig. 3c), suggesting down-regulation due to the presence of additional B-chromosomes. Taken together, our data suggested that in trans crosstalk might exist between the B- and A-chromosomes.
Using B-10L translocations, the +1B-related transcript B3849-212 was mapped to the B-chromosome region that contains the short arm, the centromeric knob, and the proximal euchromatin (Fig. 4b and c). Moreover, the sequences of B3849-212 are identical to the B-chromosome-specific AFLP fragment p3849, which has been mapped to the proximal euchromatin using hypoploids for 12 B-10L translocations (Peng et al. 2005). Most notably, the B3849-212-containing region also covers the regions that control the nondisjunction of the maize B-chromosome, in which the proximal euchromatin acts in trans (Lin 1978), and both the centromeric knob and the short arm carry cis-acting control elements (Carlson 1973; Lin 1979). In rye, the nondisjunction control region of B-chromosomes has found to be primarily composed of B-chromosome-enriched transcriptionally active repeats (Klemme et al. 2013). Furthermore, B3849-189 shows high homology with the maize StarkB sequence, which displays transcriptional activity and is located in the B-chromosome region that acts in trans to affect the rate of univalent B-chromosome loss during meiosis (Carlson and Roseman 1992; Lamb et al. 2007). Considering that B3849-212 and B3849-189 are transcribed, the B-related transcripts could be translated into proteins, or their RNA sequences could act to influence the behavior of the maize B-chromosome during cell division.
In summary, we provide evidence that the presence of the B-chromosome in maize can alter the transcription of the A genome and that the B-chromosome harbors few transcriptionally active sequences. All of the identified +1B-related transcripts are similar to sequences on A-chromosomes, supporting the hypothesis that the maize B-chromosome originated from A-chromosomes, as previously suggested (Stark et al. 1996; Page et al. 2001; Cheng and Lin 2003; Lamb et al. 2005; Peng and Cheng 2011). Additionally, various expression effects were observed in these B-related transcripts, implying the existence of transcriptional crosstalk between the B- and A-chromosomes in maize.
References
Alfenito MR, Birchler JA (1993) Molecular characterization of a maize B chromosome centric sequence. Genetics 135:589–597
Banaei-Moghaddam AM, Meier K, Karimi-Ashtiyani R, Houben A (2013) Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell 25:2536–2544
Birchler JA (1979) A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics 92:1211–1229
Birchler JA, Bhadra U, Bhadra MP, Auger DL (2001) Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234:275–288
Carchilan M, Delgado M, Ribeiro T et al (2007) Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19:1738–1749
Carchilan M, Kumke K, Mikolajewski S, Houben A (2009) Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma 118:607–616
Carlson WR (1973) A procedure for localizing genetic factors controlling mitotic nondisjunction in the B chromosome of maize. Chromosoma 42:127–136
Carlson WR (1986) The B chromosome of maize. CRC Crit Rev Plant Sci 3:201–226
Carlson WR, Roseman RR (1992) A new property of the maize B chromosome. Genetics 131:211–223
Cheng YM (2010) Evolution of the heterochromatic regions on maize B long arm based on the sequence structure of CL-repeat variants. Chromosom Res 18:605–619
Cheng YM, Lin BY (2003) Cloning and characterization of maize B chromosome sequences derived from microdissection. Genetics 164:299–310
Cheng YM, Lin BY (2004) Molecular organization of large fragments of maize B chromosome: indication of a novel repeat. Genetics 166:1947–1961
Chilton MD, McCarthy BJ (1973) DNA from maize with and without B chromosomes: a comparative study. Genetics 74:605–614
Coleman JJ, Rounsley SD, Rodriguez-Carres M et al (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5:e1000618
Dhar MK, Friebe B, Koul AK, Gill BS (2002) Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 111:332–340
Donald TM, Leach CR, Clough A, Timmis JN (1995) Ribosomal RNA genes and the B chromosome of Brachycome dichromosomatica. Heredity 74:556–561
Jones RN, Rees H (1967) Genotypic control of chromosome behavior in rye: XI. The influence of B chromosomes on meiosis. Heredity 22:333–347
Jones RN, Rees H (1982) B chromosomes. Academic Press, London
Jones RN, Viegas W, Houben A (2008) A century of B chromosomes in plants: so what? Ann Bot 101:767–775
Klemme S, Banaei-Moghaddam AM, Macas J et al (2013) High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol 199:550–558
Lamb JC, Kato A, Birchler JA (2005) Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113:337–349
Lamb JC, Riddle NC, Cheng YM, Theuri J, Birchler JA (2007) Localization and transcription of a retrotransposon-derived element on the maize B chromosome. Chromosom Res 15:383–398
Leach CR, Houben A, Field B et al (2005) Molecular evidence for transcription of genes on a B chromosome in Crepis capillaries. Genetics 171:269–278
Li B, Li N, Duan XG et al (2010) Generation of marker-free transgenic maize with improved salt tolerance using the FLP/FRT recombination system. J Biotechnol 145:206–213
Lin BY (1972) Synthesis of a set of B-A translocations involving a given segment of chromosome 10. Maize Genet Newsl 46:193–194
Lin BY (1978) Regional control of nondisjunction of the B-chromosome in maize. Genetics 90:613–627
Lin BY (1979) Two new B-10L translocations involved in the control of nondisjunction of the B chromosome in maize. Genetics 92:931–945
Lin BY, Chou HP (1997) Physical mapping of four RAPDs in the B-chromosome of maize. Theor Appl Genet 94:534–538
Lin BY, Peng SF, Chen YJ, Chen HS, Kao CF (1997) Physical mapping of RFLP markers on four chromosome arms in maize using terminal deficiencies. Mol Gen Genet 256:509–516
Lo KL, Lin YP, Chen LJ, Lin BY (2009a) Isolation and characterization of new maize B sequences from a microdissected library. Plant Mol Biol Rep 27:350–354
Lo KL, Peng SF, Chen LJ, Lin BY (2009b) Tandem organization of StarkB element (22.8 kb) in the maize B chromosome. Mol Genet Genomics 282:131–139
Longley AE (1927) Supernumerary chromosomes in Zea mays. J Agric Res 35:769–784
Maluszynska J, Schweizer D (1989) Ribosomal RNA genes in B chromosome of Crepis capillaris detected by nonradioactive in situ hybridization. Heredity 62:59–65
Martis MM, Klemme S, Banaei-Moghaddam AM et al (2012) Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci U S A 109:13343–13346
Muyle A, Zemp N, Deschamps C et al (2012) Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol 10:e1001308
Page BT, Wanous MK, Birchler JA (2001) Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159:291–302
Peng SF, Cheng YM (2011) Characterization of satellite CentC repeats from heterochromatic regions on the long arm of maize B-chromosome. Chromosom Res 19:183–191
Peng SF, Lin YP, Lin BY (2005) Characterization of AFLP sequences from regions of maize B chromosome defined by 12 B-10L translocations. Genetics 169:375–388
Pereira HS, Barão A, Caperta A et al (2009) Rye Bs disclose ancestral sequences in cereal genomes with a potential role in gametophyte chromatid segregation. Mol Biol Evol 26:1683–1697
Randolph LF (1941) Genetic characteristics of the B chromosomes in maize. Genetics 26:608–631
Rhoades MM (1968) Studies on the cytological basis of crossing over. In: Peacock WJ, Brock RD (eds) Replication and recombination of genetic material. Australian Acad Sci, Canberra, pp 229–241
Rhoades MM, Dempsey E (1972) On the mechanism of chromatin loss induced by the B chromosome of maize. Genetics 71:73–96
Roman H (1947) Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32:391–409
Roman H (1948) Directed fertilization in maize. Proc Natl Acad Sci U S A 34:36–42
Ruiz-Estévez M, López-León MD, Cabrero J, Camacho JPM (2012) B-chromosome ribosomal DNA is functional in the grasshopper Eyprepocnemis plorans. PLoS ONE 7:e36600
Stark EA, Connerton I, Bennet ST et al (1996) Molecular analysis of the structure of the maize B-chromosome. Chromosom Res 4:15–23
Staub RW (1987) Leaf striping correlated with the presence of B chromosomes in maize. J Hered 78:71–74
Trifonov VA, Dementyeva PV, Larkin DM et al (2013) Transcription of a protein-coding gene on B chromosomes of the Siberian roe deer (Capreolus pygargus). BMC Biol 11:90
Vuylsteke M, Peleman JD, Van Eijk MJY (2007) AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis. Nat Protoc 2:1399–1413
Ward E (1973) Nondisjunction: localization of the controlling site in the maize B chromosome. Genetics 73:387–391
Yoshida K, Terai Y, Mizoiri S et al (2011) B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet 7:e1002203
Zhou Q, Zhu H, Huang Q et al (2012) Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13:109
Acknowledgments
This work was supported by the National Sciences Council grant of Taiwan (NSC99-2311-B-005-003-MY3 and NSC102-2311-B-005-001-).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
B-chromosome breakpoints and chromosome compositions of four B-10L translocations. (a) The figure was modified from Cheng and Lin (2003). BS, B-chromosome short arm; CK, B-centromere and centromeric knob; PE, proximal euchromatin; DH1-DH4, four distal heterochromatins; DE, distal euchromatin. (b) A translocation heterozygote contains 10, 10-B, and B-10 chromosomes; a tertiary trisomy includes 10, 10, B-10 chromosomes; and a hypoploid carries 10 and 10-B chromosomes. Breakpoints of the B-chromosome are indicated as arrows, and arrowheads indicate the centromeres of chromosome 10 or B-chromosome (GIF 211 kb)
Supplementary Figure 2
Comparison of maize B73 genome sequences and four +1B-related transcripts. Black thin lines represent sequences of the four +1B-related transcripts. Gray arrows indicate the region that shows the highest identity with sequences in the maize B73 inbred line genome database (B73 RefGen_V2 in the MaizeGDB database). The numbers of B73 sequences that have at least 80% identity with the +1B-related transcript are indicated (GIF 37 kb)
Supplementary Figure 3
Southern hybridization of four +1B-related transcripts. Genomic DNA of L289+0B (0B), L289+1B (1B), and L289+9B (9B) was digested with EcoRI, HindIII, BamHI, XbaI, and SacI, and then hybridized with the four +1B-related transcripts B3547-179 (a), B3849-147 (b), B3849-189 (c), and B3849-212(d), respectively. The hollow arrow is representative of B-specific signals, and the solid arrow is representative of signals with B-dosage dependency following the numbers of B-chromosomes. Molecular weights are marked on left (GIF 2771 kb)
Supplementary Figure 4
RT-PCR primers designation of four +1B-related transcripts. Sequences of B3547-179 (a), B3849-147 (b), B3849-189 (c), and 3849–212 (d) were aligned with their top ten homologous sequences in the maize B73 inbred line genome database (B73 RefGen_V2 in the MaizeGDB database). The consensus sequences are highlight in black, and RT-PCR primer binding sites are marked by arrows (GIF 1954 kb)
Supplementary Figure 5
Alignment of genomic and transcribed sequences of three +1B-related transcripts. PCR products of B3849-212 (a), B3849-147 (b), and B3849-189 (c) amplified from genomic DNA (L289+0B, L289+1B, and L289+6B) and cDNA (L289+0B, L289+1B, and L289+9B) were cloned, sequenced, and aligned with their corresponding +1B-related transcript sequences. Conserved nucleotides are shaded with gray and black (GIF 4616 kb)
(GIF 4589 kb)
(GIF 7338 kb)
Supplementary Table 1
(DOC 60 kb)
Supplementary Table 2
(DOC 38 kb)
Supplementary Table 3
(DOC 86 kb)
Rights and permissions
About this article
Cite this article
Lin, HZ., Lin, WD., Lin, CY. et al. Characterization of maize B-chromosome-related transcripts isolated via cDNA-AFLP. Chromosoma 123, 597–607 (2014). https://doi.org/10.1007/s00412-014-0476-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-014-0476-7