Abstract
Key message
PeTIP4;1–1, an aquaporin gene involved in bamboo shoot growth, is regulated by abiotic stresses. Overexpression of PeTIP4;1–1 confers drought and salinity tolerance in transgenic Arabidopsis.
Abstract
Aquaporins play a central role in numerous physiological processes throughout plant growth and development. PeTIP4;1–1, an aquaporin gene isolated from moso bamboo (Phyllostachys edulis), comprises an open reading frame (ORF) of 756 bp encoding a peptide of 251 amino acids. The genomic sequence corresponding to the ORF of PeTIP4;1–1 was 1777 bp and contained three exons separated by two introns. PeTIP4;1–1 was constitutively expressed at the highest level in culms, and the expression level was elevated with increasing height of the bamboo shoot. PeTIP4;1–1 was significantly up-regulated in response to drought and salinity stresses in bamboo roots and leaves. To investigate the role of PeTIP4;1–1 in response to drought and salinity stresses, transgenic Arabidopsis plants overexpressing PeTIP4;1–1 under the control of CaMV 35S promoter were generated and subjected to morphological and physiological assays. Compared with Col-0, the transgenic plants showed enhanced tolerance to drought and salinity stresses and produced longer taproots, which had more green leaves, higher F v/F m and NPQ values, higher activities of SOD, POD and CAT, lower MDA concentration and higher water content. Transcript levels of three stress-related genes (AtP5CS, AtNHX1 and AtLEA) were enhanced. These results indicated that PeTIP4;1–1 might play an important function in response to drought and salinity stresses, and is a candidate gene for breeding of stress tolerance in other crops through genetic engineering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aquaporins (AQPs), which belong to the ancient family of major intrinsic proteins, play an important role in maintaining water and hydraulic conductivity balance in plants (Maurel et al. 2008). Recent studies show that AQPs constitute a large gene family in monocots. For instance, 35 AQP homologues were identified in wheat (Forrest and Bhave 2008), 33 in maize (Yue et al. 2012), 34 in rice (Nguyen et al. 2013), 40 in barley (Hove et al. 2015), and 26 in moso bamboo (Sun et al. 2016a), respectively. Based on the sequence of amino acids, membrane localization and substrate specificity, AQPs are classified into five major subfamilies, consisting of plasma membrane intrinsic proteins, tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins, small basic intrinsic proteins and uncharacterized X intrinsic proteins. The TIPs of Arabidopsis thaliana are further classified into five subgroups (Johanson et al. 2001; Schüssler et al. 2008). TIPs are mainly localized in the vacuole or plasma membrane and are known to increase the permeability of membranes to water and other small molecules, such as urea, ammonia and hydrogen peroxide (Schüssler et al. 2008; Yin et al. 2014). Numerous studies have showed that TIPs perform multiple functions throughout plant growth and development, including water transport, embolism repair and response to various abiotic stresses (Maurel et al. 2008).
Abiotic stresses, such as drought, waterlogging and salinity, are known to disturb the water balance and induce osmotic stress in plants, which may result in substantial losses in plant production throughout the world (Boyer 1982). The relationship between AQPs and osmotic stress has been investigated (Yin et al. 2014; Khan et al. 2015; Li et al. 2015; Chang et al. 2016). Investigation of the response of all known AQP genes of moso bamboo (Phyllostachys edulis) to drought and salinity stresses has demonstrated that most TIP genes show no significant transcript changes in roots and leaves during stress treatments, whereas PeTIP4;1 and PeTIP4;2 in roots and PeTIP1;2 and PeTIP4;1 in leaves were up-regulated briefly during drought stress. It was suggested that the inducible expression of PeTIP4;1 in roots and leaves might compensate for the severe drought stress at an early stage (Sun et al. 2016a). However, the function of PeTIP genes requires further investigation.
As an important group of forest, bamboo is mainly distributed at low latitudes, where climatic and environmental conditions are unfavorable for growth, and in particular high osmotic stress seriously affects the growth and development of bamboo. Although AQP genes in bamboo have been subjected to genome-wide analysis (Sun et al. 2016a), the functions of PeAQPs involved in osmotic stress tolerance remain unclear. In the present study, the open reading frame (ORF) of PeTIP4;1–1 and its genomic sequence were isolated from the cDNA and genomic DNA of moso bamboo, respectively. The gene structure, phylogenetic relationships and expression pattern of PeTIP4;1–1 were analyzed. The ORF of PeTIP4;1–1 was transformed into Arabidopsis thaliana. The phenotype, physiological parameters, activity of antioxidant enzymes and transcript level of stress-related genes in PeTIP4;1-1-overexpressing Arabidopsis plants under drought and salinity stresses were investigated. These results provide a foundation for understanding the function of PeTIP4;1–1.
Materials and methods
Plant materials and treatments
Moso bamboo (Phyllostachys edulis) seedlings were grown in pots in a growth chamber under a light intensity of 200 μmol m−2 s−1 with a 16 h light/8 h dark photoperiod at 28 °C. To detect tissue-specific gene expression patterns, samples of roots, culms, immature leaves, mature leaves, leaf sheaths and tender shoots were collected from 3-year-old juvenile plants. To investigate the changes in gene expression levels with shoot development, samples were collected from the base of shoots of 2, 5, 15 and 30 cm height 3-year-old juvenile plants. One-year-old seedlings were subjected to drought and salinity stress treatments. After uprooting and washing with sterile water, 30 seedlings were exposed to the open air for drought treatment, whereas the roots of another 30 seedlings were submerged in 400 mM NaCl solution for salinity treatment. The roots and leaves were collected from the treated seedlings after treatment for 1, 2, 3 and 4 h. All samples were immediately frozen in liquid nitrogen after collection and stored at −80 °C for further analysis.
cDNA synthesis and genomic DNA extraction
Trizol® Reagent solution (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNAs from the above-mentioned samples. The total RNAs extract was treated with RNase-free DNase I (Tiandz, Beijing, China) for 30 min at 37 °C to remove contaminant DNA. The absence of DNA was validated by PCR (Sun et al. 2016a). The RNA quality and concentration were examined with a spectrophotometer (Nanodrop 2000, Thermo Fisher, USA). The reverse transcription system (Promega, Beijing, China) was used for first-strand cDNA synthesis, with incubation at 42 °C for 45 min. The final cDNA product was diluted fivefold prior to use. Genomic DNAs were extracted from moso bamboo leaves using the cetyltrimethylammonium bromide method (Gao et al. 2006).
Gene isolation and bioinformatics analysis
The primers TIP4;1-F and TIP4;1-R were designed based on the nucleotide sequence of PeTIP4;1–1. Both the cDNAs and genomic DNAs were used as templates in PCRs for amplification of PeTIP4;1–1 with TIP4;1-F and TIP4;1-R using PrimeSTAR® DNA polymerase (Takara, Japan). The promoter region sequence (PeTIP4;1-1pro) was amplified using the primer pair TIP4;1p-F and TIP4;1p-R. The PCR products were inserted into the pGEM®-T Easy vector (Promega, USA) and subsequently confirmed by sequencing with an ABI 3730 sequencer (Applied Biosystems, USA). The sequences of all relevant primers are listed in supplementary S1.
The gene structure of PeTIP4;1–1 was analyzed using VectorNTI software. The number and position of introns were determined with WinPlas version 2.7. Sequence homology was evaluated by searching the National Center for Biotechnology Information database using the BLASTN tool (http://www.ncbi.nlm.nih.gov). Multiple sequence alignment of PeTIP4;1–1 and TIP4;1s from other plants (Oryza sativa, Brachypodium distachyon, Zea mays, A. thaliana and Populus trichocarpa) was generated using ClustalW version 1.83. A neighbor-joining tree was constructed using complete deletion parameters, and a bootstrap analysis with 1000 replicates was performed to assess the robustness of the topology using MEGA6.0 (Tamura et al. 2013). The cis-acting regulatory elements in the promoter sequence were predicted from the PlantCARE database (Lescot et al. 2002).
Gene expression analysis in moso bamboo
Moso bamboo cDNA samples from the roots, culms, mature and immature leaves, leaf sheaths and tender shoots were used for analysis of PeTIP4;1–1 expression in the different tissues. Semi-quantitative reverse transcription PCR (RT-PCR) was performed with the primer pair TIP4;1-F and TIP4;1-R. PeActin (GenBank Accession No. GU434145) was used as an internal control under the same PCR condition with the primer pair PeActin-F and PeActin-R (Sun et al. 2016b).
The cDNAs prepared from shoots of different heights, and those from roots and leaves of plants subjected to drought and salinity treatments, were used for quantitative real-time PCR (qRT-PCR) analysis of PeTIP4;1–1 transcript levels. The qRT-PCR analysis was performed on a qTOWER2.2 real-time PCR system (Analytik Jena, Germany) using the Roche LightCycler®480 SYBR Green 1 kit, as previously described (Sun et al. 2016a). The primer pair PeTIP4;1F and PeTIP4;1R were used in the PCR reaction. All reactions were performed at least in biological triplicate with three technical replicates. The corresponding C t values were normalized using the C t value of NTB (Fan et al. 2013; Sun et al. 2016a). The ∆∆C t method was used to calculate the gene expression level (Livak and Schmittgen 2001).
Transient expression analysis
A fragment of the promoter region upstream of PeTIP4;1–1 was obtained using the gene-specific primer pair TIP4;1p-F1 and TIP4;1p-R1 harboring the XbaI and BamHI restriction sites, respectively (S1). The fragment was ligated into pBI101 vector digested with XbaI and BamHI to generate the construct PeTIP4;1-1pro::GUS for transient expression in tobacco leaves (Yang et al. 2000). The activity of β-glucuronidase (GUS) was examined after 2 days using GUS histochemical assays kit (Real-Times, China) following the manufacturer’s protocol. Images were captured with a digital camera (SONY DSC-HX50).
Transformation and transgenic plant generation
To construct the PeTIP4;1-1-overexpression vector, the coding sequence of PeTIP4;1–1 was amplified using the gene-specific primer pair TIP4;1-F1 and TIP4;1-R1 harboring the XbaI and BamHI restriction sites, respectively (S1). The resulting fragment was subcloned into the corresponding sites of the pPZP-GFP expression vector (Chen et al. 2007) under the control of the CaMV 35S promoter. The construct pPZP-PeTIP4;1–1 was introduced into competent cells of Agrobacterium tumefaciens (strain EHA105) by electroporation. Transformation of A. thaliana ecotype Columbia (Col-0) was conducted using the floral-dip method (Clough and Bent 1998). Transgenic plants were selected on Murashige and Skoog (MS) medium supplemented with kanamycin (50 mg L−1) and validated by RT-PCR using the primer pair TIP4;1-F and TIP4;1-R. The third generation of homozygous plants was used for further analysis. The expression levels of PeTIP4;1–1 in seven independent lines were investigated by RT-PCR using AtUbiquitin as an internal control (Yang et al. 2015a). Phenotypes of transgenic and Col-0 plants were observed and recorded simultaneously.
Stress tolerance analysis of transgenic plants
To test the stress tolerance of the transgenic Arabidopsis plants, seeds of PeTIP4;1-1-overexpressing transgenic lines (L6 and L7) and Col-0 (control) were sown on MS medium. After incubation at 4 °C for 48 h in darkness, the seeds were incubated under a 16 h light/8 h dark photoperiod at 22 °C. After 1 week, the seedlings were transplanted to vertically standing MS plates supplemented with mannitol (0, 50, 100, 150 and 200 mM) or NaCl (0, 50, 100 and 150 mM). The taproot length was measured and photographed under a binocular microscope after 2 weeks. In addition, another part of seedlings was transplanted into a substrate with a mixture of soil and vermiculite (1:1), and grown under normal water management for 2 weeks. Afterwards, stress treatments were applied by withholding water or irrigated with 200 mM NaCl for 8 and 14 days, respectively. To estimate the degree of stress experienced by the Arabidopsis plants, the non-photochemical quenching (NPQ) and maximum quantum yield in Photosystem II (F v/F m) of leaves were monitored using Imaging PAM chlorophyll fluorometer (Walz, Germany). After stress treatment, the plants were harvested and the activities of superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD), malondialdehyde (MDA) and water content were measured. Gene expression in the seedlings was also investigated using qRT-PCR before collection.
Measurements of antioxidant enzyme activities, malondialdehyde and water content
To determine the activities of SOD, CAT and POD, as well as the content of MDA, each 0.5 g sample was ground in liquid nitrogen and homogenized in 5 mL extraction buffer containing 1 × phosphate buffer (pH 7.4). The homogenate was centrifuged at 5000g at 4 °C for 10 min and the supernatant was collected for further analysis. For SOD, an assay kit (A001-1, Jiancheng, China) was used following the manufacturer’s instructions. After incubation in a water bath for 40 min at 37 °C, the samples were mixed well and held at room temperature for 10 min, and then the absorbance at 550 nm was measured with a 1 cm optical path cuvette. For CAT, an assay kit (A007-1, Jiancheng) was used. After treatment for 1 min at 37 °C, the absorbance at 405 nm was measured with a 0.5 cm optical path cuvette. For POD, an assay kit (A084-3, Jiancheng) was used. After treatment for 30 min at 37 °C, the samples were mixed well and centrifuged at room temperature for 10 min. The supernatant was used for absorbance measurement at 420 nm with a 1 cm optical path cuvette. For measurement of MDA concentration, an assay kit (A003-1, Jiancheng) was used. The samples were heated at 95 °C for 40 min, and after cooling were centrifuged at room temperature for 10 min. The supernatant was used for absorbance measurement at 532 nm with a 1 cm optical path cuvette. The activities of SOD, CAT and POD, as well as the MDA concentration were calculated using the formulae (S2), based on the average optical density values of three replicates.
For measurement of the water content of transgenic and Col-0 plants, the above-ground parts of the seedlings were collected and the fresh weight (FW) was determined. After placement on an open Petri dish and drying for 24 h at 80 °C, the dry weight (DW) was recorded. The water content was calculated according to the formula: \(\text{water content }\left( \% \right)\text{ }=\text{ }{\left( \text{FW}\,-\,\text{DW} \right)}/{\text{FW}}\;\times 100.\)
Expression analysis of stress-related genes
The transcript levels of three stress-related genes in the transgenic and Col-0 Arabidopsis plants were estimated by qRT-PCR. The three monitored genes were AtNHX1 (AT5G27150), AtLEA (AT1G02820) and AtP5CS (AT3G55610), and AtUBQ (NM_116771) was used as the reference gene (Chen et al. 2015). The primer sequences used for qRT-PCR are listed in supplementary S1.
Statistical methods
One-way analysis of variance was used for the evaluation of the statistical significance of differences among means for the qRT-PCR data, taproot length, chlorophyll fluorescence parameter values, water content, MDA concentration, and activities of SOD, CAT and POD. Differences were considered significant at the 5% significance level.
Results
Molecular characterization of PeTIP4;1–1
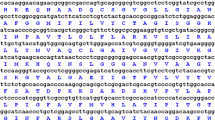
The predicted ORF of PeTIP4;1 (PH01000003G3730) was amplified from moso bamboo leaves by RT-PCR using the primer pair TIP4;1-F and TIP4;1-R. The ORF consisted of 756 bp encoding 251 amino acids with a calculated molecular mass of 25.48 kDa and a theoretical isoelectric point of 6.35. The sequence was completely consistent with that of FP096242.1 in GenBank. However, it shared only 93.25% identity with the putative protein encoded by PeTIP4;1 in the BambooGDB database at the locus PH01000003G3730 (Sun et al. 2016a), indicating that the isolated gene represented a previously uncharacterized TIP4 in moso bamboo. To avoid confusion, the gene was designated PeTIP4;1–1. The genomic sequence corresponding to the ORF of PeTIP4;1–1 was isolated using the primer TIP4;1-F and TIP4;1-R. The amplified fragment was 1777 bp in length and contained three exons and two introns (S3). The introns were fully in compliance with the intron splicing principles of GT-AG (Moore et al. 1993), in which the composition of A + T and G + C was 46.26 and 53.74%, respectively.
The promoter sequence of PeTIP4;1–1 isolated from the genomic DNAs of moso bamboo was 1497 bp. In addition to the essential promoter elements, such as TATA-box and CAAT-box, many stress-responsive elements were detected in the promoter sequence (S4), such as ATCT-motif, Box 4, GA-motif, TC-rich repeats and MBS, which indicated that PeTIP4;1–1 may be involved in response to stresses.
Alignment and phylogenetic affinities of PeTIP4;1–1
Sequence alignment of TIP4;1 homologous proteins was performed. The predicted protein encoded by PeTIP4;1–1 showed the highest homology of 88% with the homologue from rice, and more than 78% with those of other monocots, such as Setaria italica, B. distachyon and Triticum aestivum. This result indicated that TIP4;1 genes were highly conserved among monocots. Similar to other TIPs, PeTIP4;1–1 contains six transmembrane α-helices and two conserved ‘NPA’ motifs (Chaumont et al. 2001) (S5). The neighbor-joining tree showed that TIP4;1s were mainly grouped into two clusters that discriminated monocots and dicots (Fig. 1). PeTIP4;1–1 and other TIP4;1s from monocots were placed in the same cluster separate from dicots, which is consistent with the existing plant phylogeny and classification.
Phylogenetic tree constructed on the basis of PeTIP4;1–1 and TIP4;1 proteins from other plants. The number beside the branches indicated the bootstrap values based on 1000 replicates using MEGA6.0. Their GenBank Accession Numbers are as follows: Phyllostachys edulis ( PeTIP4;1–1, encoded by FP096242.1); Oryza sativa (OsTIP4;1, Q75GA5.1); Brachypodium distachyon (BdTIP4;1, XP_003568717.1); Zea mays (ZmTIP4;1, Q9ATL6.1); Arabidopsis thaliana (AtTIP4;1, NP_180152.1); Populus trichocarpa (PtTIP4;1, XP_002309548.1); Gossypium arboreum (GaTIP4;1, KHG13169.1); Hordeum vulgare (HvTIP4;1, BAI66438.1); Nicotiana tomentosiformis (NtTIP4;1, XP_009612974.1); Setaria italica (SiTIP4;1, XP_014660290.1); Triticum urartu (TuTIP4;1, EMS56264.1)
Expression patterns of PeTIP4;1–1
Transcripts of PeTIP4;1–1 in the roots, culms, mature and immature leaves, leaf sheaths and tender shoots of moso bamboo were analyzed using semi-quantitative RT-PCR. PeTIP4;1–1 was differentially expressed in all six tissues; the highest transcript level was observed in culms, with high levels detected in roots and tender shoots, and low levels detected in leaves and leaf sheaths (Fig. 2). The expression pattern of PeTIP4;1–1 indicated that it might be important for water transport during the period of rapid shoot growth, which requires supply of ample water. Therefore, we also analyzed the changes in PeTIP4;1–1 expression in shoots of different heights. The qRT-PCR analysis indicated that the level of PeTIP4;1–1 transcripts increases continuously with shoot growth. Compared with that of in 2 cm shoots, the transcript accumulation level increased by about ninefold in 30 cm shoots (Fig. 3). This finding further indicated that PeTIP4;1–1 might be involved in water transport during rapid growth of bamboo shoots.
Transcriptional analysis of PeTIP4;1–1 in shoots at different development stages during rapid growth period using qRT-PCR. The transcript of PeTIP4;1–1 demonstrated an increasing trend with the growth of shoots, in which 5, 15 and 30 cm shoots were higher than those of 2 cm shoots. Vertical bars represent standard deviation of three independent biological replicates. Asterisks indicate a significant difference at *p < 0.05, **p < 0.01
It is well known that abiotic stresses are important factors that limit the growth and development of plants, among which drought and salinity seriously affect water transport. To investigate whether PeTIP4;1–1 was responsive to drought and salinity stresses, expression of PeTIP4;1–1 in the roots and leaves of moso bamboo seedlings subjected to drought and salinity stress treatments was analyzed. The transcript level of PeTIP4;1–1 was significantly up-regulated with increasing duration of treatment, especially in roots and leaves of plants under drought stress which increased to more than 10 and 80 times than those of the control after 4 h treatment, respectively (Fig. 4). All of these results implied that PeTIP4;1–1 might be involved in drought and salinity stresses tolerance.
Transcript analysis of PeTIP4;1–1 in roots and leaves under drought and salinity stresses using qRT-PCR. a PeTIP4;1–1 expression in roots treated with drought. b PeTIP4;1–1 expression in leaves treated with drought. c PeTIP4;1–1 expression in roots treated with 400 mM NaCl. d PeTIP4;1–1 expression in leaves treated with 400 mM NaCl. Treatment for 0 h served as the control. Vertical bars represent standard deviation of three independent biological replicates. Asterisks indicate a significant difference at *p < 0.05, **p < 0.01, with respect to corresponding controls
Transient expression analysis of PeTIP4;1-1pro::GUS showed that GUS activity was detected in tobacco leaves, indicating that PeTIP4;1-1pro was able to initiate transcription of the GUS gene in tobacco leaves (S6). The role of PeTIP4;1-1pro in regulation of gene expression needs to be validated.
Stress tolerance of PeTIP4;1-1-overexpressing Arabidopsis
Given that accumulation of PeTIP4;1–1 transcripts was enhanced by drought and salinity stresses, PeTIP4;1-1-overexpressing Arabidopsis plants were used to verify whether PeTIP4;1–1 was involved in stress tolerance. Seven independent lines overexpressing PeTIP4;1–1 were obtained. Expression of PeTIP4;1–1 was detected in all seven lines but not in Col-0 plants (S7). Two T3 transgenic lines (L6 and L7) that highly expressed PeTIP4;1–1 were selected for further analysis.
The chlorophyll fluorescence parameters, such as F v/F m and NPQ, are the important indices to reflect the photoinhibition degree of plants under stresses (Maxwell and Johnson 2000). The F v/F m values and the phenotype of transgenic plants (L6 and L7) were similar to those of Col-0 plants under the non-stress condition. The phenotype of L7 was similar to that of L6 during drought and salinity stress treatments. After 8 days of drought treatment, there was no significant difference in phenotype (Fig. 5a), and the values of F v/F m between the transgenic and Col-0 plants were observed (Fig. 5e), but the NPQ values of transgenic plants were higher than those of Col-0 plants (Fig. 5e). After drought treatment for 14 days, Col-0 plants showed more severe wilting and retardation of growth as compared with the transgenic plants (Fig. 5b), and F v/F m and NPQ values of transgenic plants (L6 and L7) were significantly higher than those of Col-0 plants (F v/F m: p < 0.05; NPQ: p < 0.05) (Fig. 5e, f). Under salinity stress, Col-0 plants wilted and growth was retarded, whereas transgenic plants remained green after 8 days (Fig. 5c). After prolonged salinity treatment for 14 days, Col-0 plants were severely wilted, whereas transgenic plants showed lesser severity of wilting (Fig. 5d). The F v/F m and NPQ values of transgenic plants (L6 and L7) were significantly higher than those of Col-0 plants after salinity stress for 14 days (p < 0.01) (Fig. 5g, h).
Tolerance analysis of transgenic plants under drought and salinity stresses. Col-0, L6 and L7 plants subjected to drought stress by withholding irrigation for 8 days (a) and 14 days (b), and those subjected to salinity stress with 400 mM NaCl for 8 days (c) and 14 days (d). Comparison analyses of F v/F m (e, g) and NPQ (f, h) between L6, L7 and Col-0 plants treated with drought and salinity stresses for 8 and 14 days. Asterisks indicate a significant difference between transgenic plants and Col-0 (*p < 0.05, **p < 0.01)
To further explore the role of PeTIP4;1–1 in the conferral of stress tolerance, changes in root morphology of PeTIP4;1-1-overexpressing Arabidopsis plants were investigated. The PeTIP4;1–1 transgenic lines (L6 and L7) were cultured on vertical plates in MS medium supplemented with different concentrations of NaCl or mannitol. No difference in phenotype was observed between transgenic and Col-0 plants cultured on media lacking mannitol and NaCl. In the presence of mannitol and NaCl, no significant difference in phenotype between comparison of transgenic and Col-0 plants was observed except in taproot length (S8). The average taproot length of transgenic plants (L6 and L7) was longer than that of Col-0 plants under both mannitol and NaCl treatments (Fig. 6). These results indicated that the increase in taproot length may contribute to the enhanced tolerance of PeTIP4;1-1-overexpressing transgenic plants to drought and salinity stresses.
Comparison analysis of taproot length between PeTIP4;1-1-overexpressing transgenic plants (L6, L7) and Col-0 plants under stress treatments. a Taproot length under different concentrations of mannitol. b Taproot length under different concentrations of NaCl. The length of taproots was counted with a binocular microscope directly (n = 18) 14 days after germination. Asterisks indicate a significant difference between transgenic plants and Col-0 (*p < 0.05; **p < 0.01)
Water content and MDA concentration in PeTIP4;1-1-overexpressing Arabidopsis
The water content of the transgenic plants (L6 and L7) was significantly higher than that of Col-0 plants; the difference was much strongly significant under salinity stress (p < 0.01) as compared with that under drought stress (p < 0.05) (Fig. 7a). The MDA concentration showed the opposite trend to water content. The MDA concentration was significantly lower in transgenic plants as compared with that of Col-0 plants under both drought and salinity stresses (p < 0.01) (Fig. 7b). These physiological indices demonstrated that the transgenic plants were more tolerant to drought and salinity stresses than Col-0 plants.
Antioxidant enzyme activities in PeTIP4;1-1-overexpressing Arabidopsis
The decreased MDA concentration implied that transgenic plants may suffer less membrane damage and lipid peroxidation than Col-0 plants under drought and salinity stresses. Antioxidant enzymes play important roles in ROS scavenging. No significant difference in SOD, POD and CAT activities between transgenic plants (L6 and L7) and Col-0 plants before stress treatment was observed. However, the activities of SOD, POD and CAT in transgenic plants were higher than those in Col-0 plants after 14 days of drought and salinity stresses. The difference levels of SOD were at p < 0.05 in L6 after drought and salinity stresses and at p < 0.01 in L7 after salinity stress (Fig. 8a), those of POD were at p < 0.01 in L6 and at p < 0.05 in L7 after drought and salinity stresses (Fig. 8b), those of CAT were all at p < 0.01 in L6 and L7 after drought and salinity stresses (Fig. 8c). These changes in physiological indices indicated that overexpression of PeTIP4;1–1 conferred enhanced drought and salinity tolerance to transgenic Arabidopsis.
Expression profile analysis of stress-related genes in PeTIP4;1-1-overexpressing Arabidopsis
Ion transporters and osmotic regulators are well known two key factors contributing to plant drought and salinity tolerance. Transcript levels of Na+/H+ antiporter (NHX) gene, late embryogenesis abundant (LEA) protein gene and Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene are known as the physiological indices associated with abiotic stresses (Chen et al. 2015). In order to reveal the mechanism through which PeTIP4;1–1 involved in enhancement of stress tolerance in transgenic plants, the transcript levels of three stress-related genes (P5CS, NHX and LEA) were assayed after stress treatments. No significant differences in the transcript levels of AtP5CS, AtNHX and AtLEA were observed between transgenic lines and Col-0 plants under the non-stress condition. However, increased levels of AtP5CS, AtNHX and AtLEA transcripts were observed in transgenic plants (L6 and L7) compared with those in Col-0 plants under drought and salinity stresses. The difference levels of AtP5CS were at p < 0.01 in L6 and at p < 0.05 in L7 after drought and salinity stresses (Fig. 9a); those of AtNHX were at p < 0.05 in L6 after salinity stress and in L7 after drought stress, and at p < 0.01 in L7 after salinity stress (Fig. 9b); those of AtLEA in L6 and L7 were at p < 0.05 after drought stress and at p < 0.01 after salinity stress (Fig. 9c). These results suggested that an indirect transcriptional regulatory mechanism may be responsible for the enhanced stress tolerance of PeTIP4;1-1-overexpressing Arabidopsis plants, acting through elevation of the transcription of stress-related genes such as AtNHX, AtP5CS and AtLEA.
Discussion
Given its rapid growth rate and highly developed rhizome system, bamboo may show a strong capability for water transport (Yang et al. 2015b), which is supported by the phenomenon of shoots rapidly elongating after spring rain. The water content of bamboo shoots ranges from 88.9 to 92.8% (Zhou et al. 2013), indicating that hydraulic conductivity contributes to maintain the morphological changes of the cell during the rapid growth stage. The maximum height of bamboo shoots is determined by root pressure (Cao et al. 2012) and a strong water-potential gradient is a key factor that drives rapid growth (Yuan et al. 2015). Thus, water supply is essential for rapid growth of bamboo. AQPs regulate rapid water transport, and about 70–90% of water transportation is dependent on AQPs (Henzler and Steudie 1995). Recently, numerous of AQP genes have been identified in plants, such as pepper (Yin et al. 2014), barley (Utsugi et al. 2015) and jatropha (Khan et al. 2015). Although 26 AQP genes have been identified in moso bamboo genome, their function remains uncertain (Sun et al. 2016a). In the present study, PeTIP4;1–1 is the first aquaporin gene cloned and characterized from bamboo and one of only a small number of TIP genes studied in monocots. Based on sequence alignment and phylogenetic analysis, PeTIP4;1–1 was shown to contain six transmembrane α-helices and two conserved ‘NPA’ motifs, which indicated it may perform a similar function to that of the TIPs of other plant species.
Previous studies have demonstrated that TIPs are involved in transportation and storage of water, and control of intracellular osmotic pressure by transporting osmotic adjustment substances (Utsugi et al. 2015). The “explosively growing” shoots of moso bamboo achieve height and diameter growth by using non-structural carbohydrates (NSCs) transferred from the attached mature bamboos, which support the structural growth and metabolism of shoots (Song et al. 2016). NSCs are transported in the sap flow, which indicates that water supply is essential for maintaining normal physiological function during the rapid growth of bamboo. Some studies suggest that AQPs play an important role in cell elongation which is a vital component in the rapid growth of bamboo shoots, via the coregulation of auxin and gibberellin (Eisenbarth and Weig 2005; Peng et al. 2013b). We observed that PeTIP4;1–1 was constitutively expressed in all tissues examined, and the expression level increased with the vertical growth of the shoots. Moreover, three meristem expression elements (CAT-box) detected in the PeTIP4;1–1 promoter regions also support its crucial roles in bamboo growth and development. PeTIP4;1–1 transgenic Arabidopsis plants exhibited higher water content after exposure to drought and salinity stresses, indicating that PeTIP4;1–1 may be an important protein involved in bamboo water and NSCs transport .
AQPs can enhance tolerance to drought and high-salinity stress in pepper (Yin et al. 2014), jatropha (Khan et al. 2015) and the halophyte Atriplex canescens (Li et al. 2015). Leaf and root are the two most sensitive organs under stresses. Accumulation of PeTIP4;1–1 transcripts in bamboo roots and leaves was significantly induced by exposure to drought and salinity stress, suggesting that PeTIP4;1–1 may play a role in the response to stresses. Many stress-response elements were identified in the promoter sequence of PeTIP4;1–1, which supported its involvement in response to stresses. Interestingly, using bare-rooted bamboo seedlings exposed to the open air for drought stress treatment, we observed that the roots showed a slower and less significant response than the leaves, which may reflect the ease of water loss by bamboo leaves, into which greater quantities of water must be transported. This was supported by the significantly up-regulated expression level of PeTIP4;1–1 in leaves under drought stress. To further evaluate the biological function of PeTIP4;1–1, we generated transgenic Arabidopsis plants overexpressing PeTIP4;1–1. The transgenic plants exhibited longer taproots, more green leaves with higher F v/F m and NPQ values, and higher water content than Col-0 plants exposure to drought and salinity stress. This result was consistent with previous studies demonstrating that overexpression of TIPs can boost stress resistance (Yin et al. 2014; Khan et al. 2015).
Accumulation of ROS is characteristic of plants under stress (Apel and Hirt 2004), ROS alter aquaporin structure, thereby leading to channel closure through a direct oxidative mechanism (Kourie 1998; Zhou et al. 2011). To avoid damage to cellular components and oxidative destruction of cells, ROS must be detoxified via the ROS-scavenging pathway, which is mainly dependent on activities of antioxidant enzymes, such as SOD, POD and CAT. The activities of SOD, POD and CAT were enhanced in PeTIP4;1–1 transgenic plants under drought and salinity stress, indicating that overexpression of PeTIP4;1–1 effectively improved the antioxidant defense system, which in turn protected the plant against ROS-mediated injury under the applied stresses. This result was consistent with overexpression of SpAQP1 in tobacco, which exhibited enhanced salt tolerance through increased activities of antioxidant enzymes (Chang et al. 2016). As a product of lipid peroxidation, MDA is an indicator of salinity stress damage caused by ROS (Niu et al. 2012). The concentration of MDA was lower in PeTIP4;1–1 transgenic plants than that of Col-0 plants under drought and salinity stress, which indicated that the transgenic plants may experience less lipid peroxidation and membrane injury under salinity or drought conditions (Moore and Roberts 1998).
As one of AQPs, PeTIP4;1–1 might not be directly involved in the regulation of ion transporters and osmotic-related protein genes. To explore indirect effects of PeTIP4;1–1 overexpression, the expression profiles of three stress-related genes (P5CS, NHX and LEA) were analyzed. Our results showed that the transcript levels of AtNHX1, AtLEA and AtP5CS were up-regulated in transgenic Arabidopsis plants overexpressing PeTIP4;1-1 compared with those in Col-0 plants under both drought and salinity stresses. NHX is an important ion transporter. Overexpression of LfNHX1 confers salt and drought tolerance in tobacco plants (Rauf et al. 2014), and enhanced expression of AtNHX1 in transgenic groundnut improves salt and drought tolerance (Asif et al. 2011). In the present study, the increased accumulation of AtNHX1 transcripts implies that overexpression of PeTIP4;1–1 may improve ion transportation, which indirectly confers salt and drought tolerance in transgenic plants. Up-regulation of AtP5CS is mainly responsible for proline accumulation during salinity or drought stress (Zhang et al. 1995; Szabados and Savouré 2010), thus overexpression of PeTIP4;1–1 may have enhanced the proline content in transgenic Arabidopsis. LEAs are an ubiquitous group of polypeptides correlated with acquisition of desiccation tolerance (Pedrosa et al. 2015). Up-regulation of AtLEA transcription in PeTIP4;1-1-overexpressing Arabidopsis plants suggested that there might be an unknown mechanism for PeTIP4;1–1 interaction with LEA.
Taken together, our results demonstrated that stress tolerance conferred by PeTIP4;1–1 might be associated with a variety of pathways involved in regulation of drought and salinity responses in transgenic plants. This study represents the first attempt to elucidate the function of PeTIP4;1–1, but further research is required to better understand the function of PeTIP4;1–1 in bamboo.
Author contribution statement
ZMG and HYS designed the experiments; HYS performed experiments; HYS, LCL, LYF, and ZMG analyzed data; HSZ, YHY, and SNW contributed reagents/materials/analysis tools; ZMG and HYS wrote the paper. All authors read and approved the final manuscript.
Abbreviations
- AQPs:
-
Aquaporins
- AOD:
-
Average optical density
- CAT:
-
Catalase
- F v/F m :
-
Maximum quantum yield in photosystem II
- LEA:
-
Late embryogenesis abundant
- MDA:
-
Malondialdehyde
- MS:
-
Murashige and Skoog
- NHX:
-
Na+/H+ antiporter
- NPQ:
-
Non-photochemical quenching
- NSCs:
-
Non-structural carbohydrates
- ORF:
-
Open reading frame
- P5CS:
-
Δ1-Pyrroline-5-carboxylate synthetase
- POD:
-
Peroxidase
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- ROS:
-
Reactive oxygen species
- RT-PCR:
-
Reverse transcription PCR
- SOD:
-
Superoxide dismutase
- TIP:
-
Tonoplast intrinsic protein
References
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Asif MA, Zafar Y, Iqbal J, Iqbal MM, Rashid U, Ali GM, Arif A, Nazir F (2011) Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Mol Biotechnol 49(3):250–256
Boyer JS (1982) Plant productivity and environment. Science 218(4571):443–448
Cao KF, Yang SJ, Zhang YJ, Brodribb TJ (2012) The maximum height of grasses is determined by roots. Ecol Lett 15(7):666–672
Chang W, Liu X, Zhu J, Fan W, Zhang Z (2016) An aquaporin gene from halophyte Sesuvium portulacastrum, SpAQP1, increases salt tolerance in transgenic tobacco. Plant Cell Rep 35(2):385–395
Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125(3):1206–1215
Chen Z, Yu Y, Liu L, Xia G (2007) Isolation, characterization and functional analysis of a cdc48 homologue from tobacco BY-2 cells. Prog Nat Sci 17(2):156–162
Chen Y, Li L, Zong J, Chen J, Guo H, Guo A, Liu J (2015) Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol Biochem 91:49–55
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Eisenbarth DA, Weig AR (2005) Dynamics of aquaporins and water relations during hypocotyl elongation in Ricinus communis L. seedlings. J Exp Bot 56(417):1831–1842
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8(2):e56573
Forrest KL, Bhave M (2008) The PIP and TIP aquaporins in wheat form a large and diverse family with unique gene structures and functionally important features. Funct Integr Gen 8(2):115–133
Gao ZM, Fan SH, Gao J, Cai CJ, Peng ZH (2006) Extract genomic DNA from Phyllostachys edulis by CTAB-based method. Forest Res 19(6):725–728
Henzler T, Steudle E (1995) Reversible closing of water channels in Chara internodes provides evidence for a composite transport model of the plasma membrane. J Exp Bot 46(2):199–209
Hove RM, Ziemann M, Bhave M (2015) Identification and expression analysis of the barley (Hordeum vulgare L.) aquaporin gene family. PLoS One 10(6):e0128025
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126(4):1358–1369
Khan K, Agarwal P, Shanware A, Sane VA (2015) Heterologous expression of two jatropha aquaporins imparts drought and salt tolerance and improves seed viability in transgenic Arabidopsis thaliana. PLoS One 10(6):e0128866
Kourie JI (1998) Interaction of reactive oxygen species with ion transport mechanisms. Am J Physiol 275(1):1–24
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li J, Yu G, Sun X, Liu Y, Liu J, Zhang X, Jia C, Pan H (2015) AcPIP2, a plasma membrane intrinsic protein from halophyte Atriplex canescens, enhances plant growth rate and abiotic stress tolerance when overexpressed in Arabidopsis thaliana. Plant Cell Rep 34(8):1401–1415
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆Ct method. Methods 25(4):402–408
Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59:595–624
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51(345):659–668
Moore K, Roberts LJ (1998) Measurement of lipid peroxidation. Free Radic Res 6: 659–671
Moore MJ, Query CC, Sharp PA (1993) Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF (eds) The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 303–357
Nguyen MX, Moon S, Jung KH (2013) Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 238(4):669–681
Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ 35:1156–1170
Pedrosa AM, Martins Cde P, Gonçalves LP, Costa MG (2015) Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS One 10(12):e0145785
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, Li Y, Fan D, Guo Y, Li W, Lu Y, Weng Q, Zhou C, Zhang L, Huang T, Zhao Y, Zhu C, Liu X, Yang X, Wang T, Miao K, Zhuang C, Cao X, Tang W, Liu G, Liu Y, Chen J, Liu Z, Yuan L, Liu Z, Huang X, Lu T, Fei B, Ning Z, Han B, Jiang Z (2013a) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45(4):456–461
Peng Z, Zhang C, Zhang Y, Hu T, Mu S, Li X, Gao J (2013b) Transcriptome sequencing and analysis of the fast growing shoots of moso bamboo (Phyllostachys edulis). PLoS One 8(11):e78944
Rauf M, Shahzad K, Ali R, Ahmad M, Habib I, Mansoor S, Berkowitz GA, Saeed NA (2014) Cloning and characterization of Na+/H+ antiporter (LfNHX1) gene from a halophyte grass Leptochloa fusca for drought and salt tolerance. Mol Biol Rep 41(3):1669–1682
Schüssler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP (2008) The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J 56(5):756–767
Song X, Peng C, Zhou G, Gu H, Li Q, Zhang C (2016) Dynamic allocation and transfer of non-structural carbohydrates, a possible mechanism for the explosive growth of Moso bamboo (Phyllostachys heterocycla). Sci Rep 6:25908
Sun H, Li L, Lou Y, Zhao H, Gao Z (2016a) Genome-wide identification and characterization of aquaporin gene family in moso bamboo (Phyllostachys edulis). Mol Biol Rep 43(5):437–450
Sun H, Li L, Lou Y, Zhao H, Yang Y, Gao Z (2016b) Cloning and preliminary functional analysis of PeUGE gene from moso bamboo (Phyllostachys edulis). DNA Cell Biol. doi:10.1089/dna.2016.3389
Szabados L, SavouréA (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Utsugi S, Shibasaka M, Maekawa M, Katsuhara M (2015) Control of the water transport activity of barley HvTIP3;1 specifically expressed in seeds. Plant Cell Physiol 56(9):1831–1840
Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22(6):543–551
Yang L, Lou Y, Peng Z, Zhao H, Sun H, Gao Z (2015a) Molecular characterization and primary functional analysis of PeMPEC, a magnesium-protoporphyrin IX monomethylester cyclase gene of bamboo (Phyllostachys edulis). Plant Cell Rep 34(11):2001–2011
Yang SJ, Zhang YJ, Goldstein G, Sun M, Ma RY, Cao KF (2015b) Determinants of water circulation in a woody bamboo species: afternoon use and night-time recharge of culm water storage. Tree Physiol 35(9):964–974
Yin YX, Wang SB, Xiao HJ, Zhang HX, Zhang Z, Jing H, Zhang YL, Chen RG, Gong ZH (2014) Overexpression of the CaTIP1-1 pepper gene in tobacco enhances resistance to osmotic stresses. Int J Mol Sci 15(11):20101–20116
Yuan JL, Wen GS, Zhang MR, Zhang RM, Cai XF, Zeng YY, Li HJ, Wen X, Zhu H (2015) Water potential with Phyllostachys edulis in its fast-growth periods. J Zhejiang A F Univ 32(5):722–728
Yue X, Zhao X, Fei Y, Zhang X (2012) Correlation of aquaporins and transmembrane solute transporters revealed by genome-wide analysis in developing maize leaf. Comp Funct Genom 2012: 546930
Zhang CS, Lu Q, Verma DP (1995) Removal of feedback inhibition of delta 1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J Biol Chem 270(35):20491–20496
Zhou S, Hu W, Deng X, Ma Z, Chen L, Huang C, Wang C, Wang J, He Y, Yang G, He G (2011) Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS One 7(12):e52439
Zhou WW, He QJ, Ye CQ, Xu RY, Tong XQ, Wang B, Hua XQ, Zhou LX, Lu JS (2013) Comparative analysis of nutrients in bamboo shoot at different seasons. J Zhejiang For Sci Tech 33(4):64–67
Acknowledgements
This work received financial support from the Special Fund for Forest Scientific Research in the Public Welfare from State Forestry Administration of China (No. 201504106) and the Sub-Project of National Science and Technology Support Plan of the Twelfth Five-Year in China (No. 2015BAD04B01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Baochun Li.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, H., Li, L., Lou, Y. et al. The bamboo aquaporin gene PeTIP4;1–1 confers drought and salinity tolerance in transgenic Arabidopsis . Plant Cell Rep 36, 597–609 (2017). https://doi.org/10.1007/s00299-017-2106-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2106-3