Abstract
Purpose
Testicular torsion (TT) mainly affects boys under 18 years old. To avoid orchiectomy, TT requires an immediate operative management. The etiology of TT is still controversial. Observed familiar recurrence suggests the presence of a genetic involvement. The INSL3 gene consists of two exons, and it is specifically expressed in fetal and adult Leydig cells. In transgenic mice, deletion of this gene was observed an increased testicular mobility and testicular torsion. We have hypothesized the possible involvement of the INSL3 gene as a predisposing factor of human TT.
Methods
We performed genetic analysis in 25 pediatric patients with unilateral and intravaginal TT (left, n = 13, 56%; right, n = 12, 48%). The age of the patients ranged from 1 to 16 years (median age n = 10.4 ± 5.46 years). In this study, we included two first male cousins affected by TT. Venous peripheral blood samples was obtained after parental written informed consent.
Results
The Thr60Ala polymorphism was detected in exon 1 of INSL3 gene and other 2 rarer variants (rs1047233 and rs1003887) were identified in the 3′ untranslated region. These variants are prevalent in patients with TT instead of healthy subjects.
Conclusions
Additional studies in a larger population are needed to better understand the clinical consequence of the INSL 3 variations founded. This would allow in the future to identify the patients at risk of TT to improve clinical management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Torsion of the spermatic cord or testicular torsion (TT) represents a common surgical emergency in pediatric age, affecting, above all, prepubertal and young adult males [1, 2].
The annual incidence of TT is estimated at 3.8 per 100,000 (0.004%) for boys under 18 years old [3].
To avoid orchiectomy, TT requires an accurate, timely diagnosis; operative management within 6 h after onset of symptoms [4] allows a better salvage rate of testis [4, 5].
TT may occur during sleep or following direct trauma, increased physical activity, or sudden contraction of the cremasteric muscle, as a response to mechanical, sexual, or thermic stimulation [6].
An anatomical predisposing condition, wherein a lack of normal fixation of the testis and epididymis to the scrotum, also known as “bell clapper deformity”, continues to be the most commonly described risk factor for intravaginal TT [4].
Furthermore, a positive familiar history has been found in about 10% of children affected by TT, and it can affect multiple relatives and generations, particularly in cases of bilateral torsion [7, 8].
This evidence would suggest the presence of a genetic cause, with or without the contribution of other anatomical and environmental factors [9].
Insulin-like factor 3 (Insl3) has been characterized as a key human testicular hormone. In this regard, Insl3 and its receptor, the relaxin family peptide receptor 2 (Rxfp2), influence gubernaculum development and thickening, and are essential for trans-abdominal testis translocation during the first phase of the testicular descent process [10,11,12]. Two independent studies have demonstrated that transgenic mice with targeted deletion of INSL3 showed defects in gubernaculum growth, resulting in bilateral cryptorchidism, as well as other sexual abnormalities [13, 14].
Moreover, studies in humans have investigated the possibility that naturally occurring mutations in the INSL3 gene interfere directly with male fertility or are associated with cryptorchidism, but there was no clear contribution of INSL3 variants [11, 15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30].
In INSL3 mutant mice, increased testicular mobility and subsequent testicular torsion were found in a gene dose-dependent manner, suggesting that Insl3 is a candidate-signaling molecule involved in human torsion events [31].
In this study, we explored the potential involvement of the INSL3 gene as a predisposing risk factor inducing human TT. To test this hypothesis, we performed direct sequencing of the INSL3 gene in a group of pediatric patients who presented with unilateral and intravaginal TT. In addition, our study carried out INSL3 genotyping in a patient who reported familial testicular torsion, and thus, the analysis was extended to the first-degree family members.
Materials and methods
Venous peripheral blood samples were obtained for genetic analysis after parental written informed consent was obtained from 25 pediatric patients with unilateral TT (left, n = 13, 56%; right, n = 12, 48%). Patient ages ranged from 1 to 16 years (median age n = 10.4 ± 5.46 years).
All individuals included in this study were Caucasian, except for one of Asiatic origin, and none were family relatives; patients with other ipsi or contralateral testicular abnormalities, affected by endocrinal alterations, with a diagnosed “bell clapper deformity” or with TT as a consequence of a testicular trauma were excluded from the study. Each participant underwent surgical exploration and received contralateral testis fixation. After derotation of the spermatic cord, orchidopexy was performed in 11 patients (44%), and orchiectomy was performed in the other 14 (56%). As a patient had a familial history of TT (two first cousins), eight members of this family were included in the study for INSL3 genotyping, after written informed consent.
Control blood samples were obtained from 30 volunteers of the same age, without TT or any other abnormalities of the male genitalia, after their parents gave written informed consent. The study was approved by the local Ethical Committee of the University Hospital of Messina.
Genomic DNA was extracted from peripheral blood leukocytes with an NLM AA1001 kit (Nuclear Laser Medicine, Settala, MI, Italy) according to the manufacturer’s instructions.
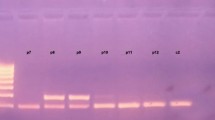
Polymerase chain reaction (PCR) primers were designed for the human gene INSL3 (reference gene ID: 3640), to cover the sequence of the two exons, as well as the intron exon boundaries. The exon 1 primer sequences were 5′-AAAGACTCGTTGCCCAGTG-3′ and 5′-CACACTCCAGTGCGGACC-3′; the exon 2 forward primer was 5′-ATGAGTGTTTGGTGGGTTAC-3′; and the exon 2 reverse primer was 5′-TGCCTCTCTAGTTATCAAGC-3′. The PCR reactions consisted of 40 ng genomic DNA, 1 µl of each of 10 µM primers, 1 µl dNTPs (10 mM), 2.5 µl 10X reaction buffer with 15 mM MgCl2, 0.2 µl StoS Taq polymerase (5 U/µl) (GeneSpin S.r.l., Lodi, Italy), and 17.8 µl sterile water in a total volume of 25 µl. The PCR thermocycler profile for exon 1 amplification was: 94 °C for 4 min; 94 °C for 1 min, 64 °C for 1 min, and 72 °C for 1 min, for 35 cycles; with a final extension at 72 °C for 4 min. PCR conditions for exon 2 amplification were: 94 °C for 4 min; 94 °C for 1 min, 59 °C for 1 min, and 72 °C for 1 min, for 35 cycles; with a final extension at 72 °C for 4 min.
PCR products generated were, respectively, 557 and 774 bp. The purified PCR fragments were sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Austin, TX, USA) using an ABI PRISM 3130 automated sequencer (Applied Biosystems, Monza, Italy) according to the manufacturer’s instructions.
Statistical analyses were carried out using Statistica 10.0 StatSoft. Fisher’s exact test was used to compare genotype frequencies of TT patients group versus healthy subjects. P values of less than 0.05 were considered significant. Hardy–Weinberg equilibrium was determined using Courtlab calculator (downloaded from http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls).
To estimate the linkage disequilibrium (LD) between polymorphisms, LD coefficients (r2 and D′) were calculated using the online tool LDlink developed by the Division of Cancer Epidemiology and Genetics (DCEG) of the National Cancer Institute (NCI), one of the National Institutes of Health (NIH) (https://analysistools.nci.nih.gov/LDlink/?tab=ldmatrix).
Results
Variations found in the INSL3 gene and clinical features of the 25 TT patients are listed in Table 1. Three common polymorphisms were detected in exon 1. Two of those are synonymous variations, so are probably not pathogenic: Ala9= (rs22866663, MAF/MinorAlleleCount: 0.1228/615) and Leu42= (rs1047233, MAF/MinorAlleleCount: 0.2572/1288). The other one was a nucleotide substitution: A to G in the C-peptide region, resulting in a threonine (Thr) to alanine (Ala) change at codon 60 (rs6523). Thr60Ala was found in 12 TT patients in homozygosis (48%) and also in a total of 12 control subjects: 4 were homozygous (13.3%) and 8 were heterozygous (26.6%).
Two other variants were identified in the 3′ untranslated region (UTR) of the INSL3 gene. The first one is a transition G>A (rs1003887) and was present in 14 TT subjects in homozygosis (56%) versus 6 controls (20%), and this was also present in heterozygosis in 7 patients (28%) versus 18 controls (60%). The second variant was a C>A substitution (rs17750642) and is a more uncommon variant (MAF/MinorAlleleCount: 0.0144/72), which was found in only 3 TT patients in heterozygosis (12%).
The rs6523 polymorphism was in strong linkage disequilibrium with the rs1003887 polymorphism (r2 = 0.861, D′ = 0.955) and in weak linkage disequilibrium with the other exon 1 polymorphism: rs1047233 (r2 = 0.374, D′ = 1). In addition, rs1047233 and rs1003887 were in weak linkage disequilibrium (r2 = 0.328, D′ = 0.964).
The statistical analysis for the last three variations described is reported in Table 2. Although Thr60Ala (rs6523) has been detected in homozygosis in significantly more TT patients compared to healthy subjects, the genotype frequencies fit the Hardy–Weinberg law only in the control group (X2 = 3.03; P = 0.08) and not in the TT group (X2 = 25; P < 0.0001). However, for rs1003887, the genotype distributions were in accordance with the equilibrium law in both the control (X2 = 1.20; P = 0.27) and patient group (X2 = 2.77; P = 0.09) and Fisher’s test result is statistically significant.
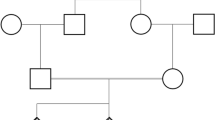
In this study, we included two first male cousins affected by unilateral TT, respectively at age of 11 (Patient ID: TT3, see Table 1) and 13 years.
We performed sequence analysis of the INSL3 gene in all members of the family to search for the predisposing allele. The genealogical tree of the two generations is outlined, and the INSL3 genotype for each member of the pedigree is reported in Fig. 1. Familiar genetic testing showed that all subjects were homozygous for Thr60Ala and rs1003887, and they carried other synonymous variations.
Discussion
Testicular torsion is responsible for long-term effects on exocrine and endocrine functions, leading to reduced fertility and compromised spermatogenesis in adulthood [32].
Genetic, hormonal, and anatomical factors influence a complex molecular mechanism leading to male gonad development, but part of this process remains unexplained. Insl3 was first described in 1990 as a novel peptide and is expressed at high levels in the human testis [33, 34]. In all mammalian species examined so far, Insl3 is a major secretory product of mature interstitial Leydig cells and is also expressed by testes in the male fetus in mid-gestation [34].
Moreover, it has been reported that Insl3 plays a crucial role in rodent testicular descent by acting on Rxfp2 receptors expressed within the gubernacular ligaments, causing these to expand and anchor the testes in the inguinal region [35]. As a consequence, numerous human mutation analyses have sought to elucidate the possible involvement of Insl3 and its specific receptor Rxfp2 in cryptorchidism [35, 36].
The analysis of sporadic and possible familial cases of undescended testis revealed several mutations, some of which are common polymorphisms, found both in healthy and affected subjects.
All patients carrying INSL3/RXFP2 mutations were heterozygous, and thus, the mutation-dependent mechanism of testicular maldescent may be caused by reduced signaling [24, 26]. However, a definitive causative role for many of these mutations in humans is still lacking [37].
The etiology of TT is still unclear and the related risk factors remain controversial; some evidence suggests that the risk of TT, a rare condition, can be inherited, particularly in cases of bilateral torsion. Furthermore, experimental animal studies suggest that the mechanism predisposing patients to TT may involve the INSL3 gene, but until now, no study has tested this hypothesis in humans [7].
Therefore, we performed a genetic analysis of the INSL3 gene in 25 pediatric patients affected by unilateral TT for the first time. We confirmed that synonymous Ala9= and Leu42= variations are common polymorphisms found in both patients and healthy subjects, as previously reported in cryptorchid subjects, probably without any pathogenic significance [36].
Our results for the Thr60Ala (rs6523) mutation are not conclusive, despite this variation seeming to be more represented in the group of TT patients in homozygosis. However, this observation could not be statistically verified due to the small sample size in analysis. Nonetheless, this missense polymorphism in the N-terminal region of the Insl3 pro-peptide can have an effect on Insl3 binding to its receptor [38, 39].
The literature, based on a larger group of cryptorchid patients, suggests that Thr60Ala is a polymorphism present at high frequency in the general population, and is probably unrelated to any particular phenotype of testicular maldescent [11, 17, 21, 24, 30, 36]. More recently, the rs6523 polymorphism of INSL3 showed significant association with increased risk of polycystic ovary syndrome in a well-characterized cohort of more than 400 Indian women [39]. Future studies in other populations are needed to establish whether Thr60Ala polymorphism in the INSL3 gene may contribute to the pathophysiology of male or female reproductive function.
The other two reported variants are less common, and both are located in the 3′ regulatory region. The G>A substitution (rs1003887) is prevalent in homozygosis in TT patients with statistical significance.
To evaluate the possible role of rs1003887 in a TT event, we analyzed the INSL3 gene in all members of a family in which two first cousins have a history of TT, and the proband (patient ID: TT3), included in our study, is homozygous for this genetic variant. The variation in analysis was found in all subjects of the family in homozygosis, in both unaffected males and females and in related affected subjects. The role of rs1003887 on Insl3 biological activity should, therefore, be better characterized.
The last variant, C>A substitution (rs17750642), was found in only three patients in heterozygosis and was not present in healthy subjects. To our knowledge, we have reported the presence of this variant for the first time, which has not been previously described in larger population screening. Additional studies are needed to confirm the possible association with suggestive phenotypes, and functional analysis is necessary to ascertain whether the mutated INSL3 is efficiently transcribed and processed as a stable mature hormone.
Limitations of this study are the small sample of patients and the lack of bilateral torsion cases, a very rare condition.
To define the role of INLS3 in the pathogenesis of TT, a multi-center study could be useful for screening a larger number of patients. This would allow to identify the patients at risk of TT and to improve future clinical management.
Furthermore, collecting more information on predisposing genotypes and clinical characteristics of suggestive phenotypes will be useful to better understand the role of endogenous factors, such as other genes involved in testicular descent and development, and possible interactions with exogenous disruptors to preserve male fertility in the future.
References
[Yang C, Song B, Tan J, Liu X, Wei GH (2011) Testicular torsion in children: a 20-year retrospective study in a single institution. Sci World J 11:362–368. https://doi.org/10.1100/tsw.2011.39
DrlÌk M, Kočvara R (2013) Torsion of spermatic cord in children: a review. J Pediatr Urol 3:259–266. https://doi.org/10.1016/j.jpurol.2012.05.016
Zhao LC, Lautz TB, Meeks JJ, Maizels M (2011) Pediatric testicular torsion epidemiology using a national database: incidence, risk of orchiectomy and possible measures toward improving the quality of care. J Urol 186(5):2009–2013. https://doi.org/10.1016/j.juro.2011.07.024
DaJusta DG, Granberg CF, Villanueva C, Baker LA (2013) Contemporary review of testicular torsion: new concepts, emerging technologies and potential therapeutics. J Pediatr Urol 9(6 Pt A):723–730. https://doi.org/10.1016/j.jpurol.2012.08.012
Arap MA, Vicentini FC, Cocuzza M, Hallak J, Athayde K, Lucon AM, Arap S, Srougi M (2007) Late hormonal levels, semen parameters, and presence of antisperm antibodies in patients treated for testicular torsion. J Androl 28(4):528–532. https://doi.org/10.2164/jandrol.106.002097
Schneck FX, Bellinger MF (1970) Abnormalities of the testes and scrotum and their surgical management. In: Campbell MF (ed) Anomalies of the kidney, vol 2, 3rd edn. WB Saunders, Philadelphia, pp 1447–1452
Shteynshlyuger A, Yu J (2013) Familial testicular torsion: a meta analysis suggests inheritance. J Pediatr Urol 9(5):683–690. https://doi.org/10.1016/j.jpurol.2012.08.002
Bowlin PR, Gatti JM, Murphy JP (2017) Pediatric testicular torsion. Surg Clin N Am 97(1):161–172. https://doi.org/10.1016/j.suc.2016.08.012
Fènichel P, Lahlou N, Coquillard P, Panaia-Ferrari P, Wagner-Mahler K, Brucker-Davis F (2015) Cord blood insulin-like peptide 3 (INSL3) but not testosterone is reduced in idiopathic cryptorchidism. Clin Endocrinol 82(2):242–247. https://doi.org/10.1111/cen.12500
Vinci G, Anjot MN, Trivin C, Lottmann H, Brauner R, McElreavey K (2004) An analysis of the genetic factors involved in testicular descent in a cohort of 14 male patients with anorchia. J Clin Endocrinol Metab 89(12):6282–6285. https://doi.org/10.1210/jc.2004-0891
Ferlin A, Simonato M, Bartoloni L, Rizzo G, Bettella A, Dottorini T, Dallapiccola B, Foresta C (2003) The INSL3-LGR8/GREAT ligand-receptor pair in human cryptorchidism. J Clin Endocrinol Metab 88:4273–4279. https://doi.org/10.1210/jc.2003-030359
Bay K, Andersson AM (2011) Human testicular insulin-like factor 3: in relation to development, reproductive hormones and andrological disorders. Int J Androl 34(2):97–109. https://doi.org/10.1111/j.1365-2605.2010.01074.x
Nef S, Parada LF (1999) Cryptorchidism in mice mutant for Insl3. Nat Genet 22:295–299. https://doi.org/10.1038/10364
Zimmermann S, Steding G, Emmen JM, Brinkmann AO, Nayernia K, Holstein AF, Engel W, Adham IM (1999) Targeted disruption of the Insl3 gene causes bilateral cryptorchidism. Mol Endocrinol 13(5):681–691. https://doi.org/10.1210/mend.13.5.0272
Huang X, Jia J, Sun M, Li M, Liu N (2016) Mutational screening of the INSL3 gene in azoospermic males with a history of cryptorchidism. Andrologia 48(7):835–839. https://doi.org/10.1111/and.12522
Koskimies P, Virtanen H, Lindstrom M, Kaleva M, Poutanen M, Huhtaniemi I, Toppari J (2000) A common polymorphism in the human relaxin-like factor (RLF) gene: no relationship with cryptorchidism. Pediatr Res 47:538–541
Krausz C, Quintana-Murci L, Fellous M, Siffroi JP, McElreavey K (2000) Absence of mutations involving the INSL3 gene in human idiopathic cryptorchidism. Mol Hum Reprod 6:298–302
Tomboc M, Lee PA, Mitwally MF, Schneck FX, Bellinger M, Witchel SF (2000) Insulin-like 3/relaxin-like factor gene mutations are associated with cryptorchidism. J Clin Endocrinol Metab 85(11):4013–4018. https://doi.org/10.1210/jcem.85.11.6935
Lim HN, Raipert-de Meyts E, Skakkebaek NE, Hawkins JR, Hughes IA (2001) Genetic analysis of the INSL3 gene in patients with maldescent of the testis. Eur J Endocrinol 144:129–137
Marin P, Ferlin A, Moro E, Garolla A, Foresta C (2001) Different insulin-like 3 (INSL3) gene mutations not associated with human cryptorchidism. J Endocrinol Invest 24:RC13-RC15. https://doi.org/10.1007/BF03343848
Takahashi I, Takahashi T, Komatsu M, Matsuda J, Takada G (2001) Ala/Thr60 variant of the Leydig insulin-like hormone is not associated with cryptorchidism in the Japanese population. Pediatr Int 43:256–258
Baker LA, Nef S, Nguyen MT, Stapleton R, Nordenskjold A, Pohl H, Parada LF (2002) The insulin-3 gene: lack of a genetic basis for human cryptorchidism. J Urol 167:2534–2537
Canto P, Escudero I, Soderlund D, Nishimura E, Carranza-Lira S, Gutierrez J, Nava A, Mendez JP (2003) A novel mutation of the insulin-like 3 gene in patients with cryptorchidism. J Hum Genet 48:86–90. https://doi.org/10.1007/s100380300012
Ferlin A, Bogatcheva NV, Gianesello L, Pepe A, Vinanzi C, Agoulnik AI, Foresta C (2006) Insulin-like factor 3 gene mutations in testicular dysgenesis syndrome: clinical and functional characterization. Mol Hum Reprod 12:401–406. https://doi.org/10.1093/molehr/gal043
Ferlin A, Zuccarello D, Zuccarello B, Chirico MR, Zanon GF, Foresta C (2008) Genetic alterations associated with cryptorchidism. JAMA 300(19):2271–2276. https://doi.org/10.1001/jama.2008.668
Feng S, Cortessis VK, Hwang A, Hardy B, Koh CJ, Bogatcheva NV, Agoulnik AI (2004) Mutation analysis of INSL3 and GREAT/ LGR8 genes in familial cryptorchidism. Urology 64:1032–1036. https://doi.org/10.1016/j.urology.2004.06.051
Foresta C, Ferlin A (2004) Role of INSL3 and LGR8 in cryptorchism and testicular functions. Reprod Biomed Online 9(3):294–298
El Houate B, Rouba H, Sibai H, Barakat A, Chafik A, Chadli el B, Imken L, Bogatcheva NV, Feng S, Agoulnik AI, McElreavey K (2007) Novel mutations involving the INSL3 gene associated with cryptorchidism. J Urol 177:1947–1951. https://doi.org/10.1016/j.juro.2007.01.002
Yamazawa K, Wada Y, Sasagawa I, Aoki K, Ueoka K, Ogata T (2007) Mutation and polymorphism analyses of INSL3 and LGR8/GREAT in 62 Japanese patients with cryptorchidism. Horm Res 67:73–76. https://doi.org/10.1159/000096089
Mamoulakis C, Georgiou I, Dimitriadis F, Tsounapi P, Giannakis I, Chatzikyriakidou A, Antypas S, Sofras F, Takenaka A, Sofikitis N (2014) Genetic analysis of the human Insulin-like 3 gene: absence of mutations in a Greek paediatric cohort with testicular maldescent. Andrologia 46(9):986–996. https://doi.org/10.1111/and.12184
Sozubir S, Barber T, Wang Y, Ahn C, Zhang S, Verma S, Lonergan D, Lorenzo AJ, Nef S, Baker LA (2010) Loss of Insl3: a potential predisposing factor for testicular torsion. J Urol 183(6):2373–2379. https://doi.org/10.1016/j.juro.2010.02.2390
Romeo C, Impellizzeri P, Arrigo T, Antonuccio P, Valenzise M, Mirabelli S, Astra Borruto F, Scalfari G, Arena F, De Luca F (2010) Late hormonal function after testicular torsion. J Pediatr Surg 45(2):411–413. https://doi.org/10.1016/j.jpedsurg.2009.10.086
Ivell R, Balvers M, Domagalski R, Ungefroren H, Hunt N, Schulze W (1997) The relaxin-like factor: a highly specific and constitutive new marker for Leydig cells in the human testis. Mol Hum Reprod 3(6):459–466
Ivell R, Anand-Ivell R (2009) Biology of insulin-like factor 3 in human reproduction. Hum Reprod Update 15(4):463–476. https://doi.org/10.1093/humupd/dmp011
Ivell R, Agoulnik AI, Anand-Ivell R (2017) Relaxin-like peptides in male reproduction—a human perspective. Br J Pharmacol 174(10):990–1001. https://doi.org/10.1111/bph.13689
Foresta C, Zuccarello D, Garolla A, Ferlin A (2008) Role of hormones, genes, and environment in human cryptorchidism. Endocr Rev 29(5):560–580. https://doi.org/10.1210/er.2007-0042
Bogatcheva NV, Truong A, Feng S, Engel W, Adham IM, Agoulnik AI (2003) GREAT/LGR8 is the only receptor for insulin-like 3 peptide. Mol Endocrinol 17(12):2639–2646. https://doi.org/10.1210/me.2003-0096
Ivell R, Heng K, Anand-Ivell R (2014) Insulin-like factor 3 and the HPG axis in the male. Front Endocrinol (Lausanne) 27(5):6. https://doi.org/10.3389/fendo.2014.00006
Shaikh N, Dadachanji R, Meherji P, Shah N, Mukherjee S (2016) Polymorphisms and haplotypes of insulin-like factor 3 gene are associated with risk of polycystic ovary syndrome in Indian women. Gene 577(2):180–186. https://doi.org/10.1016/j.gene.2015.11.033
Funding
This study did not receive any grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study. No identifying information about participants is available in the article.
Rights and permissions
About this article
Cite this article
Capra, A.P., Ferro, E., La Rosa, M.A. et al. Genetic analysis of the human insulin-like 3 gene in pediatric patients with testicular torsion. Pediatr Surg Int 34, 807–812 (2018). https://doi.org/10.1007/s00383-018-4280-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00383-018-4280-y