Abstract
Objective
To develop novel deep learning network (DLN) with the incorporation of the automatic segmentation network (ASN) for morphological analysis and determined the performance for diagnosis breast cancer in automated breast ultrasound (ABUS).
Methods
A total of 769 breast tumors were enrolled in this study and were randomly divided into training set and test set at 600 vs. 169. The novel DLNs (Resent v2, ResNet50 v2, ResNet101 v2) added a new ASN to the traditional ResNet networks and extracted morphological information of breast tumors. The accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), area under the receiver operating characteristic (ROC) curve (AUC), and average precision (AP) were calculated. The diagnostic performances of novel DLNs were compared with those of two radiologists with different experience.
Results
The ResNet34 v2 model had higher specificity (76.81%) and PPV (82.22%) than the other two, the ResNet50 v2 model had higher accuracy (78.11%) and NPV (72.86%), and the ResNet101 v2 model had higher sensitivity (85.00%). According to the AUCs and APs, the novel ResNet101 v2 model produced the best result (AUC 0.85 and AP 0.90) compared with the remaining five DLNs. Compared with the novice radiologist, the novel DLNs performed better. The F1 score was increased from 0.77 to 0.78, 0.81, and 0.82 by three novel DLNs. However, their diagnostic performance was worse than that of the experienced radiologist.
Conclusions
The novel DLNs performed better than traditional DLNs and may be helpful for novice radiologists to improve their diagnostic performance of breast cancer in ABUS.
Key Points
• A novel automatic segmentation network to extract morphological information was successfully developed and implemented with ResNet deep learning networks.
• The novel deep learning networks in our research performed better than the traditional deep learning networks in the diagnosis of breast cancer using ABUS images.
• The novel deep learning networks in our research may be useful for novice radiologists to improve diagnostic performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Breast cancer has the highest incidence and mortality rate of malignant tumors in women. Early diagnosis and treatment of this particular cancer is helpful in decreasing the mortality rates [1, 2]. Higher breast density is more common among Asian women, which is associated with a relatively higher risk of being diagnosed with breast cancer than among those with lower breast densities [3,4,5].
Automated breast ultrasound (ABUS), as a new ultrasonography technology, has been proven to overcome the shortcomings of handheld ultrasound (HHUS) and plays a pivotal role in the screening and staging of breast cancer [6, 7]. Supplemental ABUS in women with mammographically dense breasts can help radiologists detect early-stage breast cancers [8, 9]. ABUS can reconstruct three-dimensional (3D) images of the breast lesion volume, including coronal, axial, and sagittal views, in which the coronal view has been shown to improve early detection in dense breasts and diagnostic accuracy [10, 11]. However, for certain types of malignant tumors, ABUS with at least 3 volume images may lead to mistakes in diagnosis [12]. In recent years, artificial intelligence (AI) schemes have been expected to facilitate breast screening by ABUS [13, 14].
Deep learning (DL) is a subset of machine learning (ML) and AI that extracts a complex hierarchy of features from images by its self-learning ability [15]. Deep learning networks (DLNs) have a wide range of assistance in image diagnosis and prediction because of their advantages, such as being fast, accurate, and reproducible [16,17,18]. The typical representative of DLNs is convolutional neural network (CNN) which has been the most popular method for pattern recognition and computer vision applications in image analysis. The outstanding performance of CNN has been shown to increase with depth for some tasks and has been applied to ABUS imaging analysis in recent years [19,20,21]. The interpretation time of ABUS screening for dense breast tissue was decreased compared with that of radiologists [22, 23]. Some studies have used CNN for breast cancer detection and diagnosis by ABUS [24, 25]. However, the networks they used were traditional or simple, which did not consider the characteristics of ABUS itself, so that it might be difficult to extract rich and effective image features. To further explore the performance of DLN, we developed a novel DLN and compared its diagnostic performance with that of radiologists on breast cancer by ABUS.
Materials and methods
Patients
This study was approved by the Institutional Review Board of our hospital. Patients with breast lesions between November 2018 and January 2020 were retrospectively reviewed in this study. Indications of ABUS were routine screening, or patients with or without symptomatic breast lesions that did not have ulceration or protrusion that significantly affects the image quality. All patients were subjected to clinical routine conventional ABUS in combination with HHUS examination if there were breast lesions in ABUS or HHUS. All the patients underwent more than one and a half years of follow-up, or eventually underwent biopsy or surgical treatment. Complete pathological information was acquired after surgery. Clinical information of the patients and the ABUS features of the breast lesions were recorded. The inclusion criteria were as follows: (1) ABUS images with breast lesions; (2) the lesions considered benign were followed up in our hospital for more than one and a half years; (3) lesions did not undergo biopsy or any treatment prior to the ABUS examination; (4) the images were clear for image mark; and (5) complete data and clinical information. The exclusion criteria for the patients were as follows: (1) patients with pathological results neither benign nor malignant; (2) patients with no surgery and no follow-up within 2 years; (3) without clear pathological confirmation; (4) ABUS features of the breast lesions cannot be clearly seen with being affected by the shadow behind the nipple or with low-quality image; and (5) incomplete data and clinical information.
ABUS examination
The InveniaTM Automated Breast Ultrasound System (Invenia ABUS, Automated Breast Ultrasound System, GE Healthcare) with an automated 6–14-MHz linear broadband transducer (covering volumes of 15.4 × 17.0 × 5.0 cm) was used in the study. Image acquisition was performed by two experienced technologists. The patients were placed similar to those with conventional US. Thus, anteroposterior, medial, and lateral orientation items of the volume data were obtained, and if required, superior and inferior orientations were also obtained. After acquisition, all of the images were sent to the image reading workstation.
Datasets
A total of 769 breast lesions with 769 3D images from 743 patients (range 27–79 years) were enrolled in this study. Seg3D2 software (Seg3D 2-2.4.4), a free volume segmentation and processing tool, was used by three radiologists with more than 5 years of work experience to determine the boundary and shape of the lesions and to manually mark all of the lesions. The manually marked breast lesion volumes were used for the following two purposes: first, to help in network learning. The process of network learning is to simulate a doctor’s manual mark and to help in learning to extract the right shape information. Second, it is to test the accuracy of different models.
The training set and test set were selected by a random sampling method at 600 vs. 169. The training set was further randomly divided into a new training set and a validation set for model establishment at a ratio of 5:1. Each pair of optimal traditional and novel DLNs was tested with the test set. A flowchart describing the research process is shown in Fig. 1.
Development of the DLN
All of the marked lesions were divided into a training set and test set, cut into a suitable size according to the size of the lesions, and finally input into the network at a uniform size.
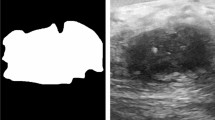
In general, CNNs consist of convolutional layers, pooling layers, and fully connected layers. The network obtained the high-level features of the input image through a multilayer convolution operation, and these features represent some important semantic information of the image. Subsequently, the fully connected layer analyzes the semantic information and completes the diagnosis. To further increase the classification accuracy, we trained an automatic separate segmentation network, according to manually mark results, to automatically extract the shape information of the lesion, and then fused the shape information of the lesion with the original data so that the network could use the shape information to improve its performance. The self-established automatic segmentation network (ASN) is shown in supplementary Fig. 2. Then, we used Dice loss for training. Dice loss was used to measure the difference between the 3D shape predicted by the ASN and that of the manually marked mass. The smaller the Dice loss, the more accurate the shape predicted by the network. After the training was completed, shape extraction was performed on the test set data through parameter migration. In Fig. 3, we list one benign case and one malignant case, the ABUS 3D images of which are processed by the segmentation network, and the visualized images of the lesions are demonstrated. The 3D volume images of the benign and malignant lesions after segmentation are shown in supplementary Fig. 3.
ResNet network, as a well-known CNN architecture, was used to implement backbone due to its strength in differentiating different classes of objects [26]. In fact, the depth of ResNet can be any layer, such as 18, 34, 50, 101, and 150 layers depending on the number of superimposed times of the residual module and the increase in depth does not always lead to better results. Considering the above reason and training time, we selected 34-, 50-, and 101-layer models as representatives of shallow, middle, and deep networks to explore the impact between depth and network performance. The newly built ASNs were merged in the traditional ResNet to create corresponding new ResNet v2 networks—namely, ResNet34 and ResNet34 v2, ResNet50 and ResNet50 v2, and ResNet101 and ResNet101 v2. For ResNet v2, the input channel was adjusted to 64 channels and the output category was adjusted to 2, indicating the probability of benign and malignant lesions.
Then, we conducted training, validation, and testing both before and after the improvement network to verify the performance of our algorithm by comparing the performance of each pair of networks. The logic diagram of the ResNet network is shown in Fig. 2.
Structural diagram of the transfer learning model for ABUS 3D images. The top subfigure shows the original ResNet model, which receives natural images as input and produces prediction categories from 1000 categories. The middle subfigure is the control group model. Compared with the original ResNet network structure, the input and output of this model are adjusted to suit the current task. The bottom subfigure is the proposed model, which extracts shape information by establishing an auxiliary task. This model integrates shape information and ABUS 3D images as ResNet network input, which pays more attention to tumor shape and makes the prediction result more accurate. ABUS = automated breast ultrasound
ABUS image and feature map visualization of breast tumor segmentation on breast lesions. A ABUS detected a hypoechoic lesion (arrows) in the upper quadrant of the right breast. The coronal view shows the proper nipple position (white point). B Feature map visualization of the breast tumor segmentation of the benign lesion. C ABUS detected a hypoechoic lesion (arrows) in the upper quadrant of the left breast. The coronal view shows the proper nipple position (white point). D Feature map visualization of the breast tumor segmentation on the malignant lesion. ABUS = automated breast ultrasound
Image analysis
Radiologist 1 (novice radiologist) with 1 year of work experience in ABUS diagnosis and radiologist 2 (experienced radiologist) with 9 years of work experience in ultrasound diagnosis and 5 years of experience in ABUS were involved in the diagnosis. The diagnostic result of each radiologist for each lesion was dichotomous (malignant or benign). Radiologists 1 and 2 were not the ones who manually marked the tumors. Only the primary ABUS image data, age, corresponding case number, and date of examination were visible to the radiologists to ensure the objectivity of the diagnosis. Image analysis in ABUS was essentially based on the evaluation of same image features than for HHUS including shape (oval, round, or irregular), margin (circumscribed, indistinct, microlobulated, angular, or spiculated), orientation (parallel or not parallel), echo pattern (hypoechoic, hyperechoic, isoechoic, anechoic, complex cystic and solid, or heterogeneous), posterior echogenicity (no posterior features, enhancement, shadowing, or combined pattern), calcification (presence or absence), and associated features (architectural distortion, ductal changes, skin changes, or edema) in Breast Imaging-Reporting and Data System (BI-RADS) lexicon [27]. Hyperechoic halo (presence or absence) and spiculation or retraction phenomenon (presence or absence) in the coronal view were added, the presence of which may be suggestive of malignancy [28]. Features of all of the lesions were analyzed by radiologist 1 and 2. Then, radiologists made BI-RADS category based on the 3D image features. BI-RADS 2–3 categories were classified as benign, and BI-RADS 4–5 categories were classified as malignant. Breast density was also visually classified by radiologists [27, 29].
Statistical analysis
SPSS 24.0 software was used for statistical analysis. The accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) results of the six models were also calculated. Since it is impossible to compare the quality of the model based on the two contradictory values (precision vs. sensitivity), F1 scores were calculated as an indicator that integrated precision and sensitivity (Supplementary Table 1 and Supplementary Fig. 1 show the calculation method). The higher the F1 score, the better the comprehensive diagnostic performance of the model. Average precision (AP) represents the accuracy of the model for the positive sample. Receiver operating characteristic (ROC) curves were drawn. By calling the function in Scikit-Learn, the AP, precision-recall curves (PRC), and areas under the ROC curve (AUCs) with 95% confidence intervals (CIs) were calculated. p < 0.05 was considered to indicate a statistically significant difference.
Results
Case inclusion and grouping
Among the 769 breast lesions, there were 454 (59.03%) malignant lesions and 315 (40.96%) benign lesions. The maximum diameter was 0.4–8.0 cm. The training set contained 600 lesions that were further divided into training set and validation set. The training set included 500 cases with 295 malignant lesions and 205 benign lesions. The validation set included 100 cases with 59 malignant lesions and 41 benign lesions. The test set included 169 cases, in which malignant and benign lesions were 100 vs. 69. The characteristics of cases in each group are shown in Supplementary Table 2.
Performance of DLN
We conducted a pairwise comparison study using the above six models to observe the diagnostic performance for benign and malignant lesions. The diagnostic results of the confusion matrices for the six models and radiologists according to the test set are shown in Table 1. According to the results in Table 2, the ResNet34 v2 model had higher specificity (76.81%) and PPV (82.22%) than the other two, and the ResNet50 v2 model had higher accuracy (78.11%) and NPV (72.86%), and the ResNet101 v2 model had higher sensitivity (85.00%). Of these values, only the specificity and PPV of the ResNet34 v2 and the NPV of the ResNet50 v2 showed significant increases compared with traditional ResNet models (p < 0.05).
According to the specificity and sensitivity, ROC curves were drawn, and AUCs were calculated. The AUCs (range 0.83–0.85) and the APs (range 0.88–0.90) are shown in Fig. 4. AP represents the accuracy of the model for the positive sample. The larger the AP is, the higher the comprehensive accuracy and recall result of the model for malignant lesions. According to the AUCs and APs, the ResNet101 v2 model produced the best results. However, it was difficult to make one kind of network perform best for all of the indicators. For accuracy, ResNet50 v2 was the highest (78.11%). For sensitivity, ResNet34 was the best (87.00%). For specificity, ResNet34 v2 performed best (76.81%). ResNet101 v2 had the highest F1 score (0.82).
Furthermore, we compared the diagnostic performance of radiologist 1 and radiologist 2 in Supplementary Table 3. The diagnostic sensitivity, specificity, PPV, and NPV of radiologist 2 were significantly higher than those of radiologist 1 (p < 0.05). Although there was no significant difference between radiologist 1 and radiologist 2 for accuracy, it was still higher in radiologist 2 than in radiologist 1. Then, we compared the three novel networks with radiologist 1 and radiologist 2. Comparing with radiologist 1, significant increases were found in accuracy and specificity (compared to ResNet34 v2), in PPV (compared to ResNet34 v2 and ResNet50 v2), and in NPV (compared to ResNet50 v2 and ResNet101 v2). ResNet34 v2 obtained significant increases with respect to radiologist 1 in accuracy, specificity, and PPV, ResNet50 v2 in PPV and NPV, and ResNet101 v2 in NPV (p < 0.05) (Table 3). The remaining results did not show significant differences. In addition, the F1 score increased from 0.77 to 0.78, 0.81, and 0.82. Comparing with radiologist 2, significantly lower values were found in accuracy (compared to ResNet34 v2), sensitivity (compared to ResNet34 v2 and ResNet50 v2), and NPV (compared to ResNet34 v2, ResNet50 v2, and ResNet101 v2). Although the remaining results did not show significant differences, the specificity was increased from 71.01 to 76.81% and 73.91% by ResNet34 v2 and ResNet50 v2. The PPVs of ResNet34 v2 and ResNet50 v2 were 82.22% and 81.82%, which were higher than that of radiologist 2 (81.65%). In general, the three novel models obtained significantly lower values of NPV than radiologist 2 and ResNet34 v2 obtained lower values of accuracy and sensitivity (Table 3).
Discussion
We successfully developed the novel DLNs and implemented incorporating the ASN to traditional network for breast cancer diagnosis based on ABUS. As reported in some studies, image segmentation is of great importance for computer-assisted breast cancer diagnosis [30,31,32,33]. Manual segmentation is time-consuming, and due to different experience, volume segmentation variations can have standard deviations up to 60% between observers [34, 35]. To improve the diagnostic performance and reduce the need for human intervention, the demand for automatic tumor segmentation is increasing [36, 37]. Tumor classification and segmentation are two important tasks for computer-aided detection (CAD) using ABUS 3D images. Y. Zhou [38] collected a clinical dataset of 170 ABUS 3D volumes from 107 patients and indicated that the proposed multitask framework could improve tumor segmentation and classification over single-task learning counterparts. In our study, we sought to develop a novel DLN that added a self-established accurate ASN to extract shape information by establishing an auxiliary task. This model integrated shape information and ABUS 3D images as ResNet network input, which paid more attention to tumor shape. As a result, the input contained richer information, so the diagnosis was more accurate than the traditional DLN.
The network uses pretraining parameters of ResNet on ImageNet, an image database organized according to the WordNet hierarchy, as initial weights and performs fine-tune on this basis. In our experiment, the model was used as a control group to verify the improvement effect of our method. By changing the backbone, different classification networks were obtained. The accuracies of the novel DLNs in the diagnosis of breast cancer were improved compared with those of traditional DLNs, and the novel DLNs performed well in the test set. The higher specificity and PPV of the ResNet34 v2 network for ABUS 3D images might help radiologists avoid unnecessary recalls of benign lesions (p < 0.05). The AUCs of the novel networks were improved to 0.83, 0.84, and 0.85. Furthermore, the APs were improved to 0.88, 0.89, and 0.90.
In many studies of breast cancer diagnosis, the accuracy of ML in ABUS is improved compared with using ABUS alone. van Zelst J.C.M. [14] established dedicated CAD software for ABUS to improve the AUC of readers in detecting more breast cancers. Without CAD, the AUC of the readers was 0.77 and improved with CAD to 0.84 (p = 0.001). The sensitivity of all readers improved by using CAD (range 5.2–10.6%), but specificity decreased for some readers (range 1.4–5.7%). Wang F. [39] used a deep CNN combined with ABUS in the classification of breast nodules and achieved a sensitivity of 87.0%, a specificity of 88.0%, and an accuracy of 87.5%. Xu X. [40] assessed the performance of radiologists in the interpretation of automated breast volume scanner (ABVS) images with the aid of a CAD system. AUCs with and without CAD were 0.833 and 0.829 for radiologists with 4 years of ABVS experience, 0.757 and 0.696 for radiologists with 5 years of experience, and 0.759 and 0.718 for radiologists with 1 year of experience. All of the AUCs were increased with the CAD system. In our study, higher accuracy, sensitivity, specificity, PPV, NPV, and F1 scores were shown in different novel DLNs than a novice radiologist. Therefore, the performance of the novel DLN based on ABUS 3D images may help in improving the diagnostic performance of a novice radiologist.
However, in our study, the novel DLNs were worse than an experienced radiologist in diagnostic performance. The reason for this may be that the training set and test set had small data, and the features of benign and malignant lesions learned by the network may be insufficient, resulting in unsatisfactory results. Experienced radiologists have studied tens of thousands of cases, and their knowledge of the number and types of malignant nodules has been enriched. Therefore, the diagnostic performance of experienced radiologists was better. Even so, its use may help to reduce time consumption and improve their reading speed as reported in some studies [13, 22, 23, 40].
There are several limitations in this study. First, this was a retrospective study with data from one center, so that the sample size of this study was small in both the training and test sets. This may make the diagnosis result of our novel DLNs not very good compared with that of an experienced radiologist. Second, this study lacked external tests. Therefore, there is still no proof that our results can be reproduced in an external cohort. Third, the comparison between novel DLNs and radiologists was made with only one novice radiologist and one experienced radiologist. In the future, we should conduct comparative studies with more radiologists with different experiences, accumulate a larger sample size, and add external test sets to validate our results and determine the value of our DLNs.
Conclusion
We developed a new segmentation network to obtain morphological information of breast lesions. Based on the traditional classification network, we added the new segmentation network to the traditional network to form novel DLNs. The novel DLNs performed better than the traditional ones in the diagnosis of breast cancer by ABUS in terms of AUC and AP values. Additionally, the novel DLNs may be helpful for novice radiologists to improve their diagnostic performance.
Abbreviations
- 3D:
-
Three dimensional
- ABUS:
-
Automated breast ultrasound
- ABVS:
-
Automated breast volume scanner
- AI:
-
Artificial intelligence
- AP:
-
Average precision
- ASN:
-
Automatic segmentation network
- AUC:
-
Area under the ROC curve
- CAD:
-
Computer-aided detection
- CI:
-
Confidence interval
- CNN:
-
Convolutional neural network
- DL:
-
Deep learning
- DLN:
-
Deep learning network
- HHUS:
-
Handheld ultrasound
- ML:
-
Machine learning
- NPV:
-
Negative predictive value
- PPV:
-
Positive predictive value
- PRC:
-
Precision-recall curves
- ROC:
-
Receiver operating characteristic
References
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
Birnbaum JK, Duggan C, Anderson BO, Etzioni R (2018) Early detection and treatment strategies for breast cancer in low-income and upper middle-income countries: a modelling study. Lancet Glob Health 6:e885–e893
Li E, Guida JL, Tian Y et al (2019) Associations between mammographic density and tumor characteristics in Chinese women with breast cancer. Breast Cancer Res Treat 177:527–536
Boyd NF, Guo H, Martin LJ et al (2007) Mammographic density and the risk and detection of breast cancer. N Engl J Med 356:227–236
Vachon CM, Pankratz VS, Scott CG et al (2007) Longitudinal trends in mammographic percent density and breast cancer risk. Cancer Epidemiol Biomark Prev 16:921–928
Jia M, Lin X, Zhou X et al (2020) Diagnostic performance of automated breast ultrasound and handheld ultrasound in women with dense breasts. Breast Cancer Res Treat 181:589–597
Yun G, Kim SM, Yun B, Ahn HS, Jang M (2019) Reliability of automated versus handheld breast ultrasound examinations of suspicious breast masses. Ultrasonography 38:264–271
Brem RF, Tabár L, Duffy SW et al (2015) Assessing improvement in detection of breast cancer with three-dimensional automated breast US in women with dense breast tissue: the SomoInsight Study. Radiology 274:663–673
Wilczek B, Wilczek HE, Rasouliyan L, Leifland K (2016) Adding 3D automated breast ultrasound to mammography screening in women with heterogeneously and extremely dense breasts: report from a hospital-based, high-volume, single-center breast cancer screening program. Eur J Radiol 85:1554–1563
Rella R, Belli P, Giuliani M et al (2018) Automated Breast Ultrasonography (ABUS) in the screening and diagnostic setting: indications and practical use. Acad Radiol 25:1457–1470
Zhang L, Bao LY, Tan YJ et al (2019) Diagnostic performance using automated breast ultrasound system for breast cancer in Chinese women aged 40 years or older: a comparative study. Ultrasound Med Biol 45:3137–3144
Kelly KM, Dean J, Comulada WS, Lee SJ (2010) Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur Radiol 20:734–742
van Zelst JCM, Tan T, Clauser P et al (2018) Dedicated computer-aided detection software for automated 3D breast ultrasound; an efficient tool for the radiologist in supplemental screening of women with dense breasts. Eur Radiol 28:2996–3006
van Zelst JCM, Tan T, Platel B et al (2017) Improved cancer detection in automated breast ultrasound by radiologists using computer aided detection. Eur J Radiol 89:54–59
Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ (2017) Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 30:449–459
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H (2018) Artificial intelligence in radiology. Nat Rev Cancer 18:500–510
Chambara N, Ying M (2019) The diagnostic efficiency of ultrasound computer-aided diagnosis in differentiating thyroid nodules: a systematic review and narrative synthesis. Cancers 11:1759
Ha T, Jung Y, Kim JY, Park SY, Kang DK, Kim TH (2019) Comparison of the diagnostic performance of abbreviated MRI and full diagnostic MRI using a computer-aided diagnosis (CAD) system in patients with a personal history of breast cancer: the effect of CAD-generated kinetic features on reader performance. Clin Radiol 74:817.e815–817.e821
Wang Y, Wang N, Xu M et al (2020) Deeply-supervised networks with threshold loss for cancer detection in automated breast ultrasound. IEEE Trans Med Imaging 39:866–876
Li Y, Wu W, Chen H, Cheng L, Wang S (2020) 3D tumor detection in automated breast ultrasound using deep convolutional neural network. Med Phys 47:5669–5680
Wang Y, Choi EJ, Choi Y, Zhang H, Jin GY, Ko SB (2020) Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning. Ultrasound Med Biol 46:1119–1132
Jiang Y, Inciardi MF, Edwards AV, Papaioannou J (2018) Interpretation time using a concurrent-read computer-aided detection system for automated breast ultrasound in breast cancer screening of women with dense breast tissue. AJR Am J Roentgenol 211:452–461
Yang S, Gao X, Liu L et al (2019) Performance and reading time of automated breast US with or without computer-aided detection. Radiology 292:540–549
Moon WK, Huang YS, Hsu CH et al (2020) Computer-aided tumor detection in automated breast ultrasound using a 3-D convolutional neural network. Comput Methods Prog Biomed 190:105360
Chiang TC, Huang YS, Chen RT, Huang CS, Chang RF (2019) Tumor detection in automated breast ultrasound using 3-D CNN and prioritized candidate aggregation. IEEE Trans Med Imaging 38:240–249
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 770-778. https://doi.org/10.1109/CVPR.2016.90
American College of Radiology (2013) ACR BI-RADS® Atlas. Breast Imaging Reporting and Data System, 5th edn. Acadmerican College of Radiology, Reston
Zheng FY, Yan LX, Huang BJ et al (2015) Comparison of retraction phenomenon and BI-RADS-US descriptors in differentiating benign and malignant breast masses using an automated breast volume scanner. Eur J Radiol 84:2123–2129
Kim WH, Moon WK, Kim SJ et al (2013) Ultrasonographic assessment of breast density. Breast Cancer Res Treat 138:851–859
Noble JA, Boukerroui D (2006) Ultrasound image segmentation: a survey. IEEE Trans Med Imaging 25:987–1010
Gu P, Lee WM, Roubidoux MA, Yuan J, Wang X, Carson PL (2016) Automated 3D ultrasound image segmentation to aid breast cancer image interpretation. Ultrasonics 65:51–58
Liu L, Li K, Qin W et al (2018) Automated breast tumor detection and segmentation with a novel computational framework of whole ultrasound images. Med Biol Eng Comput 56:183–199
Pan P, Chen H, Li Y, Cai N, Cheng L, Wang S (2021) Tumor segmentation in automated whole breast ultrasound using bidirectional LSTM neural network and attention mechanism. Ultrasonics 110:106271
Calas MJ, Almeida RM, Gutfilen B, Pereira WC (2010) Intraobserver interpretation of breast ultrasonography following the BI-RADS classification. Eur J Radiol 74:525–528
Li XA, Tai A, Arthur DW et al (2009) Variability of target and normal structure delineation for breast cancer radiotherapy: an RTOG multi-institutional and multiobserver study. Int J Radiat Oncol Biol Phys 73:944–951
Hu Y, Guo Y, Wang Y et al (2019) Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med Phys 46:215–228
Lei Y, He X, Yao J et al (2021) Breast tumor segmentation in 3D automatic breast ultrasound using Mask scoring R-CNN. Med Phys 48:204–214
Zhou Y, Chen H, Li Y et al (2021) Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med Image Anal 70:101918
Wang F, Liu X, Yuan N et al (2020) Study on automatic detection and classification of breast nodule using deep convolutional neural network system. J Thorac Dis 12:4690–4701
Xu X, Bao L, Tan Y, Zhu L, Kong F, Wang W (2018) 1000-case reader study of radiologists’ performance in interpretation of automated breast volume scanner images with a computer-aided detection system. Ultrasound Med Biol 44:1694–1702
Acknowledgements
The review was completed under the guidance of Wen Cheng, who also reviewed the final version of the manuscript. All feature analysis was done by H. Chen and Q.C. Wang. Professor G.N. Luo, Z.S. Wang, and K.Q. Wang performed image preprocessing. The collection of the cases was completed by H. Chen, B. Li, H.T. Shang, H. Shao, and S.S. Sun. Thanks to professor J.N. Li for her guidance in data analysis. All authors had read and approved the final manuscript.
Funding
There was no external funding to support the writing of this review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Dr. Wen Cheng.
Conflict of interest
No conflict of interest exists in this manuscript.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• multiple case-multiple reader study
• performed at one institution
• diagnosis study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 447 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Chen, H., Luo, G. et al. Performance of novel deep learning network with the incorporation of the automatic segmentation network for diagnosis of breast cancer in automated breast ultrasound. Eur Radiol 32, 7163–7172 (2022). https://doi.org/10.1007/s00330-022-08836-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08836-x