Abstract
Objectives
To develop a machine learning–based radiomics model based on multiparametric magnetic resonance imaging (MRI) for preoperative discrimination between central neurocytomas (CNs) and gliomas of lateral ventricles.
Methods
A total of 132 patients from two medical centers were enrolled in this retrospective study. Patients from the first medical center were divided into a training cohort (n = 74) and an internal validation cohort (n = 30). Patients from the second medical center were used as the external validation cohort (n = 28). Features were extracted from contrast-enhanced T1-weighted and T2-weighted images. A support vector machine was used for radiomics model investigation. Performance was evaluated using the sensitivity, specificity, and the area under the receiver operating characteristic curve (AUC). The model’s performance was also compared with those of three radiologists.
Results
The radiomics model achieved an AUC of 0.986 in the training cohort, 0.933 in the internal validation cohort, and 0.903 in the external validation cohort. In the three cohorts, the AUC values were 0.657, 0.786, and 0.708 for radiologist 1; 0.838, 0.799, and 0.790 for radiologist 2; and 0.827, 0.871, and 0.862 for radiologist 3. When assisted by the radiomics model, two radiologists improved their performance in the training cohort (p < 0.05) but not in the internal or external validation cohorts.
Conclusions
The machine learning radiomics model based on multiparametric MRI showed better performance for distinguishing CNs from lateral ventricular gliomas than did experienced radiologists, and it showed the potential to improve radiologist performance.
Key Points
• The machine learning radiomics model shows excellent performance in distinguishing CNs from gliomas.
• The radiomics model outweighs two experienced radiologists (area under the receiver operating characteristic curve, 0.90 vs 0.79 and 0.86, respectively).
• The radiomics model has the potential to enhance radiologist performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
According to the 2021 World Health Organization classification of tumors of the central nervous system, central neurocytoma (CN) is classified as a grade II neuronal and mixed neuronal-glial tumor [1], which nearly always occurs in the lateral ventricle, accounting for 0.25 to 0.5% of intracranial tumors [2]. CN patients often present with nonspecific symptoms and signs of intracranial hypertension caused by obstructive hydrocephalus [3, 4]. Compared to lateral ventricular glioma (such as most astrocytoma and ependymal tumors), CNs have benign biological behavior and a good prognosis after total resection [4, 5]. Different from CN, intraventricular gliomas tend to have a poor prognosis, which necessitates a more extensive resection [5]. Therefore, accurate preoperative diagnosis has a high impact on treatment planning and prognostic counseling for patients with intraventricular mass.
Magnetic resonance imaging (MRI) is the preferred imaging method for intracranial tumors. Several characteristic MRI features of CN have been reported in previous studies; these signs include scalloping [6], broad-based attachment [4, 7, 8], soap bubble [9,10,11], peripheral cysts [12], fluid-fluid level [12], and gemstone [12], which have significant value in diagnosing CN. A previous study showed that the above six MRI features can accurately distinguish CN from meningioma and choroid plexus papillary tumors [12]. However, it is still challenging to distinguish CN from intraventricular astrocytoma and ependymoma [13,14,15]. Moreover, the diagnostic accuracy of subjective evaluation depends on the experience level of a radiologist [16]. A recent study showed that the average apparent diffusion coefficient (ADC) value was potentially useful in the differentiation between CN and ependymoma [17]. However, there are many factors affecting the ADC value, such as tumor heterogeneity, the sequences and parameters, and the field strength of MRI scanners.
With the rapid development of medical image analysis methods, radiomics has become a popular focus in research [18]. Radiomics extracts a large number of quantitative features from medical images via high-throughput calculation methods. Machine learning based on radiomics can more accurately assess the heterogeneity of tumors [19,20,21]. Support vector machine (SVM) is a machine learning method based on statistical learning theory and structural risk minimization [22], which is considered a good model of statistical learning in small sample databases. By combining SVM with radiomics, this method has shown good learning ability and generalization capability, and has been widely used in brain tumor diagnosis, grading, and survival analysis [23, 24]. However, SVM has not yet been combined with radiomics for the diagnosis of lateral ventricle tumors.
This study aimed to develop a radiomics model with machine learning, using multiparametric MRI, to effectively distinguish CNs from lateral ventricular gliomas. In addition, the diagnostic performance of the model was compared with experienced neuroradiologists to determine the ability of the radiomics model to enhance radiologist performance.
Materials and methods
Study participants
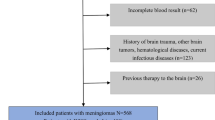
This retrospective study was approved by the Institutional Review Board of two medical centers, and written informed consent was waived. A radiology database was used to obtain MRI data of CNs and other intraventricular tumors from the picture archiving and communication system. Patients were consecutively enrolled in medical center A (Nanfang Hospital, from July 2010 to March 2020) and medical center B (Zhujiang Hospital, from September 2016 to October 2020). Figure 1 shows the inclusion and exclusion criteria. Inclusion criteria were as follows: (1) truly intraventricular tumors arise from the ventricular walls and lie within the ventricular cavity; (2) pseudo-ventricular tumors originate in the adjacent tissues but radiologically appear as intraventricular masses; (3) age 16 years or older; (4) postoperative pathological diagnosis confirmed by hematoxylin-eosin staining and immunohistochemistry. Exclusion criteria were as follows: (1) non-CN or non-glioma (such as meningioma, choroid plexus papilloma, ependymoblastoma, lymphoma, metastasis); (2) MRI images were incomplete or contained severe artifacts.
Ultimately, 104 patients with lateral ventricle tumor were enrolled from medical center A, including 50 cases of CN and 54 cases of glioma (glioma subtypes are summarized in Supplementary Table S1), which were then randomly divided into a training cohort (74 cases) and an internal validation cohort (30 cases) with a ratio of 2.5:1 by using stratified sampling on the Darwin research platform (Yizhun Medical AI Co. Ltd.). Patients from medical center B with 13 cases of CN and 15 cases of glioma were utilized as an external validation cohort.
MRI data acquisition
All imaging data included two sets of images: preoperative T2-weighted (T2W) and contrast-enhanced T1-weighted (CE-T1W). The acquisition parameters of these protocols are summarized in Supplementary Table S2.
Patients from medical center A were examined with four MRI scanners (Siemens Magnetom Vision Plus 1.5T, GE Optima MR360 1.5T, GE Signa Excite 3.0T, and Philips Achieva 3.0T) with the use of a head or head-neck coil. For these patients, contrast agent (Omniscan TM, GE Healthcare; Magnevist, Bayer Schering; gadopentetate dimeglumine, Consun) was administered at a dose of 0.2 mmol/kg at a rate of 2.0–2.5 mL/s using a power injector (Spectris Solaris EP, Medrad; TennesseeXD003, Ulrich Medical) via the antecubital vein, followed by a 20-mL sterile saline flush.
Patients from medical center B were examined with two MRI scanners (GE Optima MR360 1.5T and Philips Achieva 3.0T) with the use of a head coil. For these patients, a contrast agent (Gd-DTPA, Magnevist, Bayer Schering) was injected at a dose of 0.2 mmol/kg at a rate of 2.0–2.5 mL/s through the median cubital vein, followed by a 20-mL sterile saline flush.
Tumor segmentation
The Darwin research platform (https://arxiv.org/abs/2009.00908) was used to perform radiomic feature extraction and selection and to build machine learning models. All MRI scans were resampled to 1 × 1 × 1 mm3. Tumors were segmented manually by two radiologists (with 3 and 20 years of experience respectively) on T2W and CE-T1W images. The radiologists were blinded to all clinical information. Regions of interest (ROIs) were defined according to the visible gross tumor volume located in the ventricles, including the solid component of the tumor, necrosis, and cyst. First, ROIs were drawn on CE-T1W to carefully separate the lesion from the adjacent brain tissue, and then, they were automatically transferred to T2W images on the Darwin research platform. If the ROIs were exactly the areas that delineate the tumor boundary on T2W images, they were used directly; otherwise, they were manually drawn according to the T2W images. After all contours of the whole lesion were drawn slice by slice, three-dimensional reconstruction was carried out to get volumes of interest (VOIs). To avoid partial volume effects, the bottom and top slices of the lesion were not included. To determine the intra-observer reproducibility of tumor segmentation, the intraclass correlation coefficients (ICC) were calculated. The features that had good to excellent reliability (ICC ≥ 0.80) and significant difference between CNs and gliomas were included for developing the radiomics model.
Feature extraction
After segmentation, three groups of radiomic features were extracted: shape, first-order, and texture features. In short, shape features describe basic geometric characteristics, tumor size, shape, and surface roughness; first-order features describe the distribution of voxel intensities within the mask-defined image region using commonly used and basic metrics; texture features include gray-level co-occurrence matrices (GLCM), gray-level run-length matrices (GLRLM), gray-level size zone matrices (GLSZM), neighboring gray tone difference matrices (NGTDM), and gray-level dependence matrices (GLDM), which capture the spatial interdependence of voxels in images and show characteristics related to spatial heterogeneity such as gray change, granularity, and roughness of the image. Additionally, first-order and texture features were processed further using six filters, i.e., exponential, square, square root, logarithm, log-sigma-3-0-mm-3D, and wavelet. Wavelet filters extract features from the eight wavelet decomposition images. Altogether, 107 original features and 1209 filter-based features were collected for each patient image sequence (Supplementary Table S3). Definitions and formulas of these radiomic features are located at https://pyradiomics.readthedocs.io/en/latest/features.html. All radiomic features were normalized using minimum–maximum values to eliminate the magnitudes of different features by scaling values to [0, 1].
Feature selection and model development
In this study, three methods were used to select relevant radiomic features in the training cohort from the feature subsets of T2W or CE-T1W sequences alone and in combination. These methods include SelectKBest, the least absolute shrinkage and selection operator (LASSO) algorithm, and support vector machine recursive feature elimination (SVM-RFE). SelectKBest is a univariate feature selection method using the analysis of variance F-values to analyze the relationship between the features and the results of classification. It was used first to select the top features with the largest F-values. Second, LASSO selected relevant features according to the best parameter (alpha). Finally, SVM-RFE selected the optimal features.
Following this, machine learning models were developed based on the selected features and SVM. The kernel for training the SVM model was linear. The Rad score helps discriminate CN from intraventricular glioma and was calculated for each patient by the SVM classifier. The complete process of the radiomics pipeline is shown in Fig. 2.
Subjective evaluation
Before image evaluation, all six MRI features and their definitions were introduced to three radiologists (radiologist 1, a neuroradiology fellow; radiologist 2, with 7 years of experience in neurology imaging; and radiologist 3, with 20 years of experience in neurology imaging), including signs such as scalloping, broad-based attachment, peripheral cyst, soap bubble, fluid-fluid level, and gemstone [12]. The radiologists, blinded to all clinical information, including the CN–glioma ratio in the dataset and the radiomics model’s performance, independently reviewed the T2W and CE-T1W images of all patients from medical center A and recorded each MRI feature (present or absent). In the event of discrepancies, feature determination was decided by the majority. In addition, for each case in the three cohorts, radiologists recorded whether they thought the tumor could be classified as CN or glioma, based on their review of the images (unassisted prediction) and whether they wanted to change their initial assessment after being shown the radiomics model’s prediction (assisted prediction).
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics for Windows, Version 25.0 (IBM Corp.). The Shapiro–Wilk test was used to determine whether the data conformed to a normal distribution. For data that conformed to a normal distribution, a parameter test was used, and for data that did not conform to a normal distribution, a non-parametric test was used. A chi-squared test was used to compare differences in categorical variables. The independent t test or Mann-Whitney U test was used to compare differences in continuous variables. The repeatability of tumor segmentation was evaluated by ICC. An ICC greater than 0.80 was considered to represent good agreement. For the subjective evaluation, diagnosis agreement among the radiologists when unassisted by the radiomics model was assessed using the Fleiss kappa, with values of 0.00 to 0.20 considered slight, 0.21 to 0.40 fair, 0.41 to 0.60 moderate, 0.61 to 0.80 substantial, and 0.81 to 1.00 almost perfect agreement. The receiver operating characteristic (ROC) curve was performed to assess the diagnostic performance of these six MRI features, radiologists, and radiomics models. Areas under the curves (AUCs) were calculated. Accuracy, sensitivity, and specificity were determined from the optimal threshold by maximizing the Youden index. The AUCs of the three radiologists and the radiomics model were compared by using the DeLong method using MedCalc (version 15.0, MedCalc software). Bonferroni correction was used for multiple comparisons of AUCs. The effect of model assistance on the radiologists’ predictions was evaluated by comparing their AUCs with and without assistance, using the DeLong test. The gplot and pheatmap packages were used for a heatmap analysis to evaluate associations between radiomic and MRI features. All statistical tests were two-sided, and a p value of < 0.05 indicated a significant difference.
Results
Participant characteristics
The clinical characteristics and MRI features of 104 patients from medical center A are summarized in Table 1. There were no significant differences in sex or age distribution between the two groups (p > 0.05). The demographic characteristics of the training, internal validation, and external validation cohorts are summarized in Table 2. There were no statistically significant differences in sex, age distribution, or pathology among the three cohorts (p > 0.05). Two representative cases of CN and glioma on MRI are shown in Fig. 3.
In medical center A, six MRI features were statistically different in the two groups of tumors (all p < 0.05; Table 1). The performance of the six MRI features in diagnosing CN is shown in Supplementary Table S4. The peripheral cyst sign showed the highest AUC (0.790), followed by soap bubble (0.696) and broad-based attachment (0.673). The scalloping (0.631), fluid-fluid level (0.621), and gemstone signs (0.622) showed relatively low AUCs.
Radiomics model construction with machine learning
Three models were established based on T2W images, CE-T1W images, and a combination of both, which include nine, eight, and five features respectively (Table 3). The performances of these three models in the training cohort and internal validation cohort are summarized in Supplementary Table S5. The radiomics model combining T2W and CE-T1W yielded an AUC of 0.986 in the training cohort and 0.933 in the internal validation cohort, which indicates good performance. According to the Bayesian information criterion, the combined model was chosen as the final radiomics model. This model was evaluated in the external validation cohort, with AUC, sensitivity, and specificity values of 0.903, 0.846, and 0.933 respectively (Table 4). The radiomics score (Rad score) calculation formula is shown in the Supplementary Material, and higher Rad scores indicate higher likelihoods of glioma (Fig. 4).
Comparison between subjective evaluation and model prediction
For all cases in both centers, inter-reader agreement among the three radiologists in diagnosing CN without model assistance was κ = 0.572 (p < 0·001). The radiomics model showed better performance than the radiologists did in all three cohorts (Table 4). The results of the pairwise comparison of AUCs between the radiomics model and the radiologists are shown in Supplementary Table S6. The difference was statistically significant in the training cohort (p < 0.008) but not in the internal validation or external validation cohorts. With the aid of the radiomics model, radiologist 1 (p = 0.023) and radiologist 3 (p = 0.021) showed improvement in the training cohort but not in the internal or external cohort (all p > 0.05; Fig. 5); radiologist 2 did not show improved performance in any cohort (Supplementary Table S7).
Performance of CE-T1W+T2W radiomics model and radiologists on the reader study. Training cohort (a), internal validation cohort (b), and external validation cohort (c). Radiologist 1 represents a neuroradiology fellow with 2 years of experience, radiologist 2 represents the neuroradiologist with 7 years of experience, and radiologist 3 represents the expert neuroradiologist with 20 years of experience. The performance of each radiologist is shown, with points at the base of each arrow representing radiologist performance before viewing model’s output, and the arrow showing change in performance after viewing model’s output
Correlation between MRI features and radiomics features
Using a heatmap, the association between radiomic features and MRI features was determined (Fig. 6). The results showed that the peripheral cyst sign was correlated with all five radiomic features (p < 0.05). The scalloping, broad-based attachment, and soap bubble signs were related to four radiomic features respectively. The fluid-fluid level sign was related to three radiomic features, and the gemstone sign was only related to two radiomic features. The first important radiomic feature, T2_logarithm_fistorder_Skewness, was only related to three MRI features (broad-based attachment, peripheral cyst, and gemstone signs); while the last one, T2_logarithm_firstorder_Kurtosis, was associated with all six MRI features. The other radiomic features were related to at least four MRI features.
Heatmap of association between selected radiomic features and MR features. Radiomic features descend by the order of importance from top to bottom and MRI features descend by AUC values from left to right. Data are p values. p values < 0.05 indicates statistically associations, as determined using t tests
Discussion
CN is an important differential diagnosis of intraventricular glioma, with a different prognosis and treatment course from lateral ventricle glioma. In this study, the SVM machine learning classification algorithm was used to develop a radiomics model based on T2W and CE-T1W images. This model can reliably distinguish CN from glioma in the training cohort and internal validation cohort, with good generalization ability in the external validation cohort. Moreover, its diagnostic efficacy was better than those of experienced neuroradiologists. Furthermore, use of the radiomics model as a prospective diagnostic assistant tended to improve radiologist performance.
Several studies have shown that characteristic MRI signs, such as scalloping, peripheral bubble, and soap bubble, are specific signs for CN [12]. However, the interobserver agreement of these signs is not very satisfactory [3, 6]. At the same time, diagnosis of diseases based on qualitative analysis is susceptible to personal experience and ability [16], with suboptimal interobserver agreement as shown in our present study. Radiomics incorporates high-order features that cannot be recognized by the naked eye, which reflect tumor pathophysiological characteristics and have incremental value for diagnosis [25]. In this study, the radiomics model can accurately predict the pathological type, and its overall performance outweighed experienced experts. Notably, the models based on a single imaging sequence can already distinguish CNs from gliomas very well, which is usually very difficult for radiologists.
Compared with models based on a single imaging sequence, the combined radiomics model not only improved the diagnostic performance, but also significantly reduced the number of features, which may reduce the risk of overfitting. As expected, this model had a good generalization in the external cohort. Specifically, the selected three T2W features are first-order signal histogram features (kurtosis, skewness, and mean value), which indicate obvious difference regarding tumor cell density and distribution of the two tumor types [25, 26]. Radiologically, CN has more cysts than glioma, and manifests with peripheral bubble, scalloping, soap bubble, and fluid-fluid level signs on T2W images, which may be the physiological basis for the selection of these histogram features. The other two features of the model are texture features. Texture features reflect the heterogeneity within the tumor [25], including GLCM, GLSZM, and GLDM, which are another group of radiomic features. In previous studies, the machine learning model based on texture features showed good diagnostic performance [27, 28]. Of note, only one radiomic feature was derived from CE-T1W images in the model. Similarly, among all the previously reported MRI signs, only the gemstone sign is based on CE-T1W images [12]. However, radiomic characteristics and MRI features reflect the biological characteristics of tumors in different scales. In this study, the most important radiomic feature (T2_logarithm_fstorder_skewness) only correlated with three of six MRI features, while the least important one (T2_logarithm_fstorder_kurtosis) correlated with all six MRI features.
The potential value of artificial intelligence (AI) to clinical work is a worthy topic of investigation. Yamashita et al reported that, although deep learning models exceeded experienced gastrointestinal pathologists at detecting colorectal cancer microsatellite instability, it failed to improve their accuracy as a group [29]. In contrast, Zhang et al showed the potential of AI in assisting radiologists in predicting treatment response in rectal cancer [30]. In our study, referring to the results of the radiomics model, two out of three radiologists improved their diagnostic performance in the training cohort. However, the effect was not significant in the internal and external validation cohorts. This may be related to the small sample size or that radiologists did not have high confidence in the performance of the radiomics model. AI models are often seen as black boxes that do not provide transparency into how they arrive at their final outputs. However, a few methods, such as class activation maps [31], provide some insight into what features models are seeing, yet these do not always directly correspond to human-interpretable features. In this study, two radiologists with extensive experience in the diagnosis and differential diagnosis of CN did not significantly improve their diagnostic performance with the aid of the radiomics model. This may be due to a lack of awareness of the excellent performance of the model, which may make the expert radiologists have the tendency to trust their own judgment over that of the model [5, 12]. Efforts to increase the interpretability of AI models may help increase confidence in AI models.
This study has some limitations. First, our results clearly show the potential of the radiomics model in differentiating CNs from lateral ventricular gliomas; however, the sample size was still small due to the low occurrence of these two tumor types. This study collected cases from two medical centers in a broad period (10 years), so a larger cohort including more institutes is warranted to verify our results. Also, as a retrospective study, this study did not include other intraventricular tumors, such as meningioma or choroid plexus papilloma. Therefore, this model is not suitable for the differential diagnosis of all intraventricular tumors. Moreover, age, gender, and location of tumor were not analyzed in the study. Many studies have shown that the clinicoradiomic models integrating different category characteristics generally have better diagnostic performance [32,33,34]. Considering that CN has a predilection site and peak incidence age [35], a multi-scale model integrating radiomic and clinical features may further enhance the diagnostic performance and stability of the model. In addition, different thresholds in the Rad score among three cohorts, as a common phenomenon with the SVM algorithm, might bring a problem to clinical translation of this model [36]. Probability calibration could be employed to solve this problem [37]. Also, the drift of threshold will be less obvious with increasing sample sizes.
In conclusion, the radiomics model based on machine learning can accurately distinguish between CNs and gliomas in the lateral ventricles. Moreover, its performance exceeds that of experienced radiologists and shows potential value in assisting decision-making. Considering that the model was developed based on images from a variety of devices and different MRI protocols, the prospect for clinical application of this model is more promising, which will help improve the preoperative diagnosis of lateral ventricle tumors and guide treatment decisions.
Abbreviations
- AUC:
-
Area under the receiver operating characteristic curve
- CE-T1W:
-
Contrast-enhanced T1-weighted
- CI:
-
Confidence interval
- CN:
-
Central neurocytoma
- ICC:
-
Intraclass correlation coefficients
- ROI:
-
Region of interest
- SVM:
-
Support vector machine
- T2W:
-
T2-weighted
References
Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23:1231–1251
Koeller KK, Sandberg GD (2002) From the archives of the AFIP. Radiographics 22:1473–1505
Donoho D, Zada G (2015) Imaging of central neurocytomas. Neurosurg Clin N Am 26:11–19
Chen CL, Shen CC, Wang J, Lu CH, Lee HT (2008) Central neurocytoma: a clinical, radiological and pathological study of nine cases. Clin Neurol Neurosurg 110:129–136
Xiao X, Zhou J, Wang J et al (2017) Clinical, radiological, pathological and prognostic aspects of intraventricular oligodendroglioma: comparison with central neurocytoma. J Neurooncol 135:57–65
Niiro T, Tokimura H, Hanaya R et al (2012) MRI findings in patients with central neurocytomas with special reference to differential diagnosis from other ventricular tumours near the foramen of Monro. J Clin Neurosci 19:681–686
Goergen SK, Gonzales MF, McLean CA (1992) Interventricular neurocytoma: radiologic features and review of the literature. Radiology 182:787–792
Shin JH, Lee HK, Khang SK et al (2002) Neuronal tumors of the central nervous system: radiologic findings and pathologic correlation. Radiographics 22:1177–1189
Ramsahye H, He H, Feng X, Li S, Xiong J (2013) Central neurocytoma: radiological and clinico-pathological findings in 18 patients and one additional MRS case. J Neuroradiol 40:101–111
Smith AB, Smirniotopoulos JG, Horkanyne-Szakaly I (2013) From the radiologic pathology archives: intraventricular neoplasms: radiologic-pathologic correlation. Radiographics 33:21–43
Freund M, Jansen O, Geletneky K, Hahnel S, Sartor K (1998) Computerized tomography and magnetic resonance imaging findings in central neurocytoma. Rofo 168:502–507
Li X, Guo L, Sheng S et al (2018) Diagnostic value of six MRI features for central neurocytoma. Eur Radiol 28:4306–4313
Nishio S, Morioka T, Suzuki S, Fukui M (2002) Tumours around the foramen of Monro: clinical and neuroimaging features and their differential diagnosis. J Clin Neurosci 9:137–141
Osztie É, Hanzély Z, Áfra D (2007) Lateral ventricle gliomas and central neurocytomas in adults diagnosis and perspectives. Eur J Radiol 69:67–73
Abrol S, Kotrotsou A, Salem A, Zinn PO, Colen RR (2017) Radiomic phenotyping in brain cancer to unravel hidden information in medical images. Top Magn Reson Imaging 26:43–53
Zhang Y, Shang L, Chen C et al (2020) Machine-learning classifiers in discrimination of lesions located in the anterior skull base. Front Oncol 10:752
Sun PF, Ma L, Ye BQ, Pei YY (2020) Application of diffusion-weighted imaging combined with apparent diffusion coefficient in differential diagnosis between central neurocytoma and ependymoma. Neuroradiology 62:439–445
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Bae S, An C, Ahn SS et al (2020) Robust performance of deep learning for distinguishing glioblastoma from single brain metastasis using radiomic features: model development and validation. Sci Rep 10:12110
Kickingereder P, Bonekamp D, Nowosielski M et al (2016) Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 281:907–918
Kang D, Park JE, Kim YH et al (2018) Diffusion radiomics as a diagnostic model for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol 20:1251–1261
Cherkassky V (1997) The nature of statistical learning theory~. IEEE Trans Neural Netw 8:1564
Zacharaki EI, Wang S, Chawla S et al (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618
Van Belle V, Pelckmans K, Van Huffel S, Suykens JA (2011) Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif Intell Med 53:107–118
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Liu K, Wen G, Lv XF et al (2013) MR imaging of cerebral extraventricular neurocytoma: a report of 9 cases. AJNR Am J Neuroradiol 34:541–546
Moiseev A, Snopova L, Kuznetsov S et al (2018) Pixel classification method in optical coherence tomography for tumor segmentation and its complementary usage with OCT microangiography. J Biophotonics 11:e201700072
Huang ZS, Xiao X, Li XD et al (2021) Machine learning-based multiparametric magnetic resonance imaging radiomic model for discrimination of pathological subtypes of craniopharyngioma. J Magn Reson Imaging 54:1541–1550
Yamashita R, Long J, Longacre T et al (2021) Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol 22:132–141
Zhang XY, Wang L, Zhu HT et al (2020) Predicting rectal cancer response to neoadjuvant chemoradiotherapy using deep learning of diffusion kurtosis MRI. Radiology 296:56–64
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2020) Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis 128:336–359
Zhang B, Tian J, Dong D et al (2017) Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res 23:4259–4269
Niu J, Zhang S, Ma S et al (2019) Preoperative prediction of cavernous sinus invasion by pituitary adenomas using a radiomics method based on magnetic resonance images. Eur Radiol 29:1625–1634
Yang L, Gu D, Wei J et al (2019) A radiomics nomogram for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Liver Cancer 8:373–386
Jia L, Li S, Du Y, Li Y, Gao F (2020) Neuroimaging diagnosis of intraventricular central neurocytoma. Neurosci Lett 735:135143
Attia MH, Attia MH, Farghaly YT, Abulnoor BAES, Curate F (2022) Performance of the supervised learning algorithms in sex estimation of the proximal femur: a comparative study in contemporary Egyptian and Turkish samples. Sci Justice 62:288–309
Van Calster B, McLernon DJ, Van Smeden M, Wynants L, Steyerberg EW (2019) Calibration: the Achilles heel of predictive analytics. BMC Med 17:230
Funding
This study has received funding by the Clinical Research Program of Nanfang Hospital, Southern Medical University (2020CR004), and the Medical Research Foundation of Guangdong Province (A2021048).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Dr. Yuankui Wu, Department of Medical Imaging, Nanfang Hospital, Southern Medical University.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
MRI data of 30 patients of CN and 27 patients of glioma have been previously reported in Journal of European Radiology.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 476 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mo, H., Liang, W., Huang, Z. et al. Machine learning–based multiparametric magnetic resonance imaging radiomics model for distinguishing central neurocytoma from glioma of lateral ventricle. Eur Radiol 33, 4259–4269 (2023). https://doi.org/10.1007/s00330-022-09319-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-09319-9