Abstract
Methyl jasmonate (MeJA) has significant effects on the apple ripening process, but little is known about the molecular mechanisms by which MeJA acts in the regulation of this complicated process. To address this question, transcriptome profiles of MeJA-treated apples and mock-treated controls were generated using RNA-sequencing technology and then compared. A total of 1092 transcripts changed significantly in response to MeJA, with 684 up-regulated and 408 down-regulated. In MeJA-treated apples, genes involved in anthocyanin biosynthesis and transport were commonly up-regulated. The up-regulated genes also included genes in the biosynthetic and signal transduction pathways for the hormones jasmonic acid and ethylene. In contrast, exogenous MeJA generally down-regulated genes associated with auxin signal transduction. Furthermore, the transcript levels of 10 jasmonic acid biosynthetic genes, 7 selected ethylene biosynthetic and signal transduction genes, 5 selected auxin signal transduction genes, and 8 selected anthocyanin biosynthetic and transport genes were confirmed via real-time qPCR. These results suggest that MeJA promotes apple ripening, likely by activating ethylene signaling and inhibiting auxin action. Because MeJA-treated apples have previously been documented to have high anthocyanin concentrations, the molecular mechanism underlying this phenomenon is also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fruits are an important part of the human diet, providing vitamins, minerals, and ‘bioactive’ compounds (Miller and Rice-Evans 1997; Heim and others 2002; Briones-Labarca and others 2011). There is good evidence that fruits promote healthy aging by protecting against heart disease and cancer (Hertog and others 1993; Boyer and Liu 2004).
Ripening is the final stage of fruit development and is unique to plant species. Ripening is a complex developmental process involving a number of dramatic changes in color, texture, flavor, and aroma (Carrari and Fernie 2006). In general, fruits can be divided into two types based on ripening pattern: climacteric fruit and non-climacteric fruit (Yang and Hoffman 1984). Apples are economically important crop that are classified as climacteric fruits, in which ethylene is the main ripening trigger (Lay-Yee and others 1990; Schaffer and others 2007). In addition to ethylene, other hormones have recently been found to influence the apple ripening process (Romani and others 1989; Fan and others 1997; Kondo and others 2005; Li and others 2006). Amongst these hormones, jasmonic acid (JA) seems to play a major role.
The JA biosynthesis pathway has been well characterized. JAs are synthesized from α-linolenic acid in the octadecanoid pathway, and three key enzymes are involved in JA biosynthesis: lipoxygenase (LOX), allene oxide synthase (AOS), and 12-oxophytodienoate reductase 3 (OPR3) (Turner and others 2002). JA and its methyl ester (MeJA) are both involved in mediating fruit ripening (Concha and others 2013). For example, the levels of JA and the AOS gene in apples increase during ripening, suggesting their potential roles in the regulation of this process (Fan and others 1997; Lv and others 2015). Pre-climacteric applications of MeJA stimulate the fruit ripening process in apple; for example, MeJA stimulates ethylene and ester biosynthesis (Fan and others 1997; Kondo and others 2005), increases anthocyanin content as well as the accumulation of several phenolic compounds, and promotes red color (Pérez and others 1993; Fan and others 1998; Rudell and Mattheis 2008).
The physiological effects of JAs in the apple ripening process have been widely studied, but the precise mechanism by which JA contributes to this process remains elusive. In this study, high-throughput RNA-Seq was utilized to analyze the changes of transcription in apple with the pre-climacteric application of MeJA. This study may provide important information regarding the function of MeJA in apple ripening and may aid in the identification of genes involved in the MeJA-activated apple ripening process.
Materials and Methods
Plant Material
The fruits were collected from ‘Taishan Zaoxia’ plants in a commercial orchard, Liaocheng, Shandong, China, in June 2014, 6 days before ripening. The fruits were treated with 10−4, 10−3, and 10−2 M MeJA, and were then transferred to plant growth chambers at 25 °C and with 10/14 h light/dark cycle. The anthocyanin content was monitored daily for 6 days. Because 10−3 M MeJA concentration has beneficial effects on fruit ripening by accelerating anthocyanin synthesis (Fig. 1), this concentration was chosen for further RNA-Seq and qPCR studies. Ten fruits per time point were sampled at 2, 6, and 10 h after treatment. The peels of the fruits from each sampling time were frozen in liquid nitrogen and stored at −80 °C until use.
Anthocyanin Content Analysis
Anthocyanin content was measured using a previously described method (Wang and others 2004; Feng and others 2010). Anthocyanin was extracted with an HCl–methanol method, and was then monitored at 553 and 600 nm. Determinations were performed in triplicate. Statistical analyses were carried out using SAS software.
RNA Isolation
Total RNA was isolated using Trizol reagent (Invitrogen, USA) following the manufacturer’s recommendations. The integrity of RNA was evaluated using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with an A260/280 ratio between 1.8 and 2.0, an A260/230 ratio greater than 1.7, and an RNA integrity number (RIN) above 8.0 were defined to be high-quality RNAs. High-quality total RNAs for each sampling time were equally mixed to construct two RNA-Seq libraries (MeJA-treated apple and mock-treated control) for sequencing.
cDNA Library Construction and Illumina Deep Sequencing
mRNA was purified from total RNA using oligo (dT) magnetic beads and then broken into short fragments in the fragmentation buffer. The mRNA fragments were used as templates to synthesize the first-strand cDNA. Then, the second strands were synthesized by adding cDNA, dNTPs, RNase H, buffer, and DNA polymerase I. The double-stranded cDNAs were purified using the Qiaquick PCR extraction kit (Qiagen, Hilden, Germany) and washed with EB buffer for end repair, poly(A) addition, and PCR amplification. Each cDNA library was sequenced using the Illumina sequencing system (HiSeq™ 2000, Illumina, San Diego, CA, USA).
Bioinformatic Analysis
Quality Control
Raw reads were initially processed using the NGS QC Toolkit (Patel and Jain 2012). In this step, low-quality reads and reads containing adapters and poly-N were removed from the raw data. All of the subsequent analyses were based on clean, high-quality data.
Mapping
Sequencing reads were mapped to the apple genome (http://genomics.research.iasma.it/) using bowtie2 (Langmead and Salzberg 2012). The FPKM and count value were calculated using eXpress (Mortazavi and others 2008).
Analysis of Differential Expression and of GO and KEGG Enrichment
Differential expression analysis was performed using the DESeq (2012) R package. P < 0.05 was set as the threshold for significant differential expression, and |log 2 fold change| ≥ 1 was used to identify the genes differentially expressed between the two libraries (MeJA-treated apple and mock-treated control). Gene Ontology (GO) and KEGG (Kyoto encyclopedia of Genes and Genomes) enrichment analyses of the DEGs were performed using R based on hypergeometric distribution.
qRT-PCR Analysis
qRT-PCR for validating RNA-Seq data was performed using the same RNA mixtures as for the RNA-Seq library construction. qRT-PCR for testing the expression patterns of phytohormone- and anthocyanin-related genes at different time points (2, 6, 10 h) was performed using the RNA for each sampling time. First-strand cDNAs were synthesized using the RevertAid™ First-Strand cDNA Synthesis kit (Fermentas, USA) and used as templates for qPCR assays. qRT-PCR was conducted as described previously (Feng and others 2010). MdActin (XM_008384595.1) was used as a reference gene. The primers used in qPCR assays are listed in Supplementary Table 1. Three replications were conducted.
Results
Effect of MeJA on Anthocyanin Accumulation in Apple Skin
We observed a dose-dependent effect of MeJA on anthocyanin accumulation in apple skin (Fig. 1). Treatment with MeJA at concentrations of 10−4 and 10−3 M generally promoted anthocyanin accumulation. The rate of anthocyanin accumulation increased as the MeJA concentration increased from 10−4 to 10−3 M during the first 4 days after treatment; however, MeJA concentration did not affect the final anthocyanin content at ripening. MeJA treatment at 10−2 M strongly inhibited anthocyanin production, which is consistent with See and others (2011), who reported that a higher concentration of MeJA caused a reduction in anthocyanin production and accumulation.
Transcriptome Sequencing and Sequence Alignment
A total of 8 Gb raw data were obtained in this study. There were 37,202,822 raw reads in the MeJA-treated apple library; the Q30 percentage (percentage of bases whose quality was greater than 30 in clean reads), valid ratio (base), and GC percentage are 87.11, 99.12, and 46.00 %, respectively. There were 48,438,800 raw reads in the mock-treated apple library; the Q30 percentage, valid ratio, and GC percentage are 87.56, 99.11, and 46.00 %, respectively. After removing the low-quality reads and reads with unknown nucleotides, 36,923,726 and 48,070,300 clean reads were obtained from the MeJA-treated apple library and the mock-treated apple library, respectively. Then, 86.43 % of the MeJA-treated apple reads and 86.42 % of the mock-treated apple reads were mapped to the reference apple genome sequence (Table 1).
Alterations in the Gene Expression Profiles Between MeJA-Treated Apple and Mock-Treated Control
After discarding the genes with small expression changes (absolute log 2 of the fold change <1), a total of 1092 differentially expressed genes (DEGs) in response to MeJA were obtained (Supplementary Table 2). The log 2 of the fold change of these DEGs ranged from −8.1 to 7.8. As shown in Fig. 2, 684 DEGs showed up-regulation, more than 86 % of the up-regulated genes showed changes in the range of 2–16-fold, and only a small proportion of the DEGs were up-regulated at least 16-fold. Compared with the up-regulated DEGs, a higher percentage of the down-regulated genes showed changes of at least 16-fold (17.6 % of the 408 down-regulated DEGs).
Annotation and Functional Classification of DEGs
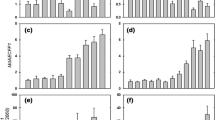
To gain insights into the biological functions of the DEGs, we performed GO analyses by comparing each DEG with the GO database. A total of 1092 DEGs were annotated in 183 GO biological processes (Supplementary Table 3). There were 121 GO terms enriched among the up-regulated DEGs (Supplementary Table 4), and 130 GO terms were enriched among the down-regulated DEGs (Supplementary Table 5). For the up-regulated DEGs, fatty acid biosynthetic process, lipid biosynthetic process, metabolic process, methylation, and protein dephosphorylation were the most enriched GO biological process terms (Fig. 3a). The most enriched GO biological process terms among the down-regulated DEGs were oxidation–reduction process, metabolic process, response to biotic stimulus, phospholipid transport, and lignin catabolic process (Fig. 3b).
The most enriched GO biological process terms analysis of the up- and down-regulated DEGs in the apple in response to MeJA treatment. a Top twenty GO biological process terms for the up-regulated DEGs in apples in response to MeJA treatment. b Top twenty GO biological process terms for the down-regulated DEGs in apples in response to MeJA treatment
To further understand the biological functions of the DEGs, we performed KEGG pathway enrichment analysis. In total, 72 and 87 regulated pathways were enriched among the down-regulated and up-regulated DEGs, respectively (Supplementary Tables 6, 7). The 20 KEGG pathways showing the strongest enrichment among the up-regulated and down-regulated DEGs are presented in Fig. 4a, b, respectively.
The most enriched KEGG pathway analysis of the up- and down-regulated DEGs in apples in response to MeJA treatment. a Top twenty enriched KEGG pathways of the up-regulated DEGs in apples in response to MeJA treatment. b Top twenty enriched KEGG pathways of the down-regulated DEGs in apples in response to MeJA treatment
Impact of MeJA on Phytohormone Pathways
The enriched DEGs associated with phytohormone pathways were mainly involved in the biosynthesis and signal transduction of jasmonic acid, ethylene, and auxin. As shown in Supplementary Table 8, there are fifteen DEGs associated with JA biosynthesis and signal transduction. Among them, thirteen well-known genes in the JA biosynthesis pathway, including OPR2 (2 members), putative OPR11 (7 members), AOS (2 members), one allene oxide cyclase 4 (AOC4), one jasmonate O-methyltransferase (JMT), and two JA-receptor proteins (protein TIFY 5A and protein TIFY 10B), were up-regulated in response to exogenous MeJA.
In ethylene biosynthesis and signal transduction, there are thirteen up-regulated DEGs and six down-regulated DEGs (Supplementary Table 9). The up-regulated DEGs include six ethylene biosynthetic genes (four 1-aminocyclopropane-1-carboxylate oxidase ACO genes, and two 1-aminocyclopropane-1-carboxylate synthase (ACS) genes) and seven ethylene signal transduction genes (one ethylene-responsive transcription factor 1B gene (ERF1), two ethylene-responsive transcription factor ERF073 genes (ERF073), one ethylene-responsive transcription factor ERF011 gene (ERF011), one ethylene-responsive transcription factor ERF071 gene (ERF071), and two ethylene-responsive transcription factor ABR1 genes (ABR1)). In contrast, ethylene signal transduction elements such as ethylene-insensitive protein 2 like (EIN2), ethylene-insensitive protein 3 like (EIN3), the ethylene-responsive transcription factor RAP2-13 like (RAP2-13), and the ethylene-responsive transcription factor ERF003 like (ERF003), as well as the AP2-like ethylene-responsive transcription factor At1g16060, were down-regulated by MeJA.
The DEGs involved in the auxin signal transduction pathway are listed in Supplementary Table 10. Among these DEGs, 18 genes were down-regulated. The down-regulated DEGs mainly function as auxin-induced proteins, auxin-responsive proteins, auxin receptors, and IAA-conjugating enzymes. The two up-regulated DEGs were ABC transporter B family member 15-like (XM_008370208.1) and IAA-amino acid hydrolase ILR1-like 4 (XM_008391362.1), which are involved in auxin transport and IAA amidohydrolysis, respectively.
Effects of MeJA on Anthocyanin Biosynthesis and Transport
Seventeen DEGs for anthocyanin biosynthesis were identified in our RNA-Seq dataset, and all of them showed up-regulated expression in response to MeJA (Supplementary Table 11). These up-regulated DEGs included key anthocyanin biosynthetic enzymes: flavonol synthase (FLS), anthocyanidin 3-O-glucosyltransferase 2 (UFGT2), putative UDP-glucose flavonoid 3-O-glucosyltransferase 3 (UFGT3), and UDP-glucose flavonoid 3-O-glucosyltransferase 7 (UFGT7). In addition, DEGs involved in anthocyanin transport were also found in the dataset, including nine glutathione S-transferase (GST), two glutathione transferase GST23-like genes, and four ABC transporter C family (ABCC) members. The expression levels of genes encoding anthocyanin transporters were all up-regulated by MeJA.
qRT-PCR Analysis
The expression patterns of a subset of the phytohormone- and anthocyanin-related genes were detected by qPCR. We determined that the expression levels changed significantly in MeJA-treated apple after 2, 6, and 10 h of treatment. Especially during the first 6 h, most genes were strongly up- or down-regulated by MeJA treatment. However, the expression levels of the majority of genes were not changed (fold change <2) in non-treated controls (Fig. 5; Supplementary Table 12). These data suggest that MeJA is an important factor in gene regulation. All the tested genes were then used to validate and verify the RNA-Seq data (Fig. 6). We observed that the qRT-PCR results showed trends similar to those for the RNA-Seq data, suggesting that the RNA-Seq data are reliable.
qPCR analysis of a subset of phytohormone- and anthocyanin-related genes at 2, 6, 10 h after MeJA treatment. Heat map showing changes in the expression levels of phytohormone- and anthocyanin-related genes in controls and MeJA-treated samples at 2, 6, and 10 h compared to 0 h after MeJA treatment. Genes in red and green represent highly and lowly expressed genes, respectively. The qPCR data are provided in Supplementary Table 12
Discussion
RNA-Seq is a high-throughput method that has been widely utilized to reveal information on the transcript profiles of model organisms, especially for discovering and identifying genes involved in the biosynthesis of various secondary metabolites and the formation of special architecture (Zhang and others 2015). In this work, we studied the transcript information of apple in response to MeJA treatment at the early stage using RNA-Seq technology. Test tissues of MeJA-treated apple and a mock-treated control at various time points were mixed into MeJA and control libraries, respectively. Finally, a total of 1092 DEGs were detected and annotated. The changes in relative expression of DEGs were further confirmed by qPCR analysis using a subset of regulated genes, indicating that the RNA-Seq data are high in quality.
Previous studies suggested that phytohormones play important roles in the apple ripening process. Among these phytohormones, ethylene is known to function as a pivotal regulation factor for fruit ripening (Giovannoni 2001; Liu and others 2016). However, the complex phenotypes related to ripening cannot be explained only by ethylene-mediated effects. Other phytohormones including JA and auxin have recently been implicated in controlling this biological process (Ohmiya 2000; Trainotti and others 2007; Li and others 2006; Schaffer and others 2013). Usually, these phytohormones function as a complex network rather than as independent linear pathways (Lv and others 2015). In this study, 55 out of 1092 DEGs were annotated for jasmonic acid, ethylene and auxin biosynthesis, and signal transduction pathways, suggesting that the promotion of fruit ripening by MeJA may be mediated directly or indirectly via regulating the transcripts of these phytohormone-related genes.
MeJA treatment of apple produced significant changes in JA-related gene expression. The JA biosynthetic genes AOC, AOS, and OPR, as well as the MeJA biosynthetic gene JMT (Turner and others 2002) were all strongly activated by MeJA treatment. This is consistent with results in Arabidopsis (Kubigsteltig and others 1999) and tomato (Wasternack 2014), in which JA biosynthesis is transcriptionally regulated by a JA-mediated positive feedback loop. According to a previous study, the levels of JAs and JA biosynthetic genes were dramatically up-regulated prior to apple ripening (Lv and others 2015), and the application of exogenous MeJA may have initiated this biological process. In addition, the JA signal receptor gene JAZ is also up-regulated in response to JA treatment. Similarly, JA induction of the expression of JAZ genes has also been observed in Arabidopsis (Yan and others 2007), poplar (Major and Constabel 2006), and tomato (Chung and others 2008), indicating that this phenomenon is conserved among plants. Because the JAZ proteins are known as repressors of JA signaling, the rapid synthesis of new JAZ proteins was suggested to attenuate the transcriptional response soon after it is initiated.
It has previously been shown that transient increases in endogenous JA concentrations occurred prior to the stage of apple ripening during which ethylene production increases rapidly. The increase in endogenous JAs was suggested to be responsible for the ethylene climacteric peak, which is a hallmark of fruit ripening (Fan and others 1998). In this study, MeJA strongly up-regulated two ethylene biosynthetic genes, ACO1 and ACS1, which have been reported to increase before and during ripening (Tan and others 2013; Bulens and others 2014). The rise of transcripts required for the ethylene climacteric likely accelerates the progression of apple ripening. A similar ethylene response to JAs has also been reported in pear and tomato, in which the expression of ACS and ACO was enhanced in pre-climacteric fruit under treatment with propyl dihydrojasmonate (PDJ) or MeJA (Yu and others 2009). These results are in agreement with the observations that suggest an acceleration of fruit ripening by JAs (Kondo and others 2009; Mukkun and Singh 2009; Yu and others 2011). In addition, MeJA treatment also up-regulated the expression of ethylene signal transduction genes, such as ERF1, ERF011, ERF071, and ERF073. Accumulating evidence suggests that ERF1 is an upstream component in both the JA and ET signaling pathways and is involved in pathogen resistance and the responses to salt and drought stress (Lorenzo and others 2003). The induction of ERF1in our study suggested that ERF1 may accelerate the fruit ripening process via ripening-specific gene regulation that integrates JA and ET signals. Unlike ERF1, EIN3 was down-regulated in MeJA-treated apple, which is consistent with the reports that EIN3 transcription is repressed by JAZ (Zhu and others 2011).
The effects of auxin on fruit ripening have been reported in several species, including peach (Ohmiya 2000), tomato (Buta and Spaulding 1994; Su and others 2015), grape (Böttcher and others 2010), and strawberry (Symons and others 2012). In peaches, increased levels of IAA are required for fruit ripening (Trainotti and others 2007). MeJA treatment strongly inhibits auxin-related genes and delays fruit ripening (Ziosi and others 2008; Soto and others 2012). GH3 inactivates IAA via conjugation, and it is considered a marker of free auxin (Staswick and others 2005). In peaches, MeJA treatment led to a decrease of GH3 gene expression. This behavior would explain the lower levels of free IAA in MeJA-treated fruits. In addition, the auxin receptor TIR1 (Dharmasiri and others 2005) was also down-regulated. This result reflected the changes of auxin reception and transport response to MeJA treatment, which further support the notion that auxin availability was low in treated peaches. The down-regulation of GH3 and TIR1 by MeJA is in accord with the ripening delay. In the present study, a similar auxin response to JAs was also found in apples. GH3, TIR1, and other auxin-responsive genes were down-regulated by MeJA, which suggest a possible decrease in free auxin availability. Unlike in peaches, decreases in IAA prior to fruit ripening have been reported in apples (Schaffer and others 2013), which is also consistent with the results in tomatoes and strawberries (Buta and Spaulding 1994; Srivastava and Handa 2005). Therefore, we propose that the positive role of MeJA in promoting apple ripening likely occurs via inhibition of auxin signaling. Surprisingly, the auxin transport-related gene ABCB15 (Matsuda and others 2011) and the IAA-amino acid hydrolase ILR1 (LeClere and others 2002) were up-regulated in treated apples. This suggests that auxin signaling is partially but not completely inhibited by MeJA. Exogenous JAs can induce a variety of phenotypical effects during fruit ripening. For example, MeJA promotes degreening of apple peel (Fan and Mattheis 1999) as well as anthocyanin accumulation (Rudell and others 2002). Anthocyanins are biosynthesized through the flavonoid pathway. Two types of genes are required for anthocyanin synthesis: biosynthetic structural genes and regulatory genes (Li 2014). Previous studies illustrated that JAs can induce both types of anthocyanin-related genes (Dombrecht and others 2007; Qi and others 2011). In our study, anthocyanin accumulation is greatly promoted, and the anthocyanin biosynthetic structural genes F3Hs and UFGTs were also up-regulated in MeJA-treated apples. However, no regulatory genes were identified among the DEGs. A recent study in apple has documented that JAs could directly mediate anthocyanin biosynthesis by removal of the JAZ repression on anthocyanin regulatory factors (An and others 2015). Considering the up-regulation of JAZ genes at this stage, the activation of regulatory genes probably occurred at the later stage. Anthocyanins are biosynthesized in the cytosol and transported into the vacuole by putative anthocyanin transporters. ABCC transporters and GSTs have been suggested to be involved in anthocyanin transport (Zhao 2015). The up-regulation of ABCC transporters and GSTs indicated that JAs also mediate anthocyanin accumulation by transcriptionally regulating anthocyanin transporters.
Conclusions
Using RNA-Seq technology, we investigated the differences in transcription between MeJA-treated and mock-treated apples. In total, 1092 DEGs were identified as showing significant responses to MeJA. The reliability of the RNA-Seq data was confirmed by qPCR. Furthermore, DEGs related to phytohormone pathways (ethylene, auxin, and JA), anthocyanin biosynthesis, and transport were identified. This study may provide important information for the identification of genes involved in MeJA-meditated apple ripening and may aid in understanding the molecular mechanisms underlying this complex biological process.
References
An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56:650–662. doi:10.1093/pcp/pcu205
Böttcher C, Keyzers RA, Boss PK, Davies C (2010) Sequestration of auxin by the indole-3-acetic acid-amino synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot 61:3615–3625. doi:10.1093/jxb/erq174
Boyer J, Liu RH (2004) Apple phytochemicals and their health benefits. J Nutr 3:5. doi:10.1186/1475-2891-3-5
Briones-Labarca V, Venegas-Cubillos G, Ortiz-Portilla S, Chacana-Ojeda M, Maureira H (2011) Effects of high hydrostatic pressure (HHP) on bioaccessibility, as well as antioxidant activity, mineral and starch contents in Granny Smith apple. Food Chem 128:520–529. doi:10.1016/j.foodchem.2011.03.074
Bulens I, Van de Poel B, Hertog ML, Cristescu SM, Harren FJ, De Proft MP, Geeraerd AH, Nicolai BM (2014) Dynamic changes of the ethylene biosynthesis in ‘Jonagold’ apple. Physiol Plant 150:161–173. doi:10.1111/ppl.12084
Buta JG, Spaulding DW (1994) Changes in indole-3-acetic acid and abscisic acid levels during tomato (Lycopersicon esculentum Mill.) fruit development and ripening. J Plant Growth Regul 13:163–166. doi:10.1007/BF00196382
Carrari F, Fernie AR (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57:1883–1897. doi:10.1093/jxb/erj020
Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis jasmonate ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146:952–964. doi:10.1104/pp.107.115691
Concha CM, Figueroa NE, Poblete LA, Oñate FA, Schwab W, Figueroa CR (2013) Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol Biochem 70:433–444. doi:10.1016/j.plaphy.2013.06.008
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9:109–119. doi:10.1016/j.devcel.2005.05.014
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245. doi:10.1105/tpc.106.048017
Fan X, Mattheis JP (1999) Methyl jasmonate promotes apple fruit degreening independently of ethylene action. HortScience 34:310–312
Fan X, Mattheis JP, Fellman JK, Patterson ME (1997) Effect of methyl jasmonate on ethylene and volatile production by summerred apples depends on fruit developmental stage. J Agric Food Chem 45:208–211. doi:10.1021/jf9603846
Fan X, Mattheis JP, Fellman JK (1998) A role for jasmonates in climacteric fruit ripening. Planta 204:444–449. doi:10.1007/s004250050278
Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255. doi:10.1007/s00425-010-1170-5
Giovannoni J (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52:725–749. doi:10.1146/annurev.arplant.52.1.725
Heim KE, Tagliaferro AR, Bobilya DJ (2002) Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J Nutr Biochem 13:572–584. doi:10.1016/S0955-2863(02)00208-5
Hertog MG, Feskens EJ, Hollman PC, Katan MB, Kromhout D (1993) Dietary antioxidant flavonoids and risk of coronary heart disease: the Zutphen Elderly Study. Lancet 342:1007–1011. doi:10.1016/0140-6736(93)92876-U
Kondo S, Setha S, Rudell DR, Buchanan DA, Mattheis JP (2005) Aroma volatile biosynthesis in apples affected by 1-MCP and methyl jasmonate. Postharvest Biol Technol 36:61–68. doi:10.1016/j.postharvbio.2004.11.005
Kondo S, Meemak S, Ban Y, Moriguchi T, Harada T (2009) Effects of auxin and jasmonates on 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase gene expression during ripening of apple fruit. Postharvest Biol Tecnol 51:281–284. doi:10.1016/j.postharvbio.2008.07.012
Kubigsteltig I, Laudert D, Weiler EW (1999) Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208:463–471. doi:10.1007/s004250050583
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Method 9:357–359. doi:10.1038/nmeth.1923
Lay-Yee M, Dellapenna D, Ross GS (1990) Changes in mRNA and protein during ripening in apple fruit (Malus domestica Borkh. cv golden delicious). Plant Physiol 94:850–853. doi:10.1104/pp.94.2.850
LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277:20446–20452. doi:10.1074/jbc.M111955200
Li S (2014) Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav 9:e27522
Li DP, Xu YF, Sun LP, Liu LX, Hu XL, Li DQ, Shu HR (2006) Salicylic acid, ethephon, and methyl jasmonate enhance ester regeneration in 1-MCP-treated apple fruit after long-term cold storage. J Agric Food Chem 54:3887–3895. doi:10.1021/jf060240j
Liu M, Gomes BL, Mila I, Purgatto E, Peres LE, Frasse P, Maza E, Zouine M, Roustan JP, Bouzayen M, Pirrello J (2016) Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato (Solanum lycopersicum). Plant Physiol 170:1732–1744. doi:10.1104/pp.15.01859
Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15:165–178. doi:10.1105/tpc.007468
Lv J, Rao J, Johnson F, Shin S, Zhu Y (2015) Genome-wide identification of jasmonate biosynthetic genes and characterization of their expression profiles during apple (Malus × domestica) fruit maturation. Plant Growth Regul 75:355–364. doi:10.1007/s10725-014-9958-0
Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172:617–635. doi:10.1111/j.1469-8137.2006.01877.x
Matsuda S, Kajizuka T, Kadota A, Nishimura T, Koshiba T (2011) NPH3- and PGP-like genes are exclusively expressed in the apical tip region essential for blue-light perception and lateral auxin transport in maize coleoptiles. J Exp Bot 62:3459–3466. doi:10.1093/jxb/err019
Miller NJ, Rice-Evans CA (1997) The relative contributions of ascorbic acid and phenolic antioxidants to the total antioxidant activity of orange and apple fruit juices and black currantdrink. Food Chem 60:331–337. doi:10.1016/S0308-8146(96)00339-1
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Method 5:621–628. doi:10.1038/nmeth.1226
Mukkun L, Singh Z (2009) Methyl jasmonate plays a role in fruit ripening of ‘Pajaro’ strawberry through stimulation of ethylene biosynthesis. Sci Hortic 123:5–10. doi:10.1016/j.scienta.2009.07.006
Ohmiya A (2000) Effects of auxin on growth and ripening of mesocarp discs of peach fruit. Sci Hortic 84:309–319. doi:10.1016/S0304-4238(99)00137-5
Patel RK, Jain M (2012) NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619. doi:10.1371/journal.pone.0030619
Pérez AG, Sanz C, Richardson DG, Olías JM (1993) Methyl jasmonate vapor promotes β-carotene synthesis and chlorophyll degradation in golden delicious apple peel. J Plant Growth Regul 12:163–167. doi:10.1007/BF00189648
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814. doi:10.1105/tpc.111.083261
Romani RJ, Hess BM, Leslie CA (1989) Salicylic acid inhibition of ethylene production by apple discs and other plant tissues. J Plant Growth Regul 8:63–69. doi:10.1007/BF02024927
Rudell DR, Mattheis JP (2008) Synergism exists between ethylene and methyl jasmonate in artificial light-induced pigment enhancement of ‘Fuji’ apple fruit peel. Postharvest Biol Technol 47:136–140. doi:10.1016/j.postharvbio.2007.05.021
Rudell DR, Mattheis JP, Fan X, Fellman JK (2002) Methyl jasmonate enhances anthocyanin accumulation and modifies production of phenolics and pigments in ‘Fuji’ apples. J Am Soc Hortic Sci 127(3):435–441
Schaffer RJ, Friel EN, Souleyre EJ, Bolitho K, Thodey K, Ledger S, Bowen JH, Ma JH, Nain B, Cohen D, Gleave AP, Crowhurst RN, Janssen BJ, Yao JL, Newcomb RD (2007) A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiol 144:1899–1912. doi:10.1104/pp.106.093765
Schaffer RJ, Ireland HS, Ross JJ, Ling TJ, David KM (2013) SEPALLATA1/2-suppressed mature apples have low ethylene, high auxin and reduced transcription of ripening-related genes. AoB Plant. doi:10.1093/aobpla/pls047
See KS, Bhatt A, Keng CL (2011) Effect of sucrose and methyl jasmonate on biomass and anthocyanin production in cell suspension culture of Melastoma malabathricum (Melastomaceae). Rev Biol Trop 59(2):597–606
Soto A, Ruiz KB, Ziosi V, Costa G, Torrigiani P (2012) Ethylene and auxin biosynthesis and signaling are impaired by methyl jasmonate leading to a transient slowing down of ripening in peach fruit. J Plant Physiol 169:1858–1865. doi:10.1016/j.jplph.2012.07.007
Srivastava A, Handa AK (2005) Hormonal regulation of tomato fruit development: a molecular perspective. J Plant Growth Regul 24:67–82. doi:10.1007/s00344-005-0015-0
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627. doi:10.1105/tpc.104.026690
Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15:114. doi:10.1186/s12870-015-0495-4
Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB (2012) Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot 63:4741–4750. doi:10.1093/jxb/ers147
Tan D, Li T, Wang A (2013) Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol Biol Rep 31:204–209. doi:10.1007/s11105-012-0490-y
Trainotti L, Tadiello A, Casadoro G (2007) The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J Exp Bot 58:3299–3308. doi:10.1093/jxb/erm178
Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14:S153–S164
Wang HC, Huang XM, Hu GB, Huang HB (2004) Studies on the relationship between anthocyanin biosynthesis and related enzymes in Litchi pericarp. Sci Agric Sin 37:2028–2032
Wasternack C (2014) Perception, signaling and cross-talk of jasmonates and the seminal contributions of the Daoxin Xie’s lab and the Chuanyou Li’s lab. Plant Cell Rep 33:707–718. doi:10.1007/s00299-014-1608-5
Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19:2470–2483. doi:10.1105/tpc.107.050708
Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35:155–189. doi:10.1146/annurev.pp.35.060184.001103
Yu M, Shen L, Fan B, Zhao D, Zheng Y, Sheng J (2009) The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol Technol 54:153–158. doi:10.1016/j.postharvbio.2009.07.001
Yu M, Shen L, Zhang A, Sheng J (2011) Methyl jasmonate-induced defense responses are associated with elevation of 1-aminocyclopropane-1-carboxylate oxidase in Lycopersicon esculentum fruit. J Plant Physiol 168:1820–1827. doi:10.1016/j.jplph.2011.05.021
Zhang Y, Chen J, Liu B, Xia M, Wang W, Shen F (2015) Transcriptome analysis of early anther development of cotton revealed male sterility genes for major metabolic pathways. J Plant Growth Regul 34:223–232
Zhao J (2015) Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci 20:576–585. doi:10.1016/j.tplants.2015.06.007
Zhu Z, An F, Feng Y, Li P, Xue L, Mu A, Jiang Z, Kim JM, To TK, Li W, Zhang X, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo H (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108:12539–12544. doi:10.1073/pnas.1103959108
Ziosi V, Bonghi C, Bregoli AM, Trainotti L, Biondi S, Sutthiwal S, Kondo S, Costa G, Torrigiani P (2008) Jasmonate-induced transcriptional changes suggest a negative interference with the ripening syndrome in peach fruit. J Exp Bot 59:563–573. doi:10.1093/jxb/erm331
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (31201593) and the Natural Science Foundation of Shandong Province, China (ZR2015CQ016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, S., Sun, J., Sun, S. et al. Transcriptional Profiles Underlying the Effects of Methyl Jasmonate on Apple Ripening. J Plant Growth Regul 36, 271–280 (2017). https://doi.org/10.1007/s00344-016-9636-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-016-9636-8