Abstract
B chromosomes (Bs) are dispensable components of the genomes of numerous species. To test whether the transcriptome of a host is influenced by Bs, we looked for differences in expression in response to additional Bs. Comparative complementary DNA amplified fragment length polymorphism experiments resulted in the identification of 16 putative B-chromosome-associated transcripts. This comprises 0.7% of the total transcript number and indicates a low activity of Bs. We also provide evidence that B chromosome influences in trans the transcription of A chromosome sequences. The B-specific transcribed sequences B1334, B8149, and B2465 belong to high-copy families with similarity to mobile elements. For all analyzed B-chromosome-derived transcripts, similar A chromosome-encoded sequences were found which supports an A-derived origin of rye B chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
B chromosomes (Bs) are dispensable components of the genomes of numerous plant and animal species. They do not pair with any of the standard A chromosomes (As) at meiosis and have irregular modes of inheritance. As they are dispensable for normal growth, Bs were considered nonfunctional and without any essential genes (reviewed in Jones and Rees 1982; Camacho et al. 2000; Jones and Houben 2003; Jones et al. 2008b). In low numbers, Bs show little or no impact on the hosts. However, increased numbers of Bs might cause phenotypic differences and reduced fertility (reviewed in Jones 1995; Jenkins and Jones 2004; Jones et al. 2008a).
Although the transcriptional activity of Bs has long been of interest, only a few detailed studies have been conducted on this property. Experiments on the transcription behavior of animal Bs support the idea that Bs are genetically inert or weakly active. Autoradiographic studies of tritiated uridine incorporated into spermatocytes of the mouse Apodemus peninsulae (Ishak et al. 1991) and the grasshoppers Myrmeleotettix maculatus and Chorthippus parallelus (Fox et al. 1974) indicated very little or no transcription of Bs. Indirect evidence for some transcription of Bs in the frog Leiopelma hochstetteri (Green 1988) and the fly Simulium juxtacrenobium (Brockhouse et al. 1989) is based on the observation that meiotic Bs form lampbrush structures. More recently, differential display reverse-transcription polymerase chain reactions (RT-PCR) were performed to compare gene expression profiles of mice (Apodemus flavicollis) with and without Bs (Tanic et al. 2005). Three complementary DNA (cDNA) fragments were additionally expressed in mice with Bs compared with animals without Bs, suggesting that the activity of some genes can be directly or indirectly associated with Bs. In canids, the proto-oncogene C-KIT has been mapped to B chromosomes (Graphodatsky et al. 2005), raising the question about its functional significance and activity.
Indirect evidence for transcriptional activity of plant Bs results from comparative analysis of esterase isozyme activity in organisms with and without Bs, for example in Scilla autumnalis (Ruiz-Rejon et al. 1980) and rye (Bang and Choi 1990). In B-positive plants, additional bands were detected by protein electrophoresis. However, in both cases, it is unclear whether the additional bands were caused by a B chromosome gene or whether Bs influenced the transcription behavior of an A-located gene. For grasshoppers, for instance, evidence exists that Bs can alter the expression activity of A-located ribosomal DNA (rDNA) genes (Cabrero et al. 1987; Teruel et al. 2007).
Except for the B-located 45S ribosomal RNA (rRNA) gene of Crepis capillaris, in which one of two B-specific members of the rRNA gene family were weakly transcribed (Leach et al. 2005), there was no direct molecular evidence for transcription of B chromosome genes in plants until the transcriptional activity of B-specific repetitive sequences has been demonstrated. In maize, a retrotransposon-derived high-copy element (Lamb et al. 2007) and, in rye, two repeat families, clustered at the long B chromosome arm, are transcriptionally active in a tissue-specific manner (Carchilan et al. 2007). The function of these B transcripts and the mechanism of transcription of B repeats are unknown at present. It has been hypothesized that these transcripts could have a structural function in the organization and regulation of Bs (Carchilan et al. 2007; Han et al. 2007).
In order to test whether the transcriptome of a host is influenced by Bs, we looked for differences in expression resulting from additional Bs and searched for transcriptionally active rye Bs sequences. Comparative cDNA-amplified fragment length polymorphism (AFLP) identified rare B-derived transcripts. Sequence similarity between A and B chromosome-derived transcripts supports the hypothesis that the rye Bs originated from As of the same species. Our analysis also provides evidence that Bs might influence the transcription of A-located sequences.
Material and methods
Plant material and plant cultivation
Plants with and without Bs from three different inbred lines of rye (Secale cereale L.), 7415, P12, and 2677 (Jimenez et al. 1994; Ortiz et al. 1996), and from hexaploid wheat cv. Lindström with and without introgressed rye Bs (Lindström 1965) were grown at the same temperature, humidity, and light conditions (16-h light, 22ºC day/16ºC night). The Bs of each line are from the same origin (Jimenez et al. 1994). Each plant was checked by chromosome counting, Southern hybridization using a B-specific probe, and PCR using primers specific for the B-specific repeat E3900 (Sandery et al. 1990).
DNA and RNA isolation
DNA was extracted according to Souza (2006) and total RNA was isolated from roots, leaves, and anthers using the Trizol method (Chomczynski and Sacchi 1987). Contaminating genomic DNA was digested with DNase (Roche). Absence of genomic DNA contamination was tested by PCR using E3900-specific primers (supplementary Table 1) on DNAse-treated RNA without reverse-transcription reaction.
cDNA-AFLP analysis and DNA cloning
The cDNA-AFLP method was performed according to Bachem et al. (1996, 1998) starting with PstI- and MseI-digested cDNA (see supplementary “Material and methods” for details). Briefly, the cDNA was synthesized starting with 1 µg of total RNA using the Super SMART cDNA synthesis kit (Clontech). For each genotype, RNA was pooled from at least eight plants. The restricted cDNA fragments were ligated with PstI and MseI adapters (supplementary Table 2). The ligated fragments were used for PCR preamplification with primers MseI 00 and PstI 03 (supplementary Table 3). Diluted preamplified DNA was used for final selective PCR amplification according to Vos et al. (1995). MseI and PstI primers containing three selective nucleotides at the 3′ end were used (supplementary Table 3). PCR products were separated in 5% acrylamide gels using a sequencing gel electrophoresis apparatus. Silver staining, performed according to Sanguinetti et al. (1994) with slight modifications, was used to visualize DNA fragments. Fragments of interest were excised from the gel and DNA was eluted and subsequently reamplified using the same PCR conditions and primer combinations as those used for selective amplification. PCR products were checked by 1.2% Tris–acetate–ethylenediaminetetraacetic acid (EDTA) agarose gel electrophoresis, cloned into pGEM-T Easy vector (Promega), and sequenced (for details, see supplementary “Material and methods”).
Sequence analysis
DNA sequences were processed using the software “Editseq” and aligned using “MegAlign” (Lasergene 6). The following databases were used for sequence comparisons, Basic Local Alignment Search Tool (NCBI, http://www.ncbi.nlm.nih.gov/BLAST/), TIGR (http://www.tigr.org/tdb/e2k1/plant.repeats/), and GrainGene (http://wheat.pw.usda.gov/GG2/blast.shtml).
Reverse transcriptase PCR and Northern blot hybridization
Seventy-five nanograms of cDNA from leaves was used for each RT-PCR reaction. Primers were designed specifically for the candidate sequences (supplementary Table 1). The PCR conditions were: 3 min 95ºC; 30 cycles: 30 s 95ºC, 30-s annealing (temperatures are indicated in supplementary Table 1), 2 min 72ºC. The products were separated on 1.2% Tris–acetate–ethylenediaminetetraacetic acid agarose gels.
For Northern hybridization, 20-μg RNA from roots, leaves, and anthers were blotted onto a Hybond-N+ membrane (GE Healthcare). Prehybridization and hybridization were done in Church buffer (7% sodium dodecyl sulfate (SDS), 10 mM EDTA, 0.5 M phosphate buffer, pH 7.2) at 64°C. Equal loading of RNA samples after spectrophotometric measurement was monitored by gel electrophoresis and ethidium bromide staining. The probes were labeled using HexaLabel™ DNA Labeling Kit (Fermentas). After overnight hybridization, the membranes were washed twice in 40 mM phosphate buffer (pH 7.0) containing 1% SDS and 2 mM EDTA at 65°C. Radioactivity was visualized using either X-ray films or a phosphoimager (Fuji).
Restriction digestion, gel electrophoresis of genomic DNA, and Southern hybridization
Southern hybridization was performed according to (Sambrook et al. 1989). Ten micrograms of genomic DNA was restricted with the restriction enzymes: EcoRI, PstI, DraI, or XbaI (Fermentas). The digested DNA was size-fractionated by gel electrophoresis and Southern blotted. Prehybridization, probe labeling, and hybridization were performed as described for Northern hybridization.
Fluorescence in situ hybridization
The probes were labeled with biotin-16-dUTP or digoxigenin-11-dUTP by nick translation. Preparation of the chromosomes and subsequent in situ hybridization were performed according to Houben et al. (2006). The fluorescent signals were recorded by a cooled sensitive charge-coupled device camera and pseudocolored using Adobe Photoshop software.
Results
B chromosomes influence the transcription of rye weakly
To determine the general transcription activity of rye Bs, comparative cDNA-AFLP analyses were done on three different near isogenic rye 0B/+B lines namely: 7415, P 12, and 2677, denoted 1, 2, and 3, respectively. These lines were generated and described by Jimenez et al. (1994) and Ortiz et al. (1996). Extraneous sources of transcription variation were minimized by using pooled cDNA samples of at least eight plants and identical growth conditions. The size of amplified cDNA fragments ranged from 80 to 1,200 base pairs. Transcription profiles were compared between plants with 2 Bs and without Bs for each individual line and across the three different rye lines.
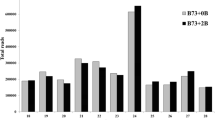
The uniform profile of the main transcripts among the three different rye lines demonstrated that the cDNA samples used for the AFLP analysis were comparable. As shown in Fig. 1, the variability of transcripts due to the presence or absence of Bs was lower than the variability among the three inbred lines. For quantification, only clearly visible DNA fragments were considered. For example, 43 bands were counted for the primer combination 1334 (Fig. 1). PCR bands in all three inbred lines were screened and additional bands present in +B or 0B material were counted (supplementary Table 5). PCR products of similar size to that of the negative control were omitted.
cDNA-AFLP fingerprint obtained after separation of PCR products amplified by primer combination 1334. 1, 2, and 3 correspond to the different rye inbred lines used without (0B) or with Bs (+B). Positions of bands unique to Bs are arrowed. Note that adaptors and primers present in the amplification reaction can result in a number of unspecific PCR products. Therefore, for each restriction ligation and amplification step, negative controls without template DNA were included (lane k)
Sixty-three primer combinations (listed in supplementary Tables 3 and 4) amplified around 2,286 clearly visible bands per rye line. A total of 112 extra bands (4.9% of the average number of bands per rye line) were found in +B material from either 1, 2, or all 3 rye lines. However, only 16 of these bands (0.7% of the average number of bands per rye line) of different intensity and size were consistently found in all three lines. These 16 bands were considered as putative B-derived transcript candidates. The extra bands associated with Bs, which were detected in one or two rye lines only, were not analyzed further. Since Bs might also influence the transcription of the A chromosome complement, the number of additional bands in 0B material was also determined. A total of 132 extra bands (5.8%) were found in material without B chromosomes of 1, 2, or all 3 rye lines. However, only four 0B extra bands (0.18% of average number of bands per rye line) were consistently found in all three lines. Thus, our cDNA-AFLP data suggest that B chromosomes are capable of weakly influencing the transcription activity of the genome.
Some B transcripts are encoded by members of mobile element families
Nine out of 16 +B extra bands, which were present in the +B material of all three inbred rye lines were isolated and sequenced (listed in Table 1). Four of the cloned B-associated transcripts (B1334, B8149, B2465, and B2565) were similar to different types of mobile elements. Fragments B1553 and B2442 displayed high similarity with zeaxanthin epoxidase and the Mal-like protein of Triticum aestivum, respectively. The remaining B extra bands (B1559, B2448, and B2453) were very similar to T. aestivum cDNA sequences of unknown function.
We then analyzed the genomic organization, chromosomal position, and transcription of the mobile element-like B-associated sequences (B1334, B8149, and B2465). As the sequences of rye A and B chromosomes are highly similar (Timmis et al. 1975), wheat–rye B chromosome addition lines (Lindström 1965) with and without Bs were included in this study.
Southern hybridization indicated that B1334 (suppl. Fig. 1), B2465 (Fig. 2, suppl. Fig. 2) and B8149 (Fig. 3, suppl. Fig. 3) sequences are members of high-copy sequence families with multiple locations in the rye genome. The hybridization patterns of B1334 and B2465 with DNA of rye with and without Bs were similar. No cross-hybridization of rye B1334 was detected with genomic DNA of wheat. However, DNA of wheat plants with additional rye Bs displayed hybridization patterns resembling those of rye, although of lower hybridization intensity (suppl. Fig. 1). Southern hybridization of B2465 revealed also cross-hybridization with 0B wheat and a few extra bands were detected in DNA of wheat with Bs restricted with BamHI (Fig. 2, arrowed). The hybridization patterns of B8149 differed among the rye and wheat lines regardless of the presence or absence of Bs (Fig. 3, suppl. Fig. 3). Furthermore, a few additional bands were consistently found in rye and wheat with Bs. The polymorphic hybridization pattern suggests transcriptional activity of B8149-like sequences in rye and wheat genomes.
Southern hybridization of rye probe B2465 with EcoRI-, BamHI-, XbaI-, and DraI-digested genomic rye and wheat DNA from plants with (+B) and without Bs (0B). 1, 2, and 3 are three different rye inbred lines. W—Lindström wheat with (+B) and without Bs (0B). Note identical hybridization patterns of 0B and +B rye and additional bands in wheat plants with B chromosomes (arrowed)
Southern hybridization of probe B8149 with EcoRI- and BamHI-digested genomic rye and wheat DNA from plants with (+B) and without Bs (0B). 1, 2, and 3 are three different rye inbred lines. W—Lindström wheat with (+B) and without Bs (0B). Note the highly polymorphic pattern. Additional B-specific restriction bands are arrowed
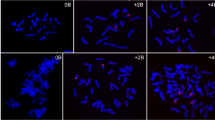
Fluorescence in situ hybridization (FISH) analysis revealed an almost uniform distribution of every candidate sequence along all rye A and B chromosomes (Fig. 4). Simultaneous in situ hybridization with the differentially labeled B-specific high-copy probe D1100 (Blunden et al. 1993) confirmed the identification of Bs. Reduced cross-hybridization of B1334 and B8149 was found in the terminal heterochromatic regions of As and Bs. The enhanced B1334-specific hybridization signals along Bs suggest an accumulation of this mobile element-like sequence in Bs. Hence, our Southern hybridization and FISH data indicate that the B-derived transcripts B1334, B2465, and B8149 are encoded by members of high-copy families with similar sequence organization on rye A and B chromosomes.
Fluorescence in situ hybridization of mitotic metaphase cells of rye containing Bs (arrowed) with the B-specific repeat D1100 (in green) and with the probes: B1334, B8149, and B2465 (in red). The chromosomes are counterstained with DAPI (in blue). The insets (in merge) show enlarged Bs and the chromosomal position of the D1100-positive region are indicated by arrowheads. Note very weak cross-hybridization of B1334 and B8149 with the D1100-positive region and stronger hybridization signals of B1334 along B chromosomes
To confirm transcription activity and B specificity of the candidate transcripts, RT-PCR and Northern analysis were conducted. RT-PCR was done on rye and wheat plants with 2Bs and without Bs using primers that are complementary to regions with the lowest level of similarity between B1334, B8149, B2465, and the corresponding published database sequences with high similarity (listed in supplementary Table 1). RT-PCR using B8149- and B1334-specific primers yielded in products of the expected size from cDNA template obtained only from pooled material with Bs of all three rye inbred lines (Fig. 5). The almost equal amounts of PCR products suggest similar levels of B chromosome transcription irrespective of the A chromosome genotype. Conversely, B1334 activity of Bs added to wheat was significantly reduced compared with B8149. Decreasing the PCR stringency resulted in the amplification of 0B products as was the case in +B material. The sequences of RT-PCR products resulting from 0B cDNA closely resembled the PCR products of +B cDNA (98–99%) but with some SNPs (Fig. 6, supplementary Fig. 4a, b). Therefore, B1334- and B8149-like sequences located on A chromosomes are transcriptionally active too.
RT-PCR analysis of B8149, B1334, and B2465 on RNA isolated from leaves of the three rye inbred lines 1, 2, and 3 and Lindström wheat (W) with (+B) and without Bs (0B). Note the faint band in Lindström wheat with Bs (in B1334, arrowed). To confirm that equal amounts of cDNA were present, RT-PCR was carried out with primers specific for the elongation factor eEF1 alpha (EF alpha) designed on a barley cDNA sequence with accession number Z50789 (Nielsen et al. 1997). Negative control using RNA of all samples without an initial reverse-transcription step to demonstrate the absence of genomic DNA contamination (no RT)
Alignment of a part of B1334-like sequences amplified from cDNA of plants with (+B) and without (0B) B chromosomes. Single-nucleotide polymorphisms are underlined. GenBank sequence AF175285 shows the highest similarity. Sequences B1334-1–3 are sequences obtained via cDNA-AFLP and sequences B1334-4–12 are sequences obtained via RT-PCR
PCR on genomic DNA was then done to test whether B8149 and B1334 transcripts are encoded by Bs. B-specific amplification was found for B8149 (Fig. 7), but, for B1334, PCR products were observed in both 0B and +B material (data not shown). This confirms that B1334-like sequences are located on both types of chromosomes.
A rye genotype-dependent transcription behavior was found for B2465. PCR products of expected size were observed for +B cDNA of rye lines 1 and 3 as well as Lindström wheat. However, PCR products were observed in 0B material of rye line 1. Most notably, B2465 in rye line 2 was much more transcribed in 0B material than in +B material. To verify the apparent downregulation of B2465 due to the presence of Bs, RT-PCR experiments were conducted on leaf RNA isolated from single 0B and +B plants of the same line. Results shown in Fig. 8 confirm downregulation of B2465 transcripts in the presence of Bs. Hence, at the transcript level, a crosstalk might exist in trans between A and B chromosomes.
RT-PCR analysis of B2465 on leaf RNA isolated from single plants of the rye inbred lines 2 with (+B) and without B chromosomes (0B). As positive control, a RT-PCR was done with a B8149-specific primer pair and with an EF1alpha-specific primer pair (EF alpha) to demonstrate equal amounts of cDNA. Negative control using RNA of all samples without an initial reverse-transcription step to demonstrate the absence of genomic DNA contamination (no RT)
To further characterize the transcribed sequences, Northern hybridization experiments were conducted on total RNA isolated from roots, leaves, and anthers of 0B and +B plants of rye line 1. The hybridization patterns of all three probes did not differ between RNA isolated from plants with and without Bs, suggesting that sequences similar to those isolated from Bs are also transcribed from A chromosomes (Fig. 9). Probe B1334 detected a continuum of transcripts ranging in size from more than 6 kb to less than 0.2 kb in all types of tissues, although of highest intensity in anther tissue. B8149- and B2465-like sequences showed highest transcriptional activity in leaf tissue and almost no or weak activity in root and anther tissue.
Discussion
In this study, the general transcription activity of Bs was analyzed by comparative cDNA-AFLP of plants with and without Bs from three isogenic rye lines. Rye is an outbreeder and additional polymorphic transcripts coming from genetically diverse As could be expected. Therefore, three lines were employed to distinguish transcripts associated with B chromosomes from those derived from A chromosome polymorphism. Since rye Bs are mainly composed of DNA sequences in common with those of the As (Rimpau and Flavell 1975; Timmis et al. 1975) apart from the terminal part of the long arm (Tsujimoto and Niwa 1992; Wilkes et al. 1995; Houben et al. 1996), the most widely accepted view is that they are derived from the As. The processes that gave rise to rye Bs during evolution remain unclear. If rye Bs were derived from A chromosomes, a high degree of sequence similarity might also exist between A- and B-encoded transcripts. Therefore, the cDNA-AFLP method (Bachem et al. 1996), which allows the discrimination of even single-nucleotide polymorphisms, was employed to characterize the transcriptome in response to the additional B chromosomes and to determine whether rye Bs have any transcriptionally active sequences. The apparent conflict between the existence of B-specific transcripts, which were identified by cDNA-AFLP, and the cross-hybridization of these AFLP-derived sequences with 0B genomic DNA is because the hybridization methods did not discriminate single-nucleotide differences between the A- and B-derived sequences.
Using 63 primer combinations, 112 extra bands (4.9% of the average number of bands per rye line) were found in plants with Bs of either 1, 2, or all 3 rye lines. However, only 16 B extra bands (0.7% of the average number of bands per rye line) of different intensity were consistently found in all three rye lines. Assuming that the transcriptome of a 0B rye plant is equally derived from all seven A chromosome pairs, each A chromosome type could encode around 14% of the transcriptome. Not unexpectedly, this number is much higher than the 0.7% of B-associated transcripts and confirms a low transcription activity of B chromosomes. However, due to the limited number of primer combinations tested for AFLP and the likely high sequence similarity of A- and B-derived transcripts, our analysis might underestimate the number of B-encoded transcripts. Furthermore, considering an A-derived origin of rye Bs, the total activity of identical A- and B-located sequences does not necessarily increase due to the presence of extra Bs since many target loci on varied chromosomal segments (like aneuploids) exhibit dosage compensation (reviewed in Birchler et al. 2005).
The number of additional cDNA-AFLP bands in 0B material was determined to test whether Bs can alter the expression of A-localized sequences as previously suggested (e.g., Ruiz-Rejon et al. 1980). A total of 132 extra bands (5.8% of the average number of bands per rye line) were found among plants without Bs, but only four of these bands (0.18%) were common to all three of these lines. The retrotransposon-like element B2465 represents an example in which transcription is downregulated in 0B plants in a genotype-dependent manner. We suggest that Bs may result in a variety of epigenetic effects, including the differential regulation of A-localized transposable elements through mechanisms such as homology-dependent RNA interference pathways (Slotkin and Martienssen 2007). Since the rye B most likely originated from the A chromosome complement, it seems to be reasonable that the transcription alterations of A-located sequences are caused by homology-dependent mechanisms (Matzke et al. 2002), as has been proposed for the remodeling of gene activity in newly formed hybrids and allopolyploids (Comai 2005).
Another hypothesis for explaining how the Bs could be exerting control on the rest of the genome is through their effects on the spatial organization of the genome itself. Recent work suggests that spatial positioning of genes and chromosomes can influence gene expression (Misteli 2007). Indeed, Delgado and colleagues (1995, 2004) observed that in interphase nuclei of rye, A chromosomal ribosomal DNA displays a more compact distribution in cells with Bs compared to cells without Bs. A more compact distribution of rDNA sites suggests a lower level of rRNA gene activity. A similar effect of an almost gene-deficient chromosome has been demonstrated for Drosophila. Lemos et al. (2008) demonstrated that the Y chromosome of Drosophila melanogaster regulates the activity of hundreds of genes located on other chromosomes.
Two out of nine candidates, B1553 and B2442, showed DNA sequence similarity to known genes such as zeaxanthin epoxidase and the Mal-like protein (T. aestivum), respectively. However, due to the very high level of sequence similarity of A- and B-located sequences, we could not confirm B specificity via Southern and Northern hybridization or RT-PCR (data not shown).
The B-associated transcripts B1334, B8149, and B2465, which revealed similarity with parts of mobile elements, are present in high-copy numbers in the rye genome. However, it is not known if transposition still occurs. For B2465 and B8149, the transcript level was much lower and tissue-type specific; probably, they are degenerated nonfunctional mobile elements. Alternatively, transcription of B2465, B1334, and B8149 could also be explained by a read through of these sequences next to other active mobile elements, as has been reported for the transcription of a repeat family in Arabidopsis thaliana that is regulated by a retrotransposon (May et al. 2005).
The transcriptionally active sequence B1334 was similar to the transposase region of the transposon-like gene “Revolver” (Tomita et al. 2008). We confirmed the data of Tomita et al. (2008) by identifying an unusually high transcription activity of this element and high-copy number in rye with an almost complete absence in 0B Lindström wheat. However, the similarity of Southern hybridization patterns between 0B and +B rye and wheat-carrying Bs suggests a similar organization of B1334-like elements on As and Bs of rye. In contrast to the data for B8149 and B2465, in situ hybridization revealed an accumulation of B1334 signals on B chromosomes. Thus, an accumulation of “Revolver”-like elements occurred on Bs. Indeed, a B-specific accumulation of mobile elements has been reported previously for other organisms such as Alburnus alburnus (Ziegler et al. 2003). The accumulation of mobile elements on Bs can have functional and evolutionary implications. Because Bs encode a low number of genes, their accumulation does not interfere with the normal life of the plant. Furthermore, a rapid accumulation of repeats and transposons on a de novo formed B chromosome could reduce its recombination ability with the A chromosome complement and, therefore, lead to a separate evolution of the B chromosome (Camacho et al. 2000).
In summary, our data demonstrate that the B chromosomes in rye encode few transcriptional active sequences and that the B chromosome also downregulates in trans the activity of sequences located on A chromosomes in genotype-dependent manner. Because all identified B transcripts were similar to sequences located on A chromosomes, the Bs of rye should have originated from A chromosomes of the same species as previously suggested (Timmis et al. 1975, Tsujimoto and Niwa 1992; Wilkes et al. 1995; Houben et al. 1996).
References
Bachem CWB, vander Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9:745–753
Bachem CWB, Oomen RJFJ, Visser RGF (1998) Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol Biol Rep 16:157–173
Bang J-W, Choi HW (1990) Genetic analysis of esterase isozymes in rye (Secale cereale L.). Korean J Genetics 12:87–94
Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21:219–226
Blunden R, Wilkes TJ, Forster JW, Jimenez MM, Sandery MJ, Karp A, Jones RN (1993) Identification of the E3900 family, a 2nd family of rye B chromosome specific repeated sequences. Genome 36:706–711
Brockhouse C, Bass JAB, Feraday RM, Straus NA (1989) Supernumerary chromosome evolution in the Simulium vernum Group (Diptera, Simuliidae). Genome 32:516–521
Cabrero J, Alche JD, Camacho JPM (1987) Effects of B chromosomes on the activity of nucleolar organizer regions in the grasshopper Eyprepocnemis plorans—activation of a latent nucleolar organizer region on a B chromosome fused to an autosome. Genome 29:116–121
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. Philos Trans Roy Soc B 355:163–178
Carchilan M, Delgado M, Ribeiro T, Costa-Nunes P, Caperta A, Morais-Cecilio L, Jones RN, Viegas W, Houben A (2007) Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19:1738–1749
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem 162:156–159
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846
Delgado M, Moraiscecilio L, Neves N, Jones RN, Viegas W (1995) The influence of B chromosomes on rDNA organization in rye interphase nuclei. Chromosome Res 3:487–491
Delgado M, Caperta A, Ribeiro T, Viegas W, Jones RN, Morais-Cecilio L (2004) Different numbers of rye B chromosomes induce identical compaction changes in distinct A chromosome domains. Cytogenet Genome Res 106:320–324
Fox DP, Hewitt GM, Hall DJ (1974) DNA-replication and RNA transcription of euchromatic and heterochromatic chromosome regions during grasshopper meiosis. Chromosoma 45:43–62
Graphodatsky AS, Kukekova AV, Yudkin DV, Trifonov VA, Vorobieva NV, Beklemisheva VR, Perelman PL, Graphodatskaya DA, Trut LN, Yang FT, Ferguson-Smith MA, Acland GM, Aguirre GD (2005) The proto-oncogene C-KIT maps to canid B-chromosomes. Chromosome Res 13:113–122
Green DM (1988) Cytogenetics of the endemic New Zealand frog, Leiopelma hochstetteri—extraordinary supernumerary chromosome variation and a unique sex chromosome system. Chromosoma 97:55–70
Han F, Lamb JC, Yu W, Gao Z, Birchler JA (2007) Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 19:524–533
Houben A, Kynast RG, Heim U, Hermann H, Jones RN, Forster JW (1996) Molecular cytogenetic characterization of the terminal heterochromatic segment of the B-chromosome of rye (Secale cereale). Chromosoma 105:97–103
Houben A, Orford SJ, Timmis JN (2006) In situ hybridization to plant tissues and chromosomes. Methods Mol Biol 326:203–218
Ishak B, Jaafar H, Maetz JL, Rumpler Y (1991) Absence of transcriptional activity of the B chromosomes of Apodemus peninsulae during pachytene. Chromosoma 100:278–281
Jenkins G, Jones RN (2004) B chromosomes in hybrids of temperate cereals and grasses. Cytogenet Genome Res 106:314–319
Jimenez MM, Romera F, Puertas MJ, Jones RN (1994) B chromosomes in inbred lines of rye (Secale cereale L).1. Vigor and fertility. Genetica 92:149–154
Jones RN (1995) Tansley review no 85—B chromosomes in plants. New Phytol 131:411–434
Jones N, Houben A (2003) B chromosomes in plants: escapees from the A chromosome genome? Trends Plant Sci 8:417–423
Jones RN, Rees H (1982) B chromosomes. London, Academic, p 255
Jones RN, Viegas W, Houben A (2008a) A century of B chromosomes in plants: so what? Ann Bot 101:767–775
Jones RN, Gonzalez-Sanchez M, Gonzalez-Garcia M, Vega JM, Puertas MJ (2008b) Chromosomes with a life of their own. Cytogenet Genome Res 120:265–280
Lamb JC, Riddle NC, Cheng YM, Theuri J, Birchler JA (2007) Localization and transcription of a retrotransposon-derived element on the maize B chromosome. Chromosome Res 15:383–398
Leach CR, Houben A, Field B, Pistrick K, Demidov D, Timmis JN (2005) Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics 171:269–278
Lemos B, Araripe LO, Hartl DL (2008) Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319:91–93
Lindström J (1965) Transfer to wheat of accessory chromosomes from rye. Hereditas 54:149–155
Matzke MA, Aufsatz W, Kanno T, Mette MF, Matzke AJ (2002) Homology-dependent gene silencing and host defense in plants. Adv Genet 46:235–275
May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA (2005) Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1:e79
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800
Nielsen PS, Kleinhofs A, Olsen OA (1997) Barley elongation factor 1 alpha: genomic organization, DNA sequence, and phylogenetic implications. Genome 40:559–565
Ortiz M, Puertas MJ, Jimenez MM, Romera F, Jones RN (1996) B-chromosomes in inbred lines of rye (Secale cereale L). 2. Effects on metaphase I and first pollen mitosis. Genetica 97:65–72
Rimpau J, Flavell RB (1975) Characterization of rye B chromosome DNA by DNA–DNA hybridization. Chromosoma 52:207–217
Ruiz-Rejon M, Posse F, Oliver JL (1980) The B-chromosome system of Scilla autumnalis (Liliaceae) - Effects at the isoenzyme level. Chromosoma 79:341–348
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sandery MJ, Forster JW, Blunden R, Jones RN (1990) Identification of a family of repeated sequences on the rye B-chromosome. Genome 33:908–913
Sanguinetti CJ, Neto ED, Simpson AJG (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:914
Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285
Souza ED (2006) Eigenes Protokoll statt DNA-Kit. Protokoll für die Extraktion von DNA aus Blättern. Labor J 7:58
Tanic N, Vujosevic M, Dedovic-Tanic N, Dimitrijevic B (2005) Differential gene expression in yellow-necked mice Apodemus flavicollis (Rodentia, Mammalia) with and without B chromosomes. Chromosoma 113:418–427
Teruel M, Cabrero J, Perfectti F, Camacho JP (2007) Nucleolus size variation during meiosis and NOR activity of a B chromosome in the grasshopper Eyprepocnemis plorans. Chromosome Res 15:755–765
Timmis JN, Ingle J, Sinclair J, Jones RN (1975) Genomic quality of rye B chromosomes. J Exp Bot 26:367–378
Tomita M, Shinohara K, Morimoto M (2008) Revolver is a new class of transposon-like gene composing the Triticeae genome. DNA Res 15:49–62
Tsujimoto H, Niwa K (1992) DNA structure of the B chromosome of rye revealed by in situ hybridization using repetitive sequences. Jpn J Genet 67:233–241
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP—a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wilkes TM, Francki MG, Langridge P, Karp A, Jones RN, Forster JW (1995) Analysis of rye B chromosome structure using fluorescence in-situ hybridization (FISH). Chromosome Res 3:466–472
Ziegler CG, Lamatsch DK, Steinlein C, Engel W, Schartl M, Schmid M (2003) The giant B chromosome of the cyprinid fish Alburnus alburnus harbours a retrotransposon-derived repetitive DNA sequence. Chromosome Res 11:23–35
Acknowledgements
We are grateful to O. Weiss (IPK, Germany) for excellent technical assistance, as well as R. Pickering and I. Schubert for critically reading the manuscript. We would like to thank M. Puertas (Spain), W. Viegas (Portugal), and R. N. Jones (UK) for providing the valuable plant material. MC and AH were supported by grants of the IPK (Gatersleben) and the DFG (HO1779/10-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Comai
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Material and Methods
cDNA-amplified fragment length polymorphism (AFLP) analysis and DNA cloning (DOC 29 kb)
Suppl Fig. 1
Southern hybridization of probe B1334 with XbaI-, DraI-, BamHI-, and EcoRI-digested genomic rye DNA from plants with (+B) and without Bs (0B). 1, 2, 3 are the three different rye inbred lines. W—Lindström wheat with (+B) and without Bs (0B). Note identical hybridization patterns of 0B and +B rye and wheat plants with B chromosomes (GIF 238 KB)
Suppl Fig. 2
Southern hybridization of probe rye B2465 with XbaI-, BamHI-, and EcoRI-digested genomic rye DNA from plants with (+B) and without Bs (0B). 1, 2, 3 are the three different rye inbred lines. Note identical hybridization patterns of 0B and +B rye plants (GIF 119 KB)
Suppl Fig. 3
Southern hybridization of probe B8149 with XbaI- and DraI-digested genomic rye and wheat DNA from plants with (+B) and without Bs (0B). 1, 2, 3 are three different rye inbred lines. W—Lindström wheat with (+B) and without Bs (0B). Note the highly polymorphic hybridization pattern. Additional B-specific restriction bands are arrowed (GIF 145 KB)
Suppl. Fig. 4a
Alignment of B1334-like sequences amplified from cDNA of plants with (+B) and without (0B) B chromosomes. Single-nucleotide polymorphisms are highlighted in red. GenBank sequence AF175285 shows the highest similarity. Sequences B1334-1–9 are sequences obtained via cDNA-AFLP and sequences B1334-10–23 are sequences obtained via RT-PCR (DOC 48.5 kb)
Suppl. Fig. 4b
Alignment of B8149-like sequences amplified from cDNA of plants with (+B) and without (0B) B chromosomes. Single-nucleotide polymorphisms are highlighted in red. Sequences B8149-1–24 are sequences obtained by RT-PCR (DOC 63 kb)
Suppl. Table 1
PCR primers used (DOC 45 kb)
Suppl. Table 2
AFLP adaptor sequences (DOC 26 kb)
Suppl. Table 3
List of AFLP primer sequences (Keygene). Selective nucleotides are highlighted in red. http://wheat.pw.usda.gov/ggpages/keygeneAFLPs.html (DOC 40 kb)
Suppl. Table 4
Primer combinations used for cDNA-AFLP analysis of 0B/+B plants. (DOC 73 kb)
Suppl. Table 5
Total number of cDNA-AFLP bands and of additional bands present in 0B- or +B-positive plants only. *weak, **middle, ***strong intensity of band (DOC 137 kb)
Rights and permissions
About this article
Cite this article
Carchilan, M., Kumke, K., Mikolajewski, S. et al. Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma 118, 607–616 (2009). https://doi.org/10.1007/s00412-009-0222-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0222-8