Abstract
A novel and effective image compression–encryption scheme based on deoxyribonucleic acid (DNA)-encoding theory and compressed sensing (CS) theory is proposed. The logistic map is applied to control key image and measurement matrices, and to control DNA encoding and decoding rules used to encode and decode each row of the plain image and the key image. The plain image first forms a disordered image through DNA encryption stage, sorts it to obtain an ordered image through sorting stage, and then passes the ordered image through CS encryption stage to form a cipher image. Simulation results and attack analysis verify the validity and feasibility of the encryption scheme, whose compression and security is acceptable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Optical encryption technology has the characteristics of high speed and parallelism, and it can encrypt information in multiple dimensions of light, so it has a natural advantage in processing image. In 1995, the double random phase encoding (DRPE) technique proposed by Refregier and Javidi is one of the most classic optical image encryption technologies, which encrypts a plain image into a smooth white noise image by two random phase masks (RPMs) placed on the input plane and the Fourier plane, respectively [1]. Researchers combine DRPE with various optical transformations to demonstrate the huge potential of optical image encryption technology [2,3,4,5,6,7,8,9,10,11]. With the continuous development of optical components, the application of optical image encryption in the field of information security is becoming wider and wider.
In the last decade, DNA computing has been found to have parallel computing performance, high storage density, and low energy consumption [12, 13]. Therefore, investigators explore the introduction of the principle of complementary base pairing in DNA theory to achieve image encryption [14,15,16,17,18,19,20,21]. The DNA-based image encryption algorithm can be divided into two steps: first, the plain image and the key image are encoded into two DNA sequences according to certain DNA encoding rules; second, specific DNA operation (\(+, -,\) XOR ) is performed on the two DNA sequences to obtain a cipher image. Since the key image is unknown and there are multiple DNA encoding rules and DNA operations to choose from, the encryption algorithm is highly secure. However, it should be noted that DNA-based image encryption schemes are often linear encryption schemes. In a hybrid encryption scheme combining DNA encoding theory and DRPE technology, to improve the ability of the encryption scheme to resist the chosen-plaintext attack, the authors use the Message-Digest Algorithm 5 (MD5) to closely link the keys with the plain image; therefore, the key changes as the plaintext changes, but the encryption scheme is still a linear encryption scheme [20].
If the attacker gains access to the encryption or decryption machine, the linear encryption algorithm is easily cracked [22,23,24,25,26,27]. Therefore, introducing nonlinearity into encryption schemes is critical to improving their security. Compressed sensing (CS) theory was first used in the field of signal acquisition, and it can reconstruct the original signal from a small amount of measurement data, and this process is nonlinear [28,29,30,31,32]. Some scholars use CS theory to achieve image encryption, because the dimensions of the sampled data are lower than the dimensions of the original data, and visually identifiable information cannot be obtained from the sampled data [33,34,35,36,37,38,39,40,41]. CS sampling process has a corresponding optical implementation, namely the single pixel camera [42]. The CS-based image encryption scheme introduces nonlinearity into the system, providing an extra key space, and then cracking the encryption system becomes more difficult.
However, for CS-based image encryption schemes, multiple measurement matrices are often required as keys. For DNA-based image encryption schemes, key image is typically employed as the key, and sometimes it is necessary to use the DNA encoding rules of each row as keys. These all add to the burden of key storage and transmission. Chaotic systems have the characteristics of pseudo-randomness, high sensitivity to initial conditions, certainty, ergodicity, and they are often used to generate some parameters used in encryption scheme. It has been shown that in the logical mapping, when the sampling distance is relatively large, the chaotic matrix arranged by the chaotic sequence satisfies the restricted isometry property (RIP), indicating that the matrix can be used as the measurement matrix in CS sampling process [43]. As long as the initial value \(x_0\) is determined, the entire matrix is determined. Similarly, one-dimensional chaotic sequence and two-dimensional key image can also be generated this way.
In this article, we propose an image encryption scheme based on DNA encoding theory and CS theory. To reduce the data of keys, the logistic map is used to control key image and measurement matrices, and to control DNA encoding and decoding rules. The encryption scheme can be divided into three stages: DNA encryption stage, sorting stage and CS encryption stage. In the first stage, the plain image and the key image are encoded row by row to obtain two sequences, which are XORed to obtain a disordered image. In the second stage, the disordered image is arranged into an ordered image, and an index matrix is recorded. Due to the existence of the index matrix, the sorting stage is reversible. In the third stage, the ordered image is sampled by a single-pixel camera to obtain multiple measured values, which are arranged into a two-dimensional matrix form, that is, a cipher image. During the decryption process, the index matrix is used to arrange the ordered image recovered from the cipher image by the orthogonal matching pursuit (OMP) algorithm [44] into a disordered image, and then DNA decryption is performed on the disordered image to reconstruct the plain image. Simulation results and attack analysis verify the validity and feasibility of the encryption scheme, whose compression and security is acceptable .
2 Fundamental knowledge
2.1 DNA-based cryptography
There are four different nucleic acid bases, A (adenine), C (cytosine), G (guanine) and T (thymine), in a DNA sequence. According to the principle of complementary base pairing, A and T are complementary, G and C are complementary. This principle is similar to the binary system’s complementary relationship. For example, binary 1 and 0 are complementary, and expanding to two binary numbers can get 11 and 00 are complementary, 10 and 01 are also complementary. There are 24 encoding rules to encode 00, 01, 10 and 11 by A, C, G and T. However, only 8 encoding rules satisfy the principle of complementary base pairing, as shown in Table 1. Each base represents a two-bit binary sequence, therefore, each pixel value of an eight-bit grayscale image can be encoded as a DNA sequence of length 4. For example, a pixel value of 142 in decimal can be recorded as a binary sequence [10001110], and after encoding it by rule 1 in Table 1, we obtain DNA sequence [CATC]. By decoding the DNA sequence [CATC] with the same rule, the correct binary sequence [10001110] can be obtained. However, if incorrect DNA encoding rules (for instance, rule 5) are applied to decode [CATC], we will get incorrect binary sequence (for instance, [11100111]). The above analysis shows that to get the correct pixel value, the DNA rules used in decoding must be the same as those used in encoding.
While DNA encoding theory is developing, scholars have also studied some DNA-based operations, such as addition (+), subtraction (−) and exclusive-OR (XOR). The XOR operation is consistent with the XOR operation execution rules in conventional binary systems because it does not involve carry. Since there are 8 kinds of DNA encoding rules, there are eight kinds of XOR operations, and Table 2 shows one of them. We also explored the optical implementation of XOR operation in our previous work [20].
2.2 Compressive sensing
For a one-dimensional real-valued signal x of length N, consider a linear measurement process that calculates the \(M<N\) inner products between the signal x and the measurement matrix \(\varPhi \), which can be recorded as [30, 31]
where y is an \(M \times 1\) measurement value, and \(\varPsi \) is a \(N \times N\) sparse matrix. K-sparse vector \(\alpha = \varPsi ^T x\) is the sparse representation of x. Sensor matrix \(\varTheta = \varPhi \varPsi \) is an \(M \times N\) matrix. If the sensor matrix \(\varTheta \) satisfies the restricted isometry property (RIP), the original signal x can be reconstruct from undersampled data y by minimizing \(\left\| \alpha \right\| _{l_1}\) [30, 31]
where \(\left\| \alpha \right\| _{l_1}\) is the \(l_1\) norm of \(\alpha \), and \(\alpha \) can be reconstructed with high probability if the sampling number \(M \ge cK\log (N/K)\), and c is a small constant [30, 31]. The sampling rate is defined as \( \eta = M/N \). For a two-dimensional image of size \(N = n \times n\), there is \( \eta = M/N = M/n^2 \). The lower the sampling rate, the stronger the compression capability.
2.3 Logistic map
We use logistic map to control the key image and the measurement matrix, and to control the DNA encoding/decoding rules, which will be used to encode/decode each row of the plain image and the key image. The logistic map can be denoted as
When chaotic parameter \(\mu \subseteq [3.57,4]\) and iteration initial value \(x_0 \subseteq (0,1)\), the generated sequence will be chaotic. To further reduce the correlation between adjacent chaotic element values, a sampling distance d can be introduced. Research shows that when control parameter \(\mu = 4\) and the sampling distance \(d \ge 15\), the sequence generated according to Eq. 3 is approximately independent [43].
Figure 1 investigates the sensitivity of the logistic map to the iteration initial value \( x_0 \) and gives the first 30 iterations of the chaotic sequence when the sampling distance is \(d=15\). The circles ( \(\circ \) ) and asterisks ( \(*\) ) in the figure represent the iteration values generated according to Eq. 3 using the correct (0.52) and incorrect (\(0.52 + 10^{ - 15}\)) iteration initial values, respectively. It can be seen that even if the deviation of the initial value \(10^{-15}\) is very small, the deviation of the chaotic sequence generated according to the logical map will be very large, and only the deviation of the first three iteration values are small. From the fourth iteration value onwards, the following iteration values are completely different.
3 Description of the method
3.1 Key image and measurement matrix generation
We use the logistic map to generate the key image, which will be used in the DNA encryption stage and the measurement matrices, which will be used in the CS sampling stage. To construct the key image, the initial value \(x_0^{k \_ \mathrm{image}}\) is used to generate a chaotic sequence of length \(l = 15 \times n \times n\) according to Eq. 3, and one point is sampled at each interval of 15 points, and then the one-dimensional sequence is arranged into a two-dimensional matrix of size \(n \times n\). The pixel value \(\mathrm{pixel}(r,c)\) of the key image located in the r row and c column is determined by
where \(x(r,c) \in \left( {0,1} \right) \) is the pixel value of the chaotic matrix located in the r row and c column, and \(\left\lfloor \bullet \right\rfloor \) denotes rounding down function. Processing each element of the chaotic matrix by Eq. 4, a key image of \( n \times n \) we obtain, which is an eight-bit grayscale image.
The chaotic matrix generated by the logistic map can be directly used as the measurement matrix \(\varPhi \), but it should be noted that the size of the measurement matrix \(\varPhi \) is \(m \times n\), so the length of the chaotic sequence should be \(l = 15 \times m \times n\). \(M = m \times n\) sampled data are required in the CS decryption stage, so M initial values \(x_0^1,x_0^2,\ldots ,x_0^M\) are required to generate M measurement matrices.
3.2 Encryption steps
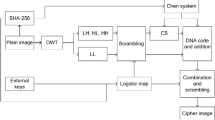
The proposed compression–encryption scheme consists of three stages, DNA encryption stage, sorting stage and CS encryption stage, as shown in Fig. 2. The plain image is processed by the DNA encryption stage to form a disordered image, which is sorted and then subjected to the CS encryption stage to obtain a sampled data. M sample values are obtained by M sampling, and they are arranged into a two-dimensional matrix form, that is, the cipher image. The details are as follows:
Step1: DNA encoding is performed on the plain image and the key image row by row, and the encoding rule is determined by
where Rule(r) is the specific DNA rule of r row, which shown in Table 1. \(x(r) \in \left( {0,1} \right) \) is rth iteration value of logistic map, and \(\left\lceil \bullet \right\rceil \) denotes rounding up function. Plain image and key image are encoded row by row until all rows of them are encoded. This step encodes two images and requires two numbers ( \(x_0^\mathrm{plain}\) and \(x_0^\mathrm{key}\) ) as keys.
Step 2: The two DNA matrices obtained in step 1 are decoded to obtain two binary matrices. The different rows use different DNA decoded rules, which are determined by Eq. 5. To ensure the pixel values are encrypted, the keys controlling the DNA decoded rules should be different from the keys controlling the DNA encoded rules. This step decodes two DNA matrices and requires two numbers ( \(x_0^{\mathrm{plain}^\prime }\) and \(x_0^{\mathrm{key}^\prime }\) ) as keys.
Step 3: Use the two binary matrices obtained in the previous step as the input of the XOR gate, and then convert the binary matrix output by the XOR gate into an eight-bit grayscale image, that is, the disordered image in Fig. 2.
Step 4: The disordered image is arranged into an ordered image according to certain orders (here is ascending order), and an index matrix is recorded as a decryption key. Because of the existence of the index matrix, this process is reversible.
Step 5: The product of the measurement matrix and the ordered image is displayed on a spatial light modulator (SLM), which is then focused on a single-pixel photodiode (PD) using a lens (L) and the recorded light intensity is transmitted to a personal computer (PC). This step corresponds to \(\textcircled {5} \sim \textcircled {8}\) in Fig. 2, that is, one sampling is performed with single pixel camera.
Step 6: Change the measurement matrix M times and record the corresponding light intensity. Here, the measurement matrix is generated with the initial value according to the logistic map, then only the initial value needs to be changed.
Step 7: M measurement values are arranged into a two-dimensional matrix form, that is, the cipher image.
The decryption process includes CS reconstruction stage, sorting stage, and DNA decryption stage. First, M measurement matrices are generated with the initial values \(x_0^1,x_0^2,\ldots , x_0^M\), and then the ordered image is restored using the CS recovery algorithm [44]. Second, the index matrix is used to arrange ordered image into disordered image. Third, the key image is calculated with \(x_0^{k \_ \mathrm{image}}\), and the DNA encoding and decoding rules of each row are, respectively, calculated with \(x_0^\mathrm{key}, x_0^{\mathrm{key}^\prime }\) and \(x_0^\mathrm{plain}\), \(x_0^{\mathrm{plain}^\prime }\), and then the plain image is calculated by DNA decryption.
In this encryption scheme, encrypting an image requires a total of \(M + 5\) numbers as encryption key, namely M initial values of measurement matrix \(x_0^1,x_0^2,\ldots ,x_0^M\), the initial value of key image \(x_0^{k \_ \mathrm{image}}\), and the initial values \(x_0^\mathrm{key}, x_0^{\mathrm{key}^\prime }\) and \(x_0^\mathrm{plain}\), \(x_0^{\mathrm{plain}^\prime }\) that controls the DNA encoding and decoding rules of the plain image and the key image. When an disordered image is arranged into an ordered image, an index matrix is recorded, which is a decryption key. Therefore, the decryption keys have one more index matrix than the encryption keys.
4 Numerical simulations and discussion
4.1 Simulated results
To verify the validity and feasibility of the encryption scheme, we encrypt an eight-bit grayscale image “boat” (Fig. 3a) with size of \(256 \times 256\). Figure 3b is the key image needed in the DNA encryption stage, and it is generated according to the method described in Sect. 3.1. Figure 3a, b is processed according to the encryption steps 1 to 3 to obtain the disordered image (Fig. 3c). If the signal is sparse or it is sparse in a certain domain, it can be sampled and reconstructed by using CS theory. However, for the disordered image (Fig. 3c), it is not regular, therefore it is impossible to directly sampling and reconstruction it with CS theory. We propose to sort the disordered image into an ordered image (Fig. 3d), and then use CS theory to sample and reconstruct it. An index matrix is recorded during the sorting stage, which is reversible.
The product of the measurement matrix and the ordered image is displayed on the SLM. After SLM being irradiated by the coherent light, the output light is concentrated by the lens onto the single-pixel detector, and then the measurement value is transmitted to the PC, that is, a sampling process is completed. Each time the measurement matrix is changed, an measurement value is obtained. The measurement data with sampling rate \( \eta = 25\% \) after multiple measurements is shown in Fig. 3e. Any information related to the plain image is not seen from the cipher image, and due to the use of CS, the dimension of the cipher image is smaller than that of the plain image.
Figure 3f is a decrypted image obtained by decrypting Fig. 3e. It can be seen that the quality of Fig. 3f is very good. To quantitatively calculate the reconstruction quality of the decrypted image, peak-to-peak signal-to-noise ratio (PSNR) and correlation coefficient (CC) are described as
where R is the reconstructed image and P is the plain image, respectively. cov(R,P) denotes cross covariance of two images. \(\sigma _R\) and \(\sigma _P\) stand for the standard deviation of reconstructed image and plain image, respectively. When the sampling rate is \(25 \%\), the PSNR and CC of the decrypted image are 45.04 dB and 0.9995, this shows that the compression–encryption scheme almost perfectly reconstructs the plain image. The encryption scheme can be divided into three stages: DNA encryption stage, sorting stage, and CS encryption stage. The DNA encryption stage (\(\textcircled {1} \sim \textcircled {3}\) in Fig. 2) and the sorting stage (\(\textcircled {4}\) in Fig. 2) process image in the integer domain, and these stage do not cause any data deviation. At the same time, the sorting stage completes the sparse representation of the disordered image. For the CS encryption stage (\(\textcircled {5} \sim \textcircled {8}\) in Fig. 2), since the input image has completed the sparse representation and the measurement matrix is a chaotic matrix generated by the logistic map, the sampled data can be reconstructed well by the OMP algorithm.
4.2 Compression performance
To quantitatively evaluate the effect of sampling rate \(\eta \) on the quality of the decrypted image, we calculate the PSNRs and CCs of the decrypted image when the sampling rates are \(25\%, 12.5\%, 6.25 \%\) and \(3.125 \% \), as shown in Figs. 3e and 4a1–a3, and the corresponding decrypted image is shown in Figs. 3f and 4b1–b3. With the sampling rate reduced, the data of the cipher image is reduced, and the quality of the decrypted image is gradually reduced. Even if the sampling rate is \(6.25\%\) (Fig. 4a2), the quality of the decrypted image (Fig. 4b2) is still very good, PSNR = 32.15 dB, CC = 0.9910, which means that the compressed encryption algorithm can compress the image to the original 1/16. However, when the sampling rate is reduced to 3.125 %, we can only see the outline of the decrypted image (Fig. 4b3), but the details are almost unrecognizable.
4.3 Sensitivity to logistic map’s initial values
For the logistic map’s initial values, which generate the key image and control the DNA encoding and decoding rules, a small deviation will cause a huge change in the generated chaotic sequence, which in turn leads to a correct decrypted image cannot be obtained. Figure 5a shows the decrypted image obtained using the incorrect logistic map’s initial value \(x_0 ^ {k \_ \mathrm{image}}+10^{-15}\). Even if the initial value \(x_0 ^ {k \_ \mathrm{image}}\) only deviates by \(10^{ - 15}\), the chaotic sequence generated by the logistic map changes greatly. Only the first three iteration values are roughly equal, and the subsequent iteration values are completely different, as shown in Fig. 1. Incorrect chaotic sequence can cause key image errors during DNA decryption stage, which in turn results in the correct decrypted image can not be obtained. When \( x_0 ^ \mathrm{key}, x_0 ^ {\mathrm{key}^\prime } \) and \( x_0 ^ \mathrm{plain} \), \( x_0 ^ {\mathrm{plain}^\prime } \), which control the DNA encoding and decoding rules of the plain image and the key image, are wrong, it will lead to similar results, as shown in Fig. 5b–e.
4.4 Sensitivity to index matrix
In the encryption process, when the disordered image (Fig. 3c) is arranged into an ordered image (Fig. 3d), an index matrix is recorded. In the decryption process, if the wrong index matrix is used, the correct disordered image cannot be obtained, which results in the DNA decryption stage cannot obtain the correct plain image. The effect of the index matrix with different error rates on the decryption result is shown in Fig. 6. When there is 25% element errors in the index matrix, noise will be generated in the decrypted result, but it is still clear, as shown in Fig. 6a. When there is 50% element errors in the index matrix, the decryption result is no longer clear, as shown in Fig. 6b. As the index matrix error rate increases, when the 75% element is wrong, only the outline of the decryption image can be seen, and the details are not recognized, as shown in Fig. 6c.
Since the DNA encryption part belongs to the stream cipher, it is characterized in that each bit of the input corresponds to each bit of the output. When there is 25% element error in the index matrix, there will be 25% of the element position errors in the reconstructed disordered image (the result of DNA encryption part). And then there will be 25% pixel value error in the decrypted image, and the visually seen decrypted image will generate noise, as shown in Fig. 6a. Due to the nature of the stream cipher, the amount of bit errors in the index matrix will lead to the same amount of bit errors obtained in the decrypted image. It can be inferred that in Fig. 6b, c, the error rates of the decrypted results are 50% and 75%, respectively, so the decrypted images are no longer clear. To get a completely correct decrypted image, the index matrix must be completely correct.
4.5 Noise attack
To quantitatively calculate the influence of noise on the decryption result, Gaussian random noise with different intensities is added to the cipher image according to the following formula
where C and \(C^\prime \) are the cipher image and the noisy cipher image respectively, G represents the Gaussian random noise with zero-mean and unit standard deviation, and q is the intensity of noise. The decrypted results with different noise intensities q are shown in Fig. 7. It can be seen that when \( q = 0.1 \), the obtained decrypted image is very clear, PSNR = 25.48 dB, CC = 0.9595. When \(q = 1.0\), the obtained decrypted image is still identifiable. As q increases, the quality of the decrypted image decreases. When \(q=10.0\), the outline of the decrypted image can still be seen, but the details are not recognized.
4.6 Chosen-plaintext attack and differential attack
Because the encryption scheme contains linear encryption part, the attacker can try to attack the encryption scheme by combining chosen-plaintext attack and differential attack. The results obtained by the delta image (entirely black except for one single pixel) processed by the DNA encryption stage, the sorting stage, and the CS encryption stage are shown in Fig. 8a1–a3, respectively. The corresponding results obtained by the three-part encryption of the all-zero image (entirely black) are shown in Fig. 8b1–b3, respectively. (ci) = (ai) − (bi) is the difference between (ai) and (bi).
The two disordered images (Fig. 8a1, b1), obtained by encrypting two plain images (delta image and all-zero image) with only one difference, seem to be disorderly, but because the DNA encryption part is a linear encryption process, the two disordered images will also differ by one, as shown in Fig. 8c1, which is also a delta image. In the sorting stage, the two disordered images (Fig. 8a1, b1) are arranged in ascending order to obtain two ordered images (Fig. 8a2, b2). When we subtract the two ordered images, the result is still only one not-zero value, but the position of the value has moved, as shown in Fig. 8c2. A simple example of the sorting stage that can move non-zero value location is shown in Table 3. This shows that the sorting stage can move the non-zero value position, but it still is a linear process. After the two ordered images (Fig. 8a2, b2) pass through the CS encryption part, the two cipher images (Fig. 8a3, b3) are obtained, respectively. The difference between the two final cipher images is no longer a delta image, but a disorganized image, as shown in Fig. 8c3. This indicates that the CS encryption part is a non-linear process and the security of the encryption scheme is improved.
5 Conclusion
Based on DNA encoding theory and CS theory, an efficient image compression–encryption scheme is investigated. To reduce the data of the keys, logistic map is used to generate key image and measurement matrices. The plain image first is processed into a disordered image through DNA encryption stage, the disordered image is sorted to obtain an ordered image through sorting stage, and then the ordered image is encrypted to obtain a cipher image through CS encryption stage. The sorting stage arranges the disordered image into ordered image and records an index matrix that can be used as a decryption key. At the same time, sorting stage completes the sparse representation of the image. In the decryption process, the index matrix is used to arrange the ordered image recovered from the cipher image by the OMP algorithm into a disordered image, and then DNA decryption is performed on the disordered image to reconstruct the plain image. Simulation results and attack analysis show that the proposed image compression–encryption scheme has good encryption effect, high sensitivity to keys, good compression performance, and the scheme can resist noise attack, chosen-plaintext attack and differential attack.
References
Philippe Refregier, Bahram Javidi, Optical image encryption based on input plane and fourier planerandom encoding. Opt. Lett. 20(7), 767–769 (1995)
Takanori Nomura, Bahram Javidi, Optical encryption using a joint transform correlator architecture. Opt. Eng. 39(8), 2031–2036 (2000)
Guohai Situ, Jingjuan Zhang, Double random-phase encoding in the fresnel domain. Opt. Lett. 29(14), 1584–1586 (2004)
Xin Zhou, Sheng Yuan, Shengwei Wang, Jian Xie, Affine cryptosystem of double-random-phase encryption based on the fractional fourier transform. Appl. Opt. 45(33), 8434–8439 (2006)
Yan Zhang, Bo Wang, Optical image encryption based on interference. Opt. Lett. 33(21), 2443–2445 (2008)
Zhengjun Liu, Qing Guo, Xu Lie, Muhammad Ashfaq Ahmad, Shutian Liu, Double image encryption by using iterative random binary encoding in gyrator domains. Opt. Express 18(11), 12033–12043 (2010)
Wen Chen, Xudong Chen, Colin J.R. Sheppard, Optical image encryption based on diffractive imaging. Opt. Lett. 35(22), 3817–3819 (2010)
Nanrun Zhou, Yixian Wang, Lihua Gong, Novel optical image encryption scheme based on fractional mellin transform. Opt. Commun. 284(13), 3234–3242 (2011)
Lihua Gong, Xingbin Liu, Fen Zheng, Nanrun Zhou, Flexible multiple-image encryption algorithm based on log-polar transform and double random phase encoding technique. J. Mod. Opt. 60(13), 1074–1082 (2013)
Zhuhong Shao, Yuping Duan, Gouenou Coatrieux, Wu Jiasong, Jinyu Meng, Huazhong Shu, Combining double random phase encoding for color image watermarking in quaternion gyrator domain. Opt. Commun. 343, 56–65 (2015)
Sheng Yuan, Yangrui Yang, Xuemei Liu, Xin Zhou, Zhenzhuo Wei, Optical image transformation and encryption by phase-retrieval-based double random-phase encoding and compressive ghost imaging. Opt. Lasers Eng. 100, 105–110 (2018)
Tom Head, Grzegorz Rozenberg, Reno S. Bladergroen, C.K.D. Breek, P.H.M. Lommerse, Herman P. Spaink, Computing with dna by operating on plasmids. Biosystems 57(2), 87–93 (2000)
Xuedong Zheng, Xu Jin, Wu Li, Parallel dna arithmetic operation based on n-moduli set. Appl. Math. Comput. 212(1), 177–184 (2009)
Qiang Zhang, Ling Guo, Xiaopeng Wei, Image encryption using dna addition combining with chaotic maps. Math. Comput. Modell. 52(11–12), 2028–2035 (2010)
Xiaopeng Wei, Ling Guo, Qiang Zhang, Jianxin Zhang, Shiguo Lian, A novel color image encryption algorithm based on dna sequence operation and hyper-chaotic system. J. Syst. Softw. 85(2), 290–299 (2012)
Lili Liu, Qiang Zhang, Xiaopeng Wei, A rgb image encryption algorithm based on dna encoding and chaos map. Comput. Electr. Eng. 38(5), 1240–1248 (2012)
Hongjun Liu, Xingyuan Wang, Abdurahman Kadir, Image encryption using dna complementary rule and chaotic maps. Appl. Soft Comput. 12(5), 1457–1466 (2012)
Xingyuan Wang, Chuanming Liu, A novel and effective image encryption algorithm based on chaos and dna encoding. Multimed. Tools Appl. 76(5), 6229–6245 (2017)
Junxin Chen, Zhiliang Zhu, Libo Zhang, Yushu Zhang, Benqiang Yang, Exploiting self-adaptive permutation-diffusion and dna random encoding for secure and efficient image encryption. Sig. Process. 142, 340–353 (2018)
Dongming Huo, Dingfu Zhou, Sheng Yuan, Shaoliang Yi, Luozhi Zhang, Xin Zhou, Image encryption using exclusive-or with dna complementary rules and double random phase encoding. Phys. Lett. A 383(9), 915–922 (2019)
Zhentao Liu, Wu Chunxiao, Jun Wang, Hu Yuhen, A color image encryption using dynamic dna and 4-d memristive hyper-chaos. IEEE Access 7, 78367–78378 (2019)
Arturo Carnicer, Mario Montes-Usategui, Sergio Arcos, Ignacio Juvells, Vulnerability to chosen-cyphertext attacks of optical encryption schemes based on double random phase keys. Opt. Lett. 30(13), 1644–1646 (2005)
Yann Frauel, Albertina Castro, Thomas J. Naughton, Bahram Javidi, Resistance of the double random phase encryption against various attacks. Opt. Express 15(16), 10253–10265 (2007)
Yuansheng Liu, Jie Tang, Tao Xie, Cryptanalyzing a rgb image encryption algorithm based on dna encoding and chaos map. Opt. Laser Technol. 60, 111–115 (2014)
Yushu Zhang, Di Xiao, Wenying Wen, Kwok-Wo Wong, On the security of symmetric ciphers based on dna coding. Inf. Sci. 289, 254–261 (2014)
Houcemeddine Hermassi, Akram Belazi, Rhouma Rhouma, Safya Mdimegh Belghith, Security analysis of an image encryption algorithm based on a dna addition combining with chaotic maps. Multimed. Tools Appl. 72(3), 2211–2224 (2014)
Lei Wang, Wu Quanying, Guohai Situ, Chosen-plaintext attack on the double random polarization encryption. Opt. Express 27(22), 32158–32167 (2019)
Emmanuel J. Candes, Justin Romberg, Terence Tao, Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans. Inf. Theory 52(2), 489–509 (2006)
David L. Donoho, Compressed sensing. IEEE Trans. Inf. Theory 52(4), 1289–1306 (2006)
Emmanuel J. Candes, Michael B. Wakin, An introduction to compressive sampling. IEEE Signal Process. Mag. 25(2), 21–30 (2008)
Richard G. Baraniuk, Compressive sensing [lecture notes]. IEEE Signal Process. Mag. 24(4), 118–121 (2007)
Fernando Soldevila, Esther Irles, V. Durán, P. Clemente, Mercedes Fernández-Alonso, Enrique Tajahuerce, Jesús Lancis, Single-pixel polarimetric imaging spectrometer by compressive sensing. Appl. Phys. B 113(4), 551–558 (2013)
Deepan Balakrishnan, Chenggen Quan, Y. Wang, Cho Jui Tay, Multiple-image encryption by space multiplexing based on compressive sensing and the double-random phase-encoding technique. Appl. Opt. 53(20), 4539–4547 (2014)
Xingbin Liu, Wenbo Mei, Du Huiqian, Optical image encryption based on compressive sensing and chaos in the fractional fourier domain. J. Mod. Opt. 61(19), 1570–1577 (2014)
Nitin Rawat, Byoungho Kim, Inbarasan Muniraj, G. Situ, Byung-Geun Lee, Compressive sensing based robust multispectral double-image encryption. Appl. Opt. 54(7), 1782–1793 (2015)
Nanrun Zhou, Haolin Li, Di Wang, Shumin Pan, Zhihong Zhou, Image compression and encryption scheme based on 2d compressive sensing and fractional mellin transform. Opt. Commun. 343, 10–21 (2015)
Jiaosheng Li, Liyun Zhong, Qinnan Zhang, Yunfei Zhou, Jiaxiang Xiong, Jindong Tian, Lu Xiaoxu, Optical image hiding based on dual-channel simultaneous phase-shifting interferometry and compressive sensing. Appl. Phys. B 123(1), 4 (2016)
Nanrun Zhou, Hao Jiang, Lihua Gong, Xinwen Xie, Double-image compression and encryption algorithm based on co-sparse representation and random pixel exchanging. Opt. Lasers Eng. 110, 72–79 (2018)
Jun Wang, Qionghua Wang, Hu Yuhen, Image encryption using compressive sensing and detour cylindrical diffraction. IEEE Photon. J. 10(3), 1–14 (2018)
Lihua Gong, Kaide Qiu, Chengzhi Deng, Nanrun Zhou, An image compression and encryption algorithm based on chaotic system and compressive sensing. Opt. Laser Technol. 115, 257–267 (2019)
Kanglei Zhou, Jingjing Fan, Haiju Fan, Ming Li, Secure image encryption scheme using double random-phase encoding and compressed sensing. Opt. Laser Technol. 121, 105769–105780 (2020)
Dharmpal Takhar, Jason N. Laska, Michael B. Wakin, Marco F. Duarte, Dror Baron, Shriram Sarvotham, Kevin F. Kelly, Richard G. Baraniuk, A new compressive imaging camera architecture using optical-domain compression. Comput. Imaging IV 6065, 606509–606518 (2006)
Yu. Lei, Jean Pierre Barbot, Gang Zheng, Hong Sun, Compressive sensing with chaotic sequence. IEEE Signal Process. Lett. 17(8), 731–734 (2010)
Joel A. Tropp, Anna C. Gilbert, Signal recovery from random measurements via orthogonal matching pursuit. IEEE Trans. Inf. Theory 53(12), 4655–4666 (2007)
Acknowledgements
This study was supported in part by the Anhui Polytechnic University Research Startup Foundation (Grant 2019YQQ007), and in part by the National Natural Science Foundation of China (Grant 61475104 and 61177009).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huo, D., Zhu, X., Dai, G. et al. Novel image compression–encryption hybrid scheme based on DNA encoding and compressive sensing. Appl. Phys. B 126, 45 (2020). https://doi.org/10.1007/s00340-020-7397-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00340-020-7397-3