Abstract
The spatial chromatin organisation and molecular interactions within and between chromatin domains and chromosome territories (CTs) are essential for fundamental processes such as replication, transcription and DNA repair via homologous recombination. To analyse the distribution and interaction of whole CTs, centromeres, (sub)telomeres and ~100-kb interstitial chromatin segments in endopolyploid nuclei, specific FISH probes from Arabidopsis thaliana were applied to 2–64C differentiated leaf nuclei. Whereas CTs occupy a distinct and defined volume of the nucleus and do not obviously intermingle with each other in 2–64C nuclei, ~100-kb sister chromatin segments within these CTs become more non-cohesive with increasing endopolyploidy. Centromeres, preferentially located at the nuclear periphery, may show ring- or half-moon like shapes in 2C and 4C nuclei. Sister centromeres tend to associate up to the 8C level. From 16C nuclei on, they become progressively separated. The higher the polyploidy level gets, the more separate chromatids are present. Due to sister chromatid separation in highly endopolyploid nuclei, the centromeric histone variant CENH3, the 180-bp centromeric repeats and pericentromeric heterochromatin form distinct subdomains at adjacent but not intermingling positions. The (sub)telomeres are frequently associated with each other and with the nucleolus and less often with centromeres. The extent of chromatid separation and of chromatin decondensation at subtelomeric chromatin segments varies between chromosome arms. A mainly random distribution and similar shapes of CTs even at higher ploidy levels indicate that in general no substantial CT reorganisation occurs during endopolyploidisation. Non-cohesive sister chromatid regions at chromosome arms and at the (peri)centromere are accompanied by a less dense chromatin conformation in highly endopolyploid nuclei. We discuss the possible function of this conformation in comparison to transcriptionally active regions at insect polytene chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interactions between chromatin segments in interphase nuclei are required for such basic biological processes as transcription, replication and DNA repair. Both transcription and replication are thought to proceed in distinct transcription and replication factories which require chromatin fibre movements. Depending on tissue and developmental stage, these processes may reorganise the 3D architecture of interphase nuclei (Chakalova et al. 2005; Chakalova and Fraser 2010; Ferrai et al. 2010; Misteli and Soutoglou 2009; Papantonis and Cook 2010). Additionally, the DNA quantity reflected by the ploidy level may also influence the interphase nuclei architecture. Specialised tissues in plants and animals may contain endopolyploid nuclei, however, even if endoploidisation was hypothesised to provide a mechanism for increasing cell size and gene transcription (Barow 2006; Sugimoto-Shirasu and Roberts 2003) the biological significance of endoreduplication is still under debate. Therefore, examination of interphase chromatin arrangement may contribute to better understand this phenomenon.
In all eukaryotes analysed so far by cytological methods, interphase chromosomes are arranged in distinct chromosome territories (CTs) (Cremer and Cremer 2010). Although mainly compactly organised, CTs may show fuzzy boarders and a certain degree of overlapping and/or intermingling with adjacent CTs (Branco and Pombo 2006) which may allow interchromosomal interactions.
Recent molecular studies based on three-dimensional genome-wide mapping of chromatin interactions (chromosome conformation capture, 3–5C) confirm the presence of CTs and interactions within and between them (Cope et al. 2010; Dekker et al. 2002; Lieberman-Aiden et al. 2009; Dostie et al. 2007; Zhao et al. 2006).
The fractal globule has been proposed as a model of chromatin architecture in interphase nuclei. It is the only statistical polymer model consistent with chromosome conformational capture data and observations obtained by fluorescence in situ hybridisation (FISH). It explains the formation of CTs and the occurrence of intra- and interchromosomal short- and long-range interactions (Mirny 2011).
Although chromosome organisation varies between different cell types (Parada et al. 2004), a radial arrangement of CTs with gene-dense chromosomes located more internally and gene-poor ones closer to the nuclear periphery was described for spherical nuclei (Boyle et al. 2001; Cremer et al. 2001; Croft et al. 1999). Interestingly, this organisation seems to be conserved in most vertebrates (Berchtold et al. 2011; Küpper et al. 2007; Koehler et al. 2009; Neusser et al. 2007). For mammals and Arabidopsis thaliana, the relative positioning of chromosomes was found to be partially transmitted through mitosis and maintained at least transiently in a mirror-symmetrical pattern in sister nuclei (Berr and Schubert 2007; Essers et al. 2005; Gerlich et al. 2003; Thomson et al. 2004; Walter et al. 2003).
Whether gene density reflects transcriptional activity and therefore influences CT positioning is still a matter of debate. Whereas several authors describe an internal positioning of actively transcribed genes (Kozubek et al. 2002; Lukasova et al. 2002; Scheuermann et al. 2004; Zink et al. 2004), the location of highly expressed genes was also found at the nuclear periphery (Brown et al. 2006; Küpper et al. 2007).
Several studies in mammals and plants indicate that transcriptional activation induces chromatin decondensation and out-looping of chromatin fibres from their CTs (Wegel and Shaw 2005; Wegel et al. 2005, 2009).
In addition to functional constraints, topological factors may influence chromosome configurations in interphase nuclei. In many eukaryotes as a relic of anaphase movement, centromeres cluster at one pole whereas telomeres localise at the opposite pole forming the so-called Rabl orientation (Rabl 1885). Rabl orientation as found in meristems and even in differentiated tissue of several Triticeae species (Dong and Jiang 1998; Schubert and Shaw 2011; Schubert et al. 2011) could mediate interaction of distinct homologous and heterologous chromatin regions.
Random but also preferential associations between homologues were reported in many eukaryotes. A close spatial association of homologues was found in somatic cells of Drosophila (Csink and Henikoff 1998; Fung et al. 1998; Hiraoka et al. 1993) possibly as a prerequisite for transvection (Coulthard et al. 2005; Duncan 2002). A non-random association was reported for a pair of barley substitution chromosomes in wheat tapetum and premeiotic nuclei (Aragon-Alcaide et al. 1997) and for homologues in specific differentiated human cell types (Chandley et al. 1996; Nagele et al. 1999). Furthermore, in murine hematopoietic cells, homologues also tend to associate (Rajapakse et al. 2009). Conversely, rye chromosome pairs added to hexaploid wheat are mostly not associated in root tip cells (Corredor et al. 2005), and homologous CTs in human cancer cells are clearly apart from each other (Heride et al. 2010).
In Arabidopsis species, CT arrangement and somatic homologous pairing in interphase nuclei occur mainly at random. Only the NOR-bearing CTs associate more often than random due to the formation of a joint nucleolus (Berr et al. 2006; Pecinka et al. 2004).
Berr and Schubert (2007) demonstrated a similar arrangement of whole Arabidopsis CTs in differentiated and meristematic cells, that it is not significantly influenced by nuclear shape, nucleolar volume and/or the level of endopolyploidy.
A random positional homologous pairing of single copy homologous sequences along euchromatic chromatin segments has been reported for somatic cells in both A. thaliana and Arabidopsis lyrata (Berr et al. 2006; Pecinka et al. 2004). Pericentromeric regions containing 5S rRNA genes are also randomly distributed in A. thaliana nuclei (Saez-Vasquez and Gadal 2010).
The collinear alignment of sister chromatids defined as cohesion (Maguire 1990; Miyazaki and Orr-Weaver 1994) is required for correct chromosome segregation during cell division as well as for DNA recombination repair and transcription (Onn et al. 2008; Uhlmann 2008). In yeast, the close distances between cohesion sites (~11 kb) along chromosomes (Glynn et al. 2004; Laloraya et al. 2000) do not allow to distinguish sister chromatids by FISH at the resolution of light microscopy (Guacci et al. 1994). In contrast, in human nuclei allelic loci of sister chromatids may occupy distant positions when probed by FISH (Selig et al. 1992; Volpi et al. 2001).
Similar observations were made in Arabidopsis. The high frequency (more than 30 %) of positional sister chromatid separation at ~100-kb mid-arm positions, the absence of preferential cohesion sites along a ~1.2-Mb euchromatic segment and the variable extension of cohesion or separation (<500 kb to 14.2 Mb) along sister chromatid arms, suggest that sister chromatid cohesion in higher plants is highly dynamic and may therefore influence the interphase chromatin architecture (Berr et al. 2006; Schubert et al. 2006, 2007, 2008).
Based on chromosome conformation capture techniques, maps of spatial chromosome interactions in interphase nuclei have already been constructed for human and yeast. These allow to analyse dynamic and functional conformations of whole genomes (Duan et al. 2010; Lieberman-Aiden et al. 2009). For plants such tools are not yet available. Therefore, we used specific DNA sequences to label whole CTs and distinct eu- and heterochromatic segments along chromosomes by FISH. Homologous and/or heterologous associations, the extend of sister chromatid cohesion and chromatin condensation allow to trace the behaviour of distinct chromatin domains. We tested whether or not CTs and various chromatin domains behave similar or different in specific differentiated 2C and 4C nuclei versus highly endopolyploid (up to 64C) nuclei, to see whether endopolyploidy levels have an impact on chromatin organisation, similar or different from that of polyteny.
The biological significance of endopolyploidy is still under debate. The understanding of changes in chromatin organisation that may occur during endoreduplication can be helpful to clarify the phenomenon. For example, in Drosophila salvary gland cells endoreduplicated sister chromatids stay cohesive and form polytene chromosomes whose centromeres associate (Zhimulev et al. 2004). In contrast, we found that in Arabidopsis with rising endopolyploidisation sister chromatids become separated. Non-cohesive chromatids, possibly corresponding to puffs of polytene chromosomes, are probably more accessible, e.g. for the transcription machinery, than strictly cohered ones.
We also show that, in addition to a random CT arrangement, similar preferential homologous and heterologous associations and degrees of chromatin condensation may occur at identical chromatin domains in differentiated leaf nuclei of different endopolyploidy levels. In this respect, no obvious chromatin rearrangements occur during endopolyploidisation. The findings suggest that structural similarities are essential to maintain similar functions within a tissue of varying endopolyploidy.
Materials and methods
Preparation of nuclei, probe labelling, immunostaining and FISH
A. thaliana (accession Columbia) and A. lyrata plants were grown under short-day conditions (8-h light/16-h dark) at 21 °C.
Differentiated endopolyploid nuclei which no longer perform mitosis were isolated and flow-sorted from rosette leaves after formaldehyde fixation using a FACS Aria (BD Biosciences) according to their ploidy level as described by Pecinka et al. (2004).
The A. thaliana BACs were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA).
The 45S rDNA-specific probe was prepared from the A. thaliana BAC clone T15P10 bearing the 45S rDNA genes.
The 180-bp centromeric repeat probe (pAL) (Martinez-Zapater et al. 1986) was generated by PCR as previously described (Kawabe and Nasuda 2005). The telomere-specific probe was generated by PCR in the absence of template DNA using primers (TAAACCC)7 and (GGGTTTA)7 (Ijdo et al. 1991). Probes specific for the three centromeric repeat families (pAa, pAge1 and pAge2) of A. lyrata were prepared from PCR products (Berr et al. 2006).
For painting of the chromosome 1 top arm (CT1top) and the chromosome 3 bottom arm (CT3bottom) (Fig. 3a) 17 pools of in total 87 BACs (from T25K16 to F12K21) and 12 pools of in total 46 BACs (from T5C2 to F16M2), respectively, were labelled with biotin-dUTP and digoxigenin-dUTP as described (Pecinka et al. 2004).
BAC DNA from positions along chromosomes 1, 3 and 5 (Fig. 1) was labelled by nick translation with digoxigenin-dUTP, biotin-dUTP, or Cy3-dUTP according to Ward (2002). Biotin was detected by avidin conjugated with Texas Red (1:1,000; Vector Laboratories), goat-anti-avidin conjugated with biotin (1:200; Vector Laboratories) and again with avidin conjugated with Texas Red; digoxigenin by mouse-anti-digoxigenin (1:250; Roche) and goat-anti-mouse conjugated with Alexa-488 (1:200; Molecular Probes). Cy3 was observed directly.
FISH was performed according to Schubert et al. (2001). Nuclei and chromosomes were counterstained with DAPI (1 μg/ml) in Vectashield (Vector Laboratories).
For colocalisation of CENH3 immunosignals with centromeric FISH signals, immunostaining and FISH were performed subsequently. After immunostaining, nuclei were fixed in 4 % paraformaldehyde/3.6 % sucrose. Immunostaining of nuclei was performed as described (Jasencakova et al. 2000). CENH3 was detected with rabbit polyclonal antisera against A. thaliana CENH3 (1:500) (Talbert et al. 2002) and goat anti-rabbit rhodamine (1:100; Jackson Immuno Research Laboratories).
Microscopic evaluation, image processing and statistics
Analysis of FISH signals was performed with an epifluorescence microscope (Zeiss Axiophot) using a 100×/1.45 Zeiss α plan-fluar objective and a 3-chip Sony (DXC-950P) colour camera. Images were captured separately for each fluorochrome using appropriate excitation and emission filters. Images were merged using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, USA).
The DOM Laycheck software (Confovis, Jena) was used to measure 2C interphase nuclei (x = 6.7, y = 3.0 and z = 2.4 μm) and the corresponding BAC (diameter, 0.2 μm) and CT (x = 2.4, y = 1.4 and z = 1.7 μm) dimensions. On this basis, the “random spatial distribution” (RSD) model (Schubert et al. 2007) was modified to simulate round-shaped homologous and heterologous chromosome segments (corresponding to BAC and CT FISH signal areas) with coordinates determined randomly in a virtual interphase nucleus. The frequency of attachment and overlapping of two BAC areas, taken as homologous or heterologous association, is considered to be random (Fig. 2). The differences between simulated values and those obtained experimentally from the FISH experiments were compared by the two-sided Fisher exact test.
Homologous and heterologous association configurations and its computer simulation in the modified “random spatial distribution” (RSD) model. a Scheme showing the main four configurations of homologous and heterologous associations in 2C interphase nuclei. b The RSD model simulates random homologous and heterologous association of ~100-kb chromosome arm segments via two small spheres in the same colour corresponding to two homologous loci with coordinates determined randomly in a virtual interphase nucleus (blue ellipsoid) of average dimensions. It is assumed that the ~100-kb segments can occupy any position within its chromosome arm territory (large red and green sphere). Segments of other arms (yellow) can occupy any other position within the nucleus. Under these limitations the coordinates of the small spheres are calculated as random values. The frequency of overlapping of two small spheres of the same colour (green spheres) and of different colours (small red and yellow spheres) are considered as random single-point homologous and heterologous association, respectively. The small red and green spheres belong to two different arms (large red and green spheres illustrating CTs) of the same chromosome. Therefore, these small spheres are linked and pair more often with each other than with the yellow spheres
The cohesion frequency of sister CTs was calculated per homologue. One FISH signal cluster and overlapping signals per homologue were regarded as cohesion, two signal clusters as separated. The cohesion of ~100-kb BAC segments was evaluated as described (Schubert et al. 2008).
CTs covering more than 50 % of the nucleus area were regarded as completely dispersed (Fig. 3b, c).
The association frequencies of chromosome termini with centromeres and NORs, respectively, were compared with expected values using the Chi² test.
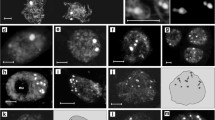
Chromosome territory (CT), ~100-kb interstitial chromatin segment and centromeric pAL DNA sequence arrangement in differentiated A. thaliana 2–64C interphase nuclei. a Scheme of differential labelling of A. thaliana chromosome regions. b Arrangement of CT1top and CT3bottom in 2–64C nuclei. 2C nuclei, both compact CT3bottom arms associate whereas the compact CT1top arms are separated (left). The right nucleus shows dispersed CT3bottom arms but the CT1top arms are compact and associated. 4C nucleus, both arms are compact and separated. 8C nucleus, association of compact CT1top arms and separation of compact CT3bottom arms. 16C nucleus, both CT1top and CT3bottom arms are compact and associated. 32C and 64C nuclei, whereas the centromeric pAL sequences stay cohesive in 4C nuclei (see Fig. 4a) signal numbers >32 and >64 indicate sister centromere separation. In both nuclei, the CT3bottom arms appear compact. c CT arrangements in 64C nuclei. From left to right: two separated compact CT1top arms; four separated compact CT1top arms; two separated compact CT3bottom arms in combination with two compact associated CT1top arms; two separated compact CT3bottom arms in combination with two partially dispersed associated CT1top arms; four separated compact CT3bottom arms in combination with two partially dispersed associated CT1top arms; two separated compact CT3bottom arms in combination with completely dispersed CT1top arms. d Compact arrangement of CT1top arms in combination with positionally separated ~100-kb mid-arm chromatin segments in 4C, 32C and 64C nuclei. Both homologous CT1top arms are separated in the 4C nucleus but cohesive in the 32C and 64C nuclei, respectively
Calculation of telomere associations with centromeres and NORs
The theoretically expected relation of telomere association with centromeres and NORs was estimated in that way that we consider only those telomeres which are associated with at least one NOR or one centromere (9.9 % in A. thaliana and 9.8 % in A. lyrata, respectively, are without any association).
In A. thaliana 2C nuclei 4 (on top arms of chromosomes 2 and 4) of the 20 telomeres (=20 %) are physically close to a NOR region. The remaining 16 telomeres (80 %) should be randomly associated to 14 chromatin segments totally, namely to 4 NORs (represented by a single nucleolus) and to 10 centromeres (4 + 10 = 14) according to their frequencies (4 of 14 of telomeres should be associated with NORs and 10 of 14 of them with centromeres). This results in 20 % (a priori localised to nucleolus) + 4/14 × 80 % = 42.9 % telomere–nucleolus association. The telomere–centromere association amounts to 10/14 × 80 % = 57.1 %.
For A. lyrata (32 telomeres, 10 of them close to the nucleolus and 16 centromeres) the analogous calculation results in an expected association frequency of 57.7 % for telomere–nucleolus association and of 42.3 % for telomere–centromere association.
Results
CTs are randomly arranged in highly endopolyploid nuclei
To analyse the organisation of CTs in 2C to 64C differentiated nuclei, we investigated by FISH the distribution of labelled BAC contigs for CT1top and CT3bottom in different colours (Fig. 3). The frequency of homologous association of ~43–60 % in 2C to 8C nuclei was similar for both CTs to that observed by Pecinka et al. (2004). We obtained now similar values for 16–64C nuclei (Table 1). Only in 64C nuclei the homologous association of CT1top was decreased to 36.7 %. This suggests a random homologous association of CTs also in highly endopolyploid nuclei.
Sister CTs, although sometimes separated are seldom dispersed and intermingled among each other in nuclei up to 64C
In A. thaliana 4C nuclei, a ~1.2-Mb chromatin segment at an interstitial position of chromosome 1 bottom arm is often (42.5 %) completely cohered. A complete sister chromatid separation of CT1top (14.2 Mb) may occur at ~4 % of homologues (Schubert et al. 2006, 2008). Here, we show that with increasing ploidy level whole CT1top and CT3bottom chromatids tend to be separated, but this separation does not exceed 15.5 % of homologues as found in 64C nuclei for CT1top (Table 1; Fig. 3c). Concurrent FISH of CT1top and the mid-arm segment T2P11 revealed that ~100-kb chromatin segments within the compact CTs are often separated in endopolyploid nuclei (Fig. 3d).
The low frequency (2.4–14.4 %) of dispersed CTs (covering >50 % of the nuclear area) in 2C to 16C nuclei even decreased to 1.0–2.3 % in 32C and 64C nuclei (Table 1; Fig. 3b, c). Interestingly, double FISH with CT1top and CT3bottom showed that this dispersion did not occur simultaneously for both CTs in all nuclei. The frequency of nuclei with only one of the homologous CTs dispersed ranged from 12.5 to 57.1 %.
In contrast to (peri)centromeres (see below), most chromosomes form one compact CT. Interstitial identical segments within these CTs are mainly separated at high endopolyploidy levels.
Centromeric and CENH3 associated repeats become separated in nuclei >16C and form distinct subdomains at adjacent positions but do not intermingle
The close cohesion of sister centromeres is essential for their bipolar orientation and subsequent segregation to opposite poles during nuclear division. The centromeric histone variant CENH3 is required to initiate the formation of kinetochores. To investigate centromere cohesion in differentiated leaf nuclei of different ploidy levels, we combined immunostaining and FISH to analyse the spatial distribution of CENH3 and of chromatin comprising 180-bp centromeric repeats (pAL).
The evaluation of 3D image stacks showed that the centromeric sequences are localised close to the nuclear periphery. In 2C, 4C and 8C nuclei, sister centromeres are often associated (Schubert et al. 2006) and in 2C and 4C nuclei the centromeres appear mainly as a compact FISH signal, suggesting tightly arranged chromatin. In some nuclei, more decondensed centromeres appear as ring- or half-moon-like structures (Fig. 4a). In 8C nuclei, centromeres start to split and are frequently separated in 32C and 64C nuclei (Figs. 3b and 4b).
Arrangement of (peri)centromeric regions in A. thaliana 2–16C interphase nuclei. a Nine (two of them associated) and ten centromeric FISH signals (pAL) in 2C and 4C nuclei, respectively. The mainly compact round shape centromeric signals are similar in 2C and in 4C nuclei. Some less compact centromeres are ring- (asterisk) or half-moon shaped (arrows). b Due to chromatin decondensation and sister chromatid separation in 8C and 16C nuclei, the centromeric histone variant CENH3 and the centromeric DNA signals form mostly contiguous subdomains within or adjacent to pericentromeric heterochromatin (bright DAPI signals). Some CENH3 (asterisks) and pAL (arrows) signals do no longer attach or overlap each other
Centromeric signal numbers higher than 32 and 64 in 32C and 64C nuclei, respectively (Fig. 3b), indicate increased sister centromere separation at higher ploidy levels. This conclusion is supported by the observation that also DAPI stained chromocenters appeared to be disintegrated.
When nuclei were hybridised with the pAL probe after immunostaining with antibodies against the centromeric histone variant CENH3, most nuclei showed co-localisation of CENH3 signals with FISH signals for pAL repeats. In nuclei, with a ploidy level higher than 4C (Fig. 4b) some 180-bp centromeric repeats were obviously not connected to CENH3 and vice versa. Limited association of CENH3 with some 180-bp repeat forming knobs along extended chromatin fibres has also been proven in A. thaliana cell cultures (Shibata and Murata 2004).
According to Nagaki et al. (2003), ~15 % of the 180-bp repeats are connected to CENH3 nucleosomes. Transposable elements, such as Athila, Tat, Tim, Copia, additionally accumulated at the centromeric regions (Arabidopsis Genome Initiative 2000) are associated to CENH3 with a frequency of up to 4.5 %. CENH3 nucleosomes not colocalising with pAL may associate with these transposable elements.
Several centromere organisation models suggest the coalescence of CENH3-containing nucleosomes to form subdomains at the inner kinetochores of both sister chromatids (Verdaasdonk and Bloom 2011). These subdomains seem to be also maintained in differentiated nuclei even at higher ploidy levels where the amount of CENH3 is not proportionally increasing with the amount of 180-bp repeats (Lermontova et al. 2006, 2007).
Similar as in A. thaliana, CENH3 forms subdomains and colocalises closely with the centromeric repeat Bilby in 2C and 4C differentiated nuclei of diploid rye (Schubert and Houben, unpublished).
Telomeres tend to associate but sister telomeres may be separated
The NORs on A. thaliana chromosomes 2 and 4 (Fig. 1) form a joint nucleolus in most of the nuclei (>90 %) surrounded by the majority of telomeres (Berr et al. 2006). To precisely define the spatial positioning of telomeres, telomeric DNA probes were localised by FISH on isolated leaf nuclei of A. thaliana (2n = 10) and A. lyrata (2n = 16) (Figs. 5 and 6).
Arrangement of (sub)telomeric regions in A. thaliana interphase nuclei. a Number of telomeric FISH signals in 2C and 4C interphase nuclei. b Number of centromere and NOR associated telomeric FISH signals in 2C nuclei. Insert shows as an example a nucleus with telomeric signals associated to 45SrDNA (NOR) and pAL (centromeric) signals. c Partial (top) and complete (bottom) association of telomeric signals with the single nucleolus in 2C and 4C nuclei. d FISH signal frequencies of subtelomeric chromatin fragments labelled by BACs T25K16, F23A5, T4P13, F16M2 and F7J8 in 2–16C nuclei. The higher than expected FISH signal numbers for F16M2, and to a lower extent for T25K16, indicate subtelomeric chromatin decondensation. e Arrangement of homologous and heterologous subtelomeric segments in 2–16C nuclei. 2C nuclei, homologous and heterologous association (see Fig. 2a) at subtelomeric segments (left) labelled by BACs F23A5, F16M2 and T7J8. The right nucleus shows homologous association but heterologous separation. The segment labelled by BAC F16M2 shows more than two signals and is therefore decondensed. Heterologous association of two (left), twice two (middle) and of all (right) subtelomeric loci labelled with BACs T4P13 and F16M2. 4C nuclei, cohesion and homologous association of subtelomeric segments of both chromosome 3 arms with that of CT5top (left) and three nuclei showing subtelomeric segment configurations of both chromosome 1 arms. 8C nucleus, arrangement of subtelomeric segments from both chromosome 1 arm ends, with the complete cohesion and homologous association for F23A5 (bottom arm) and complete separation for T25K16 (top arm). 16C nucleus, similar configuration as in the 8C nucleus but partial separation at F23A5
On average, 11.4 (4 to 14) and 15.7 (8 to 25) telomeric FISH signals were observed in A. thaliana and A. lyrata 2C interphase nuclei, respectively. In both species, telomere signal numbers varied between individual interphase nuclei, with an average number lower than the expected maximum number per species, implying that telomere association occurs (Figs. 5a and 6a). In A. thaliana, 4C nuclei showed more FISH signals than 2C nuclei (Fig. 5a) suggesting that sister telomeres can separate.
Telomeres associate more often to NORs than to centromeres
In nuclei of Arabidopsis species, we frequently observed telomeric FISH signals associated with the nucleolus (Fig. 5c). To analyse the spatial distribution of telomeres within interphase nuclei, telomeric DNA probes, 45S rDNA and centromeric probes were hybridised simultaneously to 2C interphase nuclei. In A. thaliana 62.9 % of telomeric FISH signals were found to be associated with 45SrDNA and 37.1 % with centromeres, whereas in A. lyrata 73.5 % of telomeric signals were associated with nucleoli and 26.5 % with centromeres (Figs. 5b and 6b). In A. thaliana, two of the five chromosomes (AT2 and AT4) bear NORs (Fransz et al. 1998) while in A. lyrata, five of the eight chromosomes (AL1, AL3, AL4, AL5 and AL7) contain NORs (Berr et al. 2006). Thus, considering 2C nuclei, four out of 20 in A. thaliana and ten out of 32 in A. lyrata are physically close to 45S rDNA repeats resulting in the observed preferential association of telomeric FISH signals with NORs, but the actual percentage of telomere-NOR associations is much higher in both species.
In A. thaliana, the observed associations of telomeres to the nucleolus amount to 63.3 % (719 of 1,137; expected, 42.9 %) and to centromeres 36.8 % (418 of 1,137; expected, 57.1 %). In A. lyrata, we found 73.5 % (1,133 of 1,541; expected, 57.7 %) telomere–nucleolus associations and 26.5 % (408 of 1,541; expected, 42.3 %) telomere–centromere associations.
The statistical comparison of the expected (for calculations see “Materials and methods”) with the observed frequencies of telomere associations with centromeres and NORs by the Chi² test results in highly significant (P < 0.001) differences, indicating a preferential association of telomeres to the nucleolus. The observation that ~10 % of telomeric signals were associated neither to NORs nor to centromeres in A. thaliana and A. lyrata suggest that these associations are not compulsory.
Both preferential and random chromatin associations appear at subtelomeres and at pericentromeres
To test the frequencies of homologous and heterologous associations between subtelomeric chromatin segments in A. thaliana, we performed simultaneous FISH with differently labelled probes hybridising closely to the top and bottom arm ends of chromosomes 1, 3 and 5 (Table 2; Figs. 1, 2a and 5e). A preferential (sub)telomere-specific homologous and/or heterologous association could indicate a specific arrangement of distinct chromatin domains potentially linked with their gene expression status.
Compared with the RSD model simulations assuming exemplarily a random distribution of six spheres for three arbitrarily selected subterminal regions in a virtual interphase nucleus (Fig. 2b), a significant increase of homologous association at all subtelomeric segments analysed in 2C nuclei was proven (Fig. 5e). Only the subtelomeric association at top arm of chromosome 1 (BAC T25K16) was relatively low with 9.5 %.
Also, heterologous interchromosomal associations were found to be significantly increased at most tested subtelomere combinations. Only the combination of BACs T25K16-F7J8 associated more seldom than expected at random.
A highly significant increase of intrachromosomal subtelomere association was found for chromosome 3. In contrast, for chromosome 1 the intrachromosomal subtelomeric association was less frequent than random.
Chromatin segments located at opposite arms of chromosome 1 close to the pericentromere associated in 2C nuclei more often than expected at random, while this was not true for chromosome 5. The tendency toward lower or higher homologous and heterologous association frequencies was similar in endopolyploid nuclei of 4–16C.
In summary, we conclude that in contrast to other interstitial euchromatic segments of A. thaliana and A. lyrata (Berr et al. 2006; Pecinka et al. 2004) preferential homologous and heterologous associations may occur around subtelomeres and at pericentromeres which might indicate joint gene activity patterns in these regions within nuclei of different endopolyploidy level.
Chromatin segments close to centromeres, telomeres and at mid-arm positions show a different extent of sister chromatid cohesion and chromatin condensation
Analysing the degree of condensation and sister chromatid cohesion at distinct chromatin segments could provide a hint as to the presence of transcriptionally active chromatin. If BACs of ~100-kb yield only one FISH signal per chromatid, the maximum signal number per nucleus should correspond to the ploidy level. However, depending on the chromatin segments analysed, also higher signal numbers appeared indicating chromatin decondensation. This has been found especially adjacent to the pericentromeric heterochromatin of chromosome 1 (labelled by BACs F12K21 and F2J6) and at the subtelomere of the bottom arm of chromosome 3 (labelled by BAC F16M2) in nuclei of endopolyploidy levels from 2–16C (Table 2; Fig. 5d, e). Other segments although of similar size did not show this phenomenon.
Sister chromatids are often not cohesive at mid-arm positions in higher plants (Berr et al. 2006; Schubert et al. 2006, 2007). Here, we show that also sister termini can be separated in differentiated 4C leaf nuclei. To test whether the frequency of sister chromatid separation varies between different subtelomeres and between pericentromeric chromatin segments, we calculated the separation frequencies in 4–16C nuclei. In comparison to a mid-arm segment of chromosome 1, labelled by BAC T2P11, significantly increased (BACs T25K16 and F16M2) and decreased (BACs F23A5 and T4P13) separation at subtelomeres was observed in 4C nuclei. Significantly increased separation was also evident for chromatin segments close to the pericentromeric heterochromatin of chromosome 1 but not of chromosome 5 (Table 2).
Up to 100 % sister chromatid separation was observed in 8C and 16C nuclei (Table 2; Fig. 5e). Thus, depending on the chromosomal position, the degree of chromatin condensation and sister chromatid cohesion can vary possibly in correlation with gene activity.
Discussion
There is increasing evidence of functional and topological constraints restricting a random spatial arrangement of chromatin in interphase nuclei (Lanctot et al. 2007; Misteli 2007). Chromosome size, gene density and expression during different developmental stages are factors which may constrain the random positioning of chromatin within interphase nuclei.
A network of co-regulated gene expression causing chromatin interactions during differentiation seems to result in the self-organisation of cell lineage-specific chromatin topologies. Self-organisation and fractal globule formation are promising models to explain the spatial distribution of chromatin segments and its dynamics in interphase nuclei (McNally and Mazza 2010; Misteli 2007, 2009; Rajapakse et al. 2009).
In addition to A. thaliana, a suitable model organism to analyse higher order chromatin organisation (Saez-Vasquez and Gadal 2010), we analysed comparatively the closely related species A. lyrata.
We show that in higher plants, in addition to random arrangement, preferential and dynamic chromatin association may occur within nuclei of different ploidy levels. Our data are mainly based on a defined developmental stage and tissue (nuclei isolated from rosette leaves) of Arabidopsis. To clarify whether preferential associations are connected to gene expression and whether they differ between various tissues and developmental stages, further investigations are required.
CTs and centromeric chromatin mostly maintain a compact structure even in highly endopolyploid nuclei
Fritsch and Langowski (2011) suggest that the viscoelasticity of chromatin during decondensation in interphase is changed by chromatin cross-linking and loop formation in such a way that chromatin can rapidly decondense and then consolidate to prevent its homogeneous distribution. Here, we showed that in most of the A. thaliana 2–64C nuclei the CTs maintain their distinct compact structure. Also the 180-bp centromeric and CENH3 containing chromatin segments appear as distinct adjacent sub-domains even in nuclei >16C where sister chromatids close to the centromeres start to separate. In 2C and 4C nuclei, the 180-bp centromeric repeats can form characteristic ring- or half-moon-like structures. The occurrence of CENH3 subdomains suggests that centromeric repeats form loops or solenoids with CENH3 nucleosomes always in the opposite orientation than H3-containing nucleosomes. Shibata and Murata (2004) found that on extended chromatin fibres CENH3 nucleosomes are formed only at some of the 180-bp repeats.
Experimental stretching of human and Drosophila centromeres revealed that the arrays of CENH3 nucleosomes which coalesce in nuclei are interrupted by blocks of H3-containing nucleosomes (Blower et al. 2002). This coalescence is the basic assumption of several centromere organisation models: the loop model, the solenoid model and the sinusoidal patch model (Santaguida and Musacchio 2009; Verdaasdonk and Bloom 2011). CENH3 nucleosome coalescence seems to be present also in differentiated endopolyploid A. thaliana nuclei where no further CENH3 loading occurs (Lermontova et al. 2006). However, it cannot yet been decided which of the models (if any) is true for higher plants.
The mainly random CT arrangement in Arabidopsis interphase nuclei is modified by structural and functional constraints
By computer simulation de Nooijer et al. (2009) showed that non-specific chromatin interactions in A. thaliana interphase nuclei are sufficient to explain the position of nucleoli and of chromocenters and that chromatin fibre looping might be responsible for CT formation. On the other hand, Andrey et al. (2010) conclude that conserved constraints influence the distribution of centromeres and chromocenters in nuclei of differentiated cells because they found that in distantly related species with different genome size and chromosome number such as A. thaliana (1C = 125 Mb, n = 5) and rabbit (1C = 2770 Mb, n = 22) a more regular distribution than expected at random was evident.
Previously we showed that CTs are mainly randomly distributed within interphase nuclei of two Arabidopsis species, independent of chromosome size and nuclear shape and that the formation of a single nucleolus may induce preferential CT association of NOR-bearing chromosomes (Berr et al. 2006; Pecinka et al. 2004). Homologous transgenic tandem repetitive sequences pair more often with each other and associate with chromocenters in A. thaliana nuclei than flanking euchromatin (Jovtchev et al. 2008, 2011; Pecinka et al. 2005). Similarly, sister chromatid cohesion at endogenous centromeric repetitive sequences is increased compared with euchromatic sequences in Arabidopsis. Also, more distantly related plant species (Brachycome, rye and maize) show a high frequency of cohesion at repetitive chromatin segments (Schubert et al. 2006, 2007).
Now, we demonstrate that in addition to a random CT organisation and random chromatin associations also preferential homologous and heterologous associations between euchromatic segments close to centromeric heterochromatin and at the (sub)telomeres may occur in A. thaliana.
Fang and Spector (2005) showed a cell type dependent distribution of A. thaliana centromeres in endoreduplicated nuclei with predominant clustering in root epidermal cells and dispersion in leaf epidermal cells. These authors found no precise transmission of centromere positions from the mother cell to daughter cells during mitosis, but Berr and Schubert (2007) showed transient mirror-image symmetry between meristematic daughter nuclei.
In contrast to polyploid wheat (Maestra et al. 2002; Martinez-Perez et al. 1999, 2000), in diploid rice a homologous association of centromeres and telomeres was found in root xylem and undifferentiated anther cells (Prieto et al. 2004).
Apparently, in plant species with relatively small chromosomes, different patterns of telomere distribution in interphase nuclei may occur (Fransz and De Jong 2011): in tomato at the edge of heterochromatin close to the centromere (Fransz 2004); in rice around the nuclear periphery (Prieto et al. 2004); and in budding yeast a Rabl-like conformation is present (Bystricky et al. 2005; Jin et al. 2000; Saez-Vasquez and Gadal 2010). In yeast, it was also shown that telomeres form clusters (Gotta et al. 1996) but it is not known whether these clusters include preferential association of homologous termini (as we found in A. thaliana) or not. Some telomere clustering has also been described in human cells with a higher frequency in differentiated than in cycling cells (Nagele et al. 2001).
Chromosome arm territories, similar as centromeric chromatin (see above) seem to be organised in a loop-like manner (Mateos-Langerak et al. 2009; Munkel et al. 1999). In maize, a loop of ~100 kb has been proven (Louwers et al. 2009) and in A. thaliana the size of loops emanating from heterochromatic chromocenters may vary between 100 kb and 2 Mb (Fransz et al. 2002; Fransz and De Jong 2011).
Here we show that not all chromatin fibres form loops that return to the chromocenters because in A. thaliana and A. lyrata only ~37 % and 26 % of telomeres, respectively, are located close to centromeres. The remaining telomeres mainly surround the nucleolus. Armstrong et al. (2001) suggest that nucleolus-associated telomere clustering is a prerequisite to establish synapsis during meiosis in A. thaliana. Whether the (sub)telomere associations we observed in somatic leaf nuclei are connected to transcription remains an open question.
To find out whether a transcribed gene loops out from its CT and/or influences the degree of sister chromatid cohesion, we tested a chromatin fragment of ~80 kb bearing the flowering gene FWA in nuclei of A. thaliana by FISH. Mutants where the gene is constitutively expressed in leaf tissue (Soppe et al. 2002) did not display significantly increased out-looping or decreased cohesion of the FWA region in 4C nuclei (Pecinka et al. 2004; Schubert et al. 2006) as expected if expression would be correlated with a higher degree of chromatin decondensation and an interaction with other chromatin segments. However, the frequencies of out-looping and cohesion may be influenced by the different expression levels of the other 13 genes located at the tested fragment. Here, we show that chromatin relaxation, potentially increasing interactions between gene loci, do not occur synchronously for different CTs within the same nucleus (Fig. 3b, c).
Along chromosomes different degrees of chromatin condensation may occur. We found frequent decondensation of a chromatin segment at the subtelomere of bottom arm of A. thaliana chromosome 3. Similarly, a single chromosome of Aegilops markgrafii (Greuter) Hammer contains such a stretched domain visible at an interstitial position during meiosis (Schubert 2011). In both cases it is not yet clear whether chromatin relaxation is related to transcriptional activity. In human nuclei nucleolus-associated chromatin alters its spatial distribution upon transcriptional changes (Nemeth et al. 2010). Constrained Brownian motion of chromatin could be responsible for short range chromatin movements (Chakalova and Fraser 2010). Therefore, most interactions are limited to genes on the same chromosome arm (Tolhuis et al. 2011) and occur at random. Nevertheless, long range chromatin interactions seem occasionally to play a role for regulation of gene expression (Schoenfelder et al. 2010).
Sister chromatid cohesion in A. thaliana is increased when induced double strand breaks have to be repaired (Watanabe et al. 2009). However, repair processes are not necessarily responsible for the observed variation of cohesion at subtelomeres and close to the centromeric heterochromatin. Alternatively, increased site-specific cohesion frequencies could be connected to a tissue and development-specific gene expression in transcription factories comprising the corresponding regions (Eskiw et al. 2011).
During differentiation heterochromatin becomes more condensed in mammals (Meshorer and Misteli 2006). In contrast, rye chromosomes showing Rabl orientation and a condensed string-like structure in meristematic nuclei become more relaxed in differentiated 2C and 4C nuclei (Schubert et al. 2011). Arabidopsis heterochromatin decondenses when differentiated mesophyll cells are transformed into protoplasts (Tessadori et al. 2007a). Stronger condensation of heterochromatin occurs a few days after germination (Mathieu et al. 2003; van Zanten et al. 2011) and during the floral transition (Tessadori et al. 2007b). Biotic and abiotic stress factors such as bacterial infection (Pavet et al. 2006), reduced light (Tessadori et al. 2009; van Zanten et al. 2010) and high temperature (Pecinka et al. 2010) may also induce chromatin condensation.
Regarding these observations it is important to analyse features of interphase chromatin architecture under standardised and reproducible conditions. In summary, we conclude that in nuclei of higher plants the mainly random chromatin arrangement is plastic during ontogenesis.
Endopolyploidy is accompanied by non-cohesive chromatid arrangement
Endopolyploidy occurring in plants and in animals results from amplification of sister chromatids without preceding nuclear division. It has been suggested that endopolyploidy is important to provide high DNA amounts for increased transcriptional activities in specialised cells and to compensate the lack of DNA in species with small genomes (Galitski et al. 1999; Kondorosi and Kondorosi 2004; Nagl 1976). An increased gene copy number could be helpful to protect the genome against environmental stress, e.g. the exposure with ultraviolet B light (Hase et al. 2006; Radziejwoski et al. 2011).
There are findings against the hypothesis that endoreduplication is involved in the regulation of transcription by increasing the availability of DNA templates for gene expression. Leiva-Neto et al. (2004) found that a lower degree of endoreduplication in maize endosperm did not influence the starch and protein contents and also not the corresponding transcript levels. Similarly, gene over-expression in tomato fruits could not be attributed to the degree of endoreduplication (Chevalier et al. 2011).
The widespread occurrence of endopolyploidy in seed plants and the positive correlation between DNA content and cell volume of endopolyploid cells suggest that endopolyploidy may accelerate plant growth and environmental adaption by larger cell volumes (Barow 2006; Bourdon et al. 2010; Galbraith et al. 1991; Jovtchev et al. 2006; Melaragno et al. 1993). However, recent studies in tomato revealed that cell size and fruit size can be uncoupled from the level of endopolyploidy (Chevalier et al. 2011; Nafati et al. 2011).
Here, we show that in endopolyploid A. thaliana nuclei the CTs formed by euchromatic chromosome arms maintain mostly the structure they have in 2C nuclei. This can be explained by the fractal globule model of chromatin. Only in 1–2 % of 32C and 64C nuclei we found dispersed CTs as assumed by the equilibrium globule model that describes the mixing of chromatin fibres (Lieberman-Aiden et al. 2009; Mirny 2011). Also, the occurrence of centromeric and pericentromeric sequences as small subdomains support the idea that chromatin is folded in fractal globules.
Applying ~100-kb BAC probes for FISH, Bourdon et al. (2011) describe an almost complete separation of sister chromatids in endopolyploid pericarp nuclei of up to 128C of tomato fruits. Concordantly, we demonstrate a high degree of positional sister chromatid separation in endopolyploid nuclei along chromosome arms of A. thaliana. Thus, a higher endopolyploidy in higher plants seems to be accompanied by non-cohesive chromatids acquiring a less condensed chromatin conformation which potentially makes DNA more accessible for the transcription machinery (Lieberman-Aiden et al. 2009). Whether a decondensed chromatin conformation is indeed important for a higher accessibility of genes to the transcription machinery is however still an open question. At least Kato and Lam (2003) found that endoreduplicated pavement cells display a greater range of chromatin movement than diploid guard cells in A. thaliana. Such an increased mobility could be important to bring genes together for co-expression in transcription factories. The extension of sister chromatid separation to (peri)centromeric regions is possibly tolerable in highly endopolyploid nuclei due to their mitotic inactivity.
In Drosophila, a Rabl orientation was found in endoreduplicated polytene nuclei (Hochstrasser et al. 1986) but not in other cells (Csink and Henikoff 1998). However, we show dispersed distribution of centromeric sequences in endopolyploid nuclei, indicating that endoreduplicated nuclei do not acquire Rabl configuration in A. thaliana. This is plausible because during endopolyploidisation no anaphases that mediate Rabl orientation occur.
We found a random association of homologous A. thaliana CTs in highly endopolyploid nuclei (16–64C) similar as described for 2C nuclei (Pecinka et al. 2004). Also tendencies of lower or higher frequencies of homologous or heterologous associations, sister chromatid cohesion and chromatin condensation at specific chromosome regions were similar in nuclei of different ploidy level, possibly because we analysed nuclei of the same tissue and developmental stage. Thus, large-scale chromatin rearrangements do apparently not occur during the first endopolyploidisation steps (before centromere dispersion starts).
In summary, the significant feature of endopolyploidy seems to be decreased chromatid cohesion, inducing a decondensed chromatin conformation, rather than a preferential arrangement of CTs. This extended conformation along entire Arabidopsis chromosomes could be a functional counterpart correlated to the regionally decondensed chromatid fibres in “puffs” and “Balbiani rings” of polytene chromosomes in dipterans. “Puffs” and “Balbiani rings” are the morphological manifestation of gene activity related to a specific state of differentiation (reviewed in Zhimulev et al. 2004).
Whether the separated sister chromatids of highly endoreduplicated nuclei are also organised in loops emanating from the chromocenters as suggested for nuclei of low endopolyploidy (Fransz et al. 2002; Fransz and De Jong 2011) remains to be investigated. Another interesting question is whether intrachromosomal interactions within large chromatin loops cause smaller loops therein.
Chromatin arrangement in differentiated Arabidopsis interphase nuclei- the model
Here, we achieved results concerning the behaviour (homologous and heterologous associations, sister chromatid cohesion) of pericentromeric and (sub)telomeric chromatin segments. We also tested the distribution and the degree of CT condensation at different endopolyploidy levels. Based on these results and previously obtained data, we propose models of interphase chromatin arrangement in differentiated Arabidopsis leaf nuclei of lower and higher endopolyploidy level (Fig. 7). We distinguish a varying chromatin organisation at heterochromatic (peri)centromeres, euchromatic chromosome arms and (sub)telomeres.
(Peri)centromeres
Centromeres located at the nuclear periphery may associate. Centromeric repeats, CENH3 associated repeats and pericentromeric heterochromatin form distinct co-localising subdomains. Sister centromeres separate increasingly from 16C to 64C nuclei, as their return into the mitotic cycle becomes more and more unlikely.
Euchromatic chromosome arm regions
In interphase nuclei, euchromatin seems to be organised as fractal globules forming loops of ~100 kb to 2 Mb. Euchromatic chromosome arm regions form mainly distinct CTs which do not obviously intermingle in endopolyploid nuclei. Sister chromatid cohesion/separation is variable along interphase chromosomes. Separation can reach several Mb so that whole euchromatic arm CTs may be detached. The minimum extension of cohesive sites or distances between them may fall below 500 kb in 4C nuclei. Independent of the endopolyploidy level, the chromosome arm CTs are mainly arranged randomly. Only NOR-bearing chromosomes are more often associated around a joint nucleolus. Complete dispersion of euchromatic chromosome arms appears at a low frequency and may occur not simultaneously at different CTs. Chromatin fibre out-looping from arm CTs is rare. In nuclei >8C, ~100-kb euchromatic sister chromatin segments within arm CTs are mostly not cohesive, reminiscent of transcriptionally active “puffs” along polytene chromosomes.
(Sub)telomeres
Although Arabidopsis telomeres frequently cluster (more often at NORs than at centromeres), sister telomeres can rarely be separated. Preferential as well as random association may occur between homologous and heterologous chromosome termini. The extent of positional sister chromatid separation and of chromatin condensation at subtelomeric chromatin segments varies between different chromosome arms. The frequency of homologous or heterologous association, of sister chromatid cohesion and of chromatin condensation at distinct subtelomeric segments is independent of the ploidy level. A high degree of cohesion of telomeres and of centromeres, at least in nuclei of a lower endopolyploidy level, might serve as start points for SMC5/6 complex-mediated sister chromatid cohesion when needed for recombination repair of DNA damage (Watanabe et al. 2009). A general clustering and homologous association of telomeres might reflect the potential readiness for chromosome pairing when required, e.g. for meiosis.
References
Andrey P, Kieu K, Kress C, Lehmann G, Tirichine L, Liu ZC, Biot E, Adenot PG, Hue-Beauvais C, Houba-Herin N, Duranthon V, Devinoy E, Beaujean N, Gaudin V, Maurin Y, Debey P (2010) Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Comput Biol 6(7):e1000853. doi:10.1371/journal.pcbi.1000853
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Aragon-Alcaide L, Reader S, Beven A, Shaw P, Miller T, Moore G (1997) Association of homologous chromosomes during floral development. Curr Biol 7:905–908
Armstrong SJ, Franklin FC, Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114:4207–4217
Barow M (2006) Endopolyploidy in seed plants. Bioessays 28:271–281
Berchtold D, Fesser S, Bachmann G, Kaiser A, Eilert JC, Frohns F, Sadoni N, Muck J, Kremmer E, Eick D, Layer PG, Zink D (2011) Nuclei of chicken neurons in tissues and three-dimensional cell cultures are organized into distinct radial zones. Chromosome Res 19:165–182
Berr A, Schubert I (2007) Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 176:853–863
Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I (2006) Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant Journal 48:771–783
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330
Bourdon M et al (2010) Endoreduplication and growth of fleshy fruits. Prog Bot 71:101–132
Bourdon M, Coriton O, Pirrello J, Cheniclet C, Brown SC, Poujol C, Chevalier C, Renaudin JP, Frangne N (2011) In planta quantification of endoreduplication using fluorescent in situ hybridization (FISH). Plant J 66:1089–1099
Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA (2001) The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10:211–219
Branco MR, Pombo A (2006) Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4:e138
Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ (2006) Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172:177–187
Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM (2005) Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168:375–387
Chakalova L, Fraser P (2010) Organization of transcription. Cold Spring Harb Perspect Biol. doi:10.1101/cshperspect.a000729
Chakalova L, Carter D, Debrand E, Goyenechea B, Horton A, Miles J, Osborne C, Fraser P (2005) Developmental regulation of the beta-globin gene locus. Prog Mol Subcell Biol 38:183–206
Chandley AC, Speed RM, Leitch AR (1996) Different distributions of homologous chromosomes in adult human Sertoli cells and in lymphocytes signify nuclear differentiation. J Cell Sci 109:773–776
Chevalier C, Nafati M, Mathieu-Rivet E, Bourdon M, Frangne N, Cheniclet C, Renaudin JP, Gevaudant F, Hernould M (2011) Elucidating the functional role of endoreduplication in tomato fruit development. Ann Bot 107:1159–1169
Cope NF, Fraser P, Eskiw CH (2010) The yin and yang of chromatin spatial organization. Genome Biol 11:204
Corredor E, Diez M, Shepherd K, Naranjo T (2005) The positioning of rye homologous chromosomes added to wheat through the cell cycle in somatic cells untreated and treated with colchicine. Cytogenet Genome Res 109:112–119
Coulthard AB, Nolan N, Bell JB, Hilliker AJ (2005) Transvection at the vestigial locus of Drosophila melanogaster. Genetics 170:1711–1721
Cremer T, Cremer M (2010) Chromosome territories. Cold Spring Harb Perspect Biol 2:a003889
Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T (2001) Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 9:541–567
Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
Csink AK, Henikoff S (1998) Large-scale chromosomal movements during interphase progression in Drosophila. J Cell Biol 143:13–22
de Nooijer S, Wellink J, Mulder B, Bisseling T (2009) Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucleic Acids Res 37:3558–3568
Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295:1306–1311
Dong F, Jiang J (1998) Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res 6:551–558
Dostie J, Zhan Y, Dekker J (2007) Chromosome conformation capture carbon copy technology. Curr Protoc Mol Biol 80:21.14.1–21.14.13
Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS (2010) A three-dimensional model of the yeast genome. Nature 465:363–367
Duncan IW (2002) Transvection effects in Drosophila. Ann Rev Genet 36:521–556
Eskiw CH, Cope NF, Clay I, Schoenfelder S, Nagano T, Fraser P (2011) Transcription factories and nuclear organization of the genome. Cold Spring Harb Symp Quant Biol 75:501–506
Essers J, van Cappellen WA, Theil AF, van Drunen E, Jaspers NG, Hoeijmakers JH, Wyman C, Vermeulen W, Kanaar R (2005) Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell 16:769–775
Fang Y, Spector DL (2005) Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell 16:5710–5718
Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A (2010) Gene positioning. Cold Spring Harb Perspect Biol 2:a000588
Fransz P (2004) The interphase nucleus. In: Goodman RM (ed) Encyclopedia of plant and crop science. Marcel Dekker, New York, pp 568–571
Fransz P, De Jong JH (2011) From nucleosome to chromosome: a dynamic organization of genetic information. Plant J 66:4–17
Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 13:867–876
Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 99:14584–14589
Fritsch CC, Langowski J (2011) Chromosome dynamics, molecular crowding, and diffusion in the interphase cell nucleus: a Monte Carlo lattice simulation study. Chromosome Res 19:63–81
Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW (1998) Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol 141:5–20
Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96:985–989
Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR (1999) Ploidy regulation of gene expression. Science 285:251–254
Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J (2003) Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112:751–764
Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. Plos Biology 2:1325–1339
Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134:1349–1363
Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125:517–530
Hase Y, Trung KH, Matsunaga T, Tanaka A (2006) A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J 46:317–326
Heride C, Ricoul M, Kieu K, von Hase J, Guillemot V, Cremer C, Dubrana K, Sabatier L (2010) Distance between homologous chromosomes results from chromosome positioning constraints. J Cell Sci 123:4063–4075
Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW (1993) The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol 120:591–600
Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW (1986) Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol 102:112–123
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19:4780
Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I (2000) Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087–2100
Jin QW, Fuchs J, Loidl J (2000) Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci 113:1903–1912
Jovtchev G, Schubert V, Meister A, Barow M, Schubert I (2006) Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet Genome Res 114:77–82
Jovtchev G, Watanabe K, Pecinka A, Rosin FM, Mette MF, Lam E, Schubert I (2008) Size and number of tandem repeat arrays can determine somatic homologous pairing of transgene loci mediated by epigenetic modifications in Arabidopsis thaliana nuclei. Chromosoma 117:267–276
Jovtchev G, Borisova BE, Kuhlmann M, Fuchs J, Watanabe K, Schubert I, Mette MF (2011) Pairing of lacO tandem repeats in Arabidopsis thaliana nuclei requires the presence of hypermethylated, large arrays at two chromosomal positions, but does not depend on H3-lysine-9-dimethylation. Chromosoma 120:609–619
Kato N, Lam E (2003) Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J Cell Sci 116:2195–2201
Kawabe A, Nasuda S (2005) Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol Genet Genomics 272:593–602
Koehler D, Zakhartchenko V, Froenicke L, Stone G, Stanyon R, Wolf E, Cremer T, Brero A (2009) Changes of higher order chromatin arrangements during major genome activation in bovine preimplantation embryos. Exp Cell Res 315:2053–2063
Kondorosi E, Kondorosi A (2004) Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett 567:152–157
Kozubek S, Lukasova E, Jirsova P, Koutna I, Kozubek M, Ganova A, Bartova E, Falk M, Pasekova R (2002) 3D structure of the human genome: order in randomness. Chromosoma 111:321–331
Küpper K, Kolbl A, Biener D, Dittrich S, von Hase J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, Cremer M (2007) Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma 116:285–306
Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151:1047–1056
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8:104–115
Leiva-Neto JT, Grafi G, Sabelli PA, Dante RA, Woo YM, Maddock S, Gordon-Kamm WJ, Larkins BA (2004) A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16:1854–1869
Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I (2006) Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18:2443–2451
Lermontova I, Fuchs J, Schubert V, Schubert I (2007) Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma 116:507–510
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293
Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M (2009) Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21:832–842
Lukasova E, Kozubek S, Kozubek M, Falk M, Amrichova J (2002) The 3D structure of human chromosomes in cell nuclei. Chromosome Res 10:535–548
Maestra B, De Jong JH, Shepherd K, Naranjo T (2002) Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res 10:655–667
Maguire MP (1990) Sister chromatid cohesiveness: vital function, obscure mechanism. Biochem Cell Biol 68:1231–1242
Martinez-Perez E, Shaw P, Reader S, Aragon-Alcaide L, Miller T, Moore G (1999) Homologous chromosome pairing in wheat. J Cell Sci 112(Pt 11):1761–1769
Martinez-Perez E, Shaw PJ, Moore G (2000) Polyploidy induces centromere association. J Cell Biol 148:233–238
Martinez-Zapater JM, Estelle AM, Sommerville CR (1986) A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204:4417–4423
Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, Indemans MH, Gierman HJ, Heermann DW, van Driel R, Goetze S (2009) Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci USA 106:3812–3817
Mathieu O, Jasencakova Z, Vaillant I, Gendrel AV, Colot V, Schubert I, Tourmente S (2003) Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15:2929–2939
McNally JG, Mazza D (2010) Fractal geometry in the nucleus. EMBO J 29:2–3
Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5:1661–1668
Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7:540–546
Mirny LA (2011) The fractal globule as a model of chromatin architecture in the cell. Chromosome Res 19:37–51
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800
Misteli T (2009) Self-organization in the genome. Proc Natl Acad Sci USA 106:6885–6886
Misteli T, Soutoglou E (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10:243–254
Miyazaki WY, Orr-Weaver TL (1994) Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28:167–187
Munkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J (1999) Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol 285:1053–1065
Nafati M, Cheniclet C, Hernould M, Do PT, Fernie AR, Chevalier C, Gevaudant F (2011) The specific overexpression of a cyclin-dependent kinase inhibitor in tomato fruit mesocarp cells uncouples endoreduplication and cell growth. Plant J 65:543–556
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, Lee H (1999) Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J Cell Sci 112:525–535
Nagele RG, Velasco AQ, Anderson WJ, McMahon DJ, Thomson Z, Fazekas J, Wind K, Lee H (2001) Telomere associations in interphase nuclei: possible role in maintenance of interphase chromosome topology. J Cell Sci 114:377–388
Nagl W (1976) DNA endoreduplication and polyteny understood as evolutionary strategies. Nature 261:614–615
Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G (2010) Initial genomics of the human nucleolus. PLoS Genet 6:e1000889
Neusser M, Schubel V, Koch A, Cremer T, Muller S (2007) Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma 116:307–320
Onn I, Heidinger-Pauli JM, Guacci V, Ünal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24:105–129
Papantonis A, Cook PR (2010) Genome architecture and the role of transcription. Curr Opin Cell Biol 22:271–276
Parada LA, Sotiriou S, Misteli T (2004) Spatial genome organization. Exp Cell Res 296:64–70
Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol Plant Microbe Interact 19:577–587
Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269
Pecinka A, Kato N, Meister A, Probst AV, Schubert I, Lam E (2005) Tandem repetitive transgenes and fluorescent chromatin tags alter local interphase chromosome arrangement in Arabidopsis thaliana. J Cell Sci 118:3751–3758
Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22:3118–3129
Prieto P, Santos AP, Moore G, Shaw P (2004) Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma 112:300–307
Rabl C (1885) Über Zelltheilung. Morph Jb 10:214–330
Radziejwoski A, Vlieghe K, Lammens T, Berckmans B, Maes S, Jansen MA, Knappe C, Albert A, Seidlitz HK, Bahnweg G, Inze D, De Veylder L (2011) Atypical E2F activity coordinates PHR1 photolyase gene transcription with endoreduplication onset. EMBO J 30:355–363
Rajapakse I, Perlman MD, Scalzo D, Kooperberg C, Groudine M, Kosak ST (2009) The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Natl Acad Sci USA 106:6679–6684
Saez-Vasquez J, Gadal O (2010) Genome organization and function: a view from yeast and Arabidopsis. Molecular Plant 3:678–690
Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28:2511–2531
Scheuermann MO, Tajbakhsh J, Kurz A, Saracoglu K, Eils R, Lichter P (2004) Topology of genes and nontranscribed sequences in human interphase nuclei. Exp Cell Res 301:266–279
Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42:53–61
Schubert V (2011) No neocentric activity on Aegilops markgrafii chromosome E. Cytogenet Genome Res 132:100–104
Schubert I, Shaw P (2011) Organization and dynamics of plant interphase chromosomes. Trends Plant Sci 16:273–281
Schubert I, Fransz PF, Fuchs J, De Jong JH (2001) Chromosome painting in plants. Methods Cell Sci 23:57–69
Schubert V, Klatte M, Pecinka A, Meister A, Jasencakova Z, Schubert I (2006) Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172:467–475
Schubert V, Kim YM, Berr A, Fuchs J, Meister A, Marschner S, Schubert I (2007) Random homologous pairing and incomplete sister chromatid alignment are common in angiosperm interphase nuclei. Mol Genet Genomics 278:167–176
Schubert V, Kim YM, Schubert I (2008) Arabidopsis sister chromatids often show complete alignment or separation along a 1.2-Mb euchromatic region but no cohesion “hot spots”. Chromosoma 117:261–266
Schubert V, Meister A, Tsujimoto H, Endo TR, Houben A (2011) Similar rye A and B chromosome organization in meristematic and differentiated interphase nuclei. Chromosome Res 19:645–655
Selig S, Okumura K, Ward DC, Cedar H (1992) Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J 11:1217–1225
Shibata F, Murata M (2004) Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J Cell Sci 117:2963–2970
Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21:6549–6559
Sugimoto-Shirasu K, Roberts K (2003) "Big it up": endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6:544–553
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V (2007a) Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci 120:1200–1208
Tessadori F, Schulkes RK, van Driel R, Fransz P (2007b) Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J 50:848–857
Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LA, Spillane C, Pikaard CS, Fransz P, Peeters AJ (2009) Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5:e1000638
Thomson I, Gilchrist S, Bickmore WA, Chubb JR (2004) The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol 14:166–172
Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B (2011) Interactions among polycomb domains are guided by chromosome architecture. PLoS Genet 7:e1001343
Uhlmann F (2008) Molecular biology: cohesin branches out. Nature 451:777–778
van Zanten M, Tessadori F, McLoughlin F, Smith R, Millenaar FF, van Driel R, Voesenek LA, Peeters AJ, Fransz P (2010) Photoreceptors CRYTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol 154:1686–1696
van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, Koornneef M, Fransz P, Soppe WJ (2011) Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci USA 108:20219–20224
Verdaasdonk JS, Bloom K (2011) Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12:320–332
Volpi EV, Sheer D, Uhlmann F (2001) Cohesion, but not too close. Curr Biol 11:R378
Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T (2003) Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160:685–697
Ward PB (2002) FISH probes and labelling techniques. In: Beatty BS, Mai J, Squire J (eds) FISH: a practical approach (practical approach series: 260). Oxford University Press, Oxford, pp 5–28
Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I (2009) The structural maintenance of chromosomes 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21:2688–2699
Wegel E, Shaw P (2005) Gene activation and deactivation related changes in the three-dimensional structure of chromatin. Chromosoma 114:331–337
Wegel E, Vallejos RH, Christou P, Stoger E, Shaw P (2005) Large-scale chromatin decondensation induced in a developmentally activated transgene locus. J Cell Sci 118:1021–1031
Wegel E, Koumproglou R, Shaw P, Osbourn A (2009) Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21:3926–3936
Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38:1341–1347
Zhimulev IF, Belyaeva ES, Semeshin VF, Koryakov DE, Demakov SA, Demakova OV, Pokholkova GV, Andreyeva EN (2004) Polytene chromosomes: 70 years of genetic research. Intern Rev Cytol 241:203–275
Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D (2004) Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol 166:815–825
Acknowledgements
We thank Jörg Fuchs for flow sorting of nuclei, Martina Kühne, Andrea Kunze, Joachim Bruder and Rita Schubert for excellent technical assistance, Ingo Schubert and Andreas Houben for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Peter Shaw
Rights and permissions
About this article
Cite this article
Schubert, V., Berr, A. & Meister, A. Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness. Chromosoma 121, 369–387 (2012). https://doi.org/10.1007/s00412-012-0367-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-012-0367-8