Abstract
Key message
PePIP2;7, a leaf-specific aquaporin gene in bamboo, is upregulated under abiotic stresses. Overexpressing PePIP2;7 confers abiotic stresses tolerance in transgenic Arabidopsis plant and yeast.
Abstract
Aquaporins (AQPs) participate in the regulation of water balance in plants. However, the function of AQPs in bamboo remains unclear. Here, PePIP2;7 was identified as a leaf-specific aquaporin gene in moso bamboo based on the expression analysis of transcriptome data and PCR. In situ hybridization further indicated that PePIP2;7 was mainly expressed in mesophyll cells of mature leaves, while in immature leaves it was dominant in blade edge cells followed by mesophyll cells. Interestingly, PePIP2;7 was strongly expressed in the mesophyll cells near bulliform cells of immature leaves, suggesting that PePIP2;7 might function in water transport and contribute to leaf unfolding. The transient expression assay showed that PePIP2;7 was a plasma membrane intrinsic protein. Furthermore, PePIP2;7 was upregulated under abiotic stresses such as high light, drought, and NaCl. Compared with Col-0, transgenic Arabidopsis plants overexpressing PePIP2;7 had better seed germination rate, longer taproot length, higher SOD activity, and lower MDA content under abiotic stresses. Besides, yeasts expressing PePIP2;7 also had higher tolerance to stress compared to the control. Taken together, our results show that PePIP2;7 is leaf-specific and involved in stress response, which provides new insights into aquaporin function in bamboo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aquaporins (AQPs) belong to a highly conserved superfamily of membrane proteins. AQPs, located in cell membranes as well as membrane of almost all cell organelles, play a major role in maintaining water and hydraulic balance by mediating influx and efflux of water between tissues and organs of plants (Maurel et al. 2008). Most of AQPs share a conserved structure with an α-helical bundle forming six transmembrane (TM) helices (TM1 to TM6) connected by five interhelical loops (Loop A to Loop E) (Hove and Bhave 2011). Based on the intracellular location and sequence similarities, AQPs have been categorized into seven subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), GlpF-like intrinsic proteins (GIPs), hybrid intrinsic proteins (HIPs), and uncategorized X intrinsic proteins (XIPs) (Forrest and Bhave 2007). Each subfamily could be further divided into different subgroups, and each subgroup is then again divided into different isoforms according to specialized locations and functions (Afzal et al. 2016). For example, ten members of PePIPs in moso bamboo were divided into PIP1 and PIP2 subgroups that contained three and seven different isoforms, respectively (Sun et al. 2016a).
AQPs are channel proteins that play crucial roles in water transportation across cell membranes. Plants contain a large number of AQPs involved in responding to diverse environmental stresses that disturb cell osmotic balance and nutrient homeostasis (Afzal et al. 2016). Previous research has explored intimate links between AQPs and plant growth (Maurel et al. 2015). The dynamic character of AQPs means that their transcript levels can be adjusted by environmental changes (Afzal et al. 2016; Alexandersson et al. 2005; Moshelion et al. 2015; Shivaraj et al. 2017; Sonah et al. 2017). For example, all PIP genes are downregulated in leaves under drought stress except AtPIP1;4 and AtPIP2;5, which are upregulated (Alexandersson et al. 2005). Two highly homologous genes PIP1;3 and PIP1;1 of Gossypium hirsutum have opposite expression patterns in leaves and roots (Park et al. 2012), indicating that they might play different roles in response to drought stress despite their structural similarity. PtXIP2;1 exhibited multifunction under stresses of drought, salicylic acid, and wounding, which expressed differentially in leaves and stems (Lopez et al. 2012). OsPIP2;7 enhanced the transpiration rate and tolerance to low temperature when overexpressed in rice (Li et al. 2008). Overexpression of either OsPIP1-1, PgTIP1, VfPIP1, TaNIP, MaPIP1, or PeTIP4;1-1 in Arabidopsis could increase the drought tolerance in transgenic plants (Sun et al. 2017). Similar effects were observed in transgenic tobacco with TaAQP7 and TaAQP8 (Hu et al. 2012; Zhou et al. 2012), and in transgenic tomato with SlPIP2;1, SlPIP2;7 and SlPIP2;5 (Li et al. 2016). However, the function of PIP genes in bamboo remains unclear.
Based on the genome of moso bamboo (Phyllostachys edulis) (Peng et al. 2013; Zhao et al. 2018), a total of 26 full-length aquaporin homologs have been identified, ten of which belong to the PIPs. PePIP2;7, a member of PIPs, expressed specifically in leaf blades was studied preliminarily (Sun et al. 2016a). In this study, the leaf-specific expression of PePIP2;7 was further validated using another set of transcriptome data, quantitative real-time PCR (qRT-PCR), and in situ hybridization. Moreover, as a step toward understanding the aquaporin function in bamboo under various environmental stimuli, the expression of PePIP2;7 in bamboo leaves under high light, drought, and high salinity stresses were investigated through qRT-PCR analysis. Besides, the chlorophyll fluorescence parameters of bamboo leaves under high light were measured. The phenotype, physiochemical indicator, and stress-related gene expression profiles in transgenic Arabidopsis plants overexpressing PePIP2;7 under mannitol and salinity stresses were further analyzed. Besides, the function of PePIP2;7 was further validated in yeasts. The results will lead to a better understanding of PePIP2;7 functions in moso bamboo, which will facilitate further strategies for the directional improvement in bamboo environmental adaptability by genetic engineering.
Materials and methods
Plant materials and stress treatments
Moso bamboo seeds were sown in our lab, 1-year-old potted seedlings were cultivated under a long-day condition (16 h light/8 h dark) with the light intensity of 200 μmol m−2 s−1. The growth temperature was 28 °C, and the relative air humidity was about 50% (Sun et al. 2017). Six tissues (Appendix S1), including roots (T1), stems (T2), mature leaves (T3), immature leaves (T4), leaf sheaths (T5), and shoots (T6) of six independent plants, were collected, respectively, for gene tissue-specific analysis.
For high light stress treatment, some seedlings (n = 42) were moved from normal light (200 μmol m−2 s−1) to high light (1200 μmol m−2 s−1) condition provided by cool white fluorescent tubes. The third leaf on the top of seedlings was selected for the sample collecting after 0 h, 0.5 h, 1 h, 2 h, 4 h, 8 h, and 12 h, respectively (each n = 6). Meanwhile, the chlorophyll fluorescence parameters used to analyze the photoprotection ability, such as the maximum quantum yield in Photosystem II (Fv/Fm) and the non-photochemical quenching (NPQ) were measured using a chlorophyll fluorometer Dual-PAM-100 (Walz, Germany) (Sun et al. 2017).
To simulate drought stress, some seedlings (n = 30) were taken out from pots carefully and cleaned with sterilized water, and they were later air dried at the temperature of 25 °C. Samples were collected after 0 h, 1 h, 2 h, 3 h, and 4 h, respectively (each n = 6). Additionally, the salinity stress was performed by incubating the seedling (n = 30) roots into the solution of 400 mM NaCl. Samples were collected after 0 h, 1 h, 2 h, 3 h, and 4 h, respectively (each n = 6). After freezing in liquid nitrogen, all collected samples were stored at − 80 °C for RNA extraction. Each treatment was collected with three independent biological replicates.
Tissue-specific expression profiles of AQP genes
To investigate the expression profiles of AQP genes in moso bamboo, transcriptome data generated from seven tissues, including shoots of 20 cm height (S1), shoots of 50 cm height (S2), rhizomes (S3), roots (S4), leaves (S5), panicle at early flowering stage (S6), and panicle at late flowering stage (S7), were downloaded from the European Molecular Biology Laboratory (EMBL) under the accession ERP001341 (Peng et al. 2013) and used for further analysis. The values of RPKM (Reads Per Kilobase of exon model per Million mapped reads) were uploaded into the online website (http://www.chibi.ubc.ca/matrix2png/) to generate a heat map with Matrix2png.
Nucleic acid isolation and cDNA synthesis for gene cloning
The genomic DNA of moso bamboo leaves was extracted following the CTAB method. Total RNAs were isolated from the collected samples using TRIzol Reagent Solution (Invitrogen, USA), and the cDNAs were synthesized by reverse transcription system (Promega, USA) following previous research (Sun et al. 2017). For each 20.0 μL reaction, 1000 ng of total RNA was used. According to the nucleotide sequence of PePIP2;7 (PH01000332G0830) in the bamboo genome database (http://www.bamboogdb.org) (Zhao et al. 2014), the specific primers of PePIP2;7-F and PePIP2;7-R were designed for the open reading frame (ORF) and its genomic DNA sequence isolation (Appendix S2). The cDNA and genomic DNA were used as templates in PCR respectively, which were performed using the primer pair of PePIP2;7-F and PePIP2;7-R with PrimeSTAR polymerase (Takara, Japan). The PCR amplification products were inserted into the pGEM-T easy vector (Promega, USA) and subsequently verified by sequencing (Sangon Biotech, China).
Sequence analysis
The cDNA and genomic DNA sequences of PePIP2;7 were analyzed by VectorNTI, the gene structure including the number of introns, and the position of each intron was shown using WinPlas (Version 2.7). Sequence homology searches were performed using BLASTN and BLASTX with default parameters (http://www.ncbi.nlm.nih.gov/blast/). Multiple sequence alignment of PePIP2;7 and PIPs from other plants was conducted using ClustalW arithmetic, a phylogenetic tree of the aligned PIP sequences was constructed by MEGA 7.0 (Kumar et al. 2016) using the following parameters: Neighbor-Joining tree method, complete deletion, and bootstrap with 1000 replicates. Besides, the promoter region sequence of PePIP2;7 was further analyzed, in which the putative cis-acting regulatory elements and responsive elements were predicted using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Gene expression analysis of PePIP2;7
For gene tissue-specific expression analysis, the cDNAs were synthesized using the total RNAs isolated from the roots, stems, mature and immature leaves, leaf sheaths, and tender shoots of 1-year-old bamboo seedlings. Semi-quantitative PCR was performed with the primers of semi-PePIP2;7-F and semi-PePIP2;7-R (Appendix S2). The final volume was 20.0 µL including 2.0 µL of 10 × LA Taq Buffer (Mg2+ Plus), 1.6 µL of dNTP Mixture, 1.0 µL of semi-PePIP2;7-F and semi-PePIP2;7-R, 1.0 µL cDNA, 13.2 µL ddH2O and 0.2 µL LA Taq DNA polymerase (Takara, Japan). The PCR program involved in an initial denaturation period at 95 °C for 4 min, followed by 26 cycles of 95 °C for 30 s, 62 °C for 30 s, and then 72 °C for 50 s; after the last cycle, there was a final extension period at 72 °C for 10 min. The cDNA fragment generated from PeActin (Sun et al. 2016b) was amplified as a positive control under the same PCR condition.
A qTOWER2.2 instrument (Analytik Jena, Germany) was used to perform qRT-PCR with Roche LightCycler®480 SYBR Green I Master kit, according to the procedure 95 °C for 5 min; 95 °C for 10 s, 62 °C for 8 s, 40 cycles. The reaction volume was 10.0 μL containing 5.0 μL 2 × SYBR Green I Master, 0.8 μL cDNA, 0.2 μL forward and reverse primer of qPePIP2;7-F and qPePIP2;7-R (5 μM, each) (Appendix S2) and 3.8 μL ddH2O (Sun et al. 2016a). For each condition, the qRT-PCR experiments were performed as biological triplicates. Gene expression was calculated with the 2−△△Ct method (Livak and Schmittgen 2001) using the reference gene of NTB (Fan et al. 2013; Sun et al. 2016a, 2017). Each tissue and treatment in this experiment was collected from six individual bamboo seedlings with three independent biological replicates. All steps followed the minimum information for publication of qRT-PCR experiments (MIQE) rules (Bustin et al. 2009).
In situ RNA hybridization
Both the mature leaves (fully unfolded blades) and immature leaves (unexpanded blades) were collected and fixed in FAA solution (50% ethanol, 5% acetic acid and 3.7% formaldehyde) at 4 °C overnight. The middle part of the blades was used for crosscutting to prepare for cross-sections. A specific probe (5′-GUCUUUCCAAGCUUUGGGCUGGUUGUACAGGACGG-3′) was synthesized for hybridization, which was complementary to the corresponding messenger RNAs of PePIP2;7. The labeled probes were produced using the Cy3 RNA labeling kit (Roche, Switzerland). A blue-fluorescent DNA stain of 4′,6-diamidino-2-phenylindole (DAPI) was used to stain double-stranded DNA (dsDNA) as a control. RNA in situ hybridization, and immunological detection were performed as previously described (Zhao et al. 2009). The images were captured with an IX71 fluorescence inverted microscope (Olympus, Japan).
Transient expression analysis
The coding sequence of PePIP2;7 was amplified using the gene-specific primer pair of PePIP2;7-F1 and PePIP2;7-R1 (Appendix S2), an XbaI site upstream of the start codon and a BamHI site downstream of ORF without stop codon was introduced. The PCR fragments digested with XbaI and BamHI were subcloned into the corresponding sites of the pPZP-GFP expression vector under the control of the CaMV 35S promoter. The construct of PePIP2;7 fused with GFP was verified by sequencing. The validated plasmids harboring PePIP2;7::GFP were used to prepare the particles for bombardment according to the PDS-1000/He system (Bio-Rad, USA). The pPZP vector containing CaMV 35S::GFP was used as a control. The onion inner epidermis was stripped and stretched on Murashige and Skoog (MS) medium in the clean bench. The bombardment was performed using the gene gun at 1100 p.s.i. with DNA-coated gold particles, and the bombarded samples were transferred into a chamber and incubated at 25 °C overnight. The GFP fluorescence was induced by excitation at 488 nm with an argon laser; the images were taken using a laser scanning confocal microscope (Leica TCS SP2, Germany).
Transformation and transgenic plant generation
The coding sequence of PePIP2;7 was subcloned into the multiple cloning sites of pPZP-GFP vector under the control of the CaMV 35S promoter. The vector was introduced into Agrobacterium tumefaciens strain (EHA105) by electroporation and then transformed into Arabidopsis thaliana (Col-0) using the floral-dip method (Clough and Bent 1998). Transgenic lines were selected on 1/2MS medium supplemented with kanamycin (50 mg L−1) and finally validated by RT-PCR. The expression levels of PePIP2;7 in the independent lines were investigated by RT-PCR using AtUbiquitin (NM_116771) as an internal control (Yang et al. 2015). The third generation (T3) of homozygous plants was used for further analysis.
Seed germination assay and phenotype of transgenic Arabidopsis plants under stresses
For the seed germination assays, more than 200 seeds from each of the three selected T3 PePIP2;7 transgenic lines and Col-0 plants were sown on 1/2MS medium and then vernalized for 2 days at 4 °C. The percentage of germinated seeds was counted after 8 days of normal culture (Alavilli et al. 2016; Wang et al. 2019). For root length assay, the Arabidopsis seedlings at the cotyledon stage were transferred to the medium with 300 mM mannitol or 150 mM NaCl, respectively, and the taproot lengths of 30 seedlings of each transgenic lines and Col-0 were measured and analyzed after another 8 days. Phenotypes of transgenic and Col-0 plants were observed and recorded simultaneously. All germination and stress treatment assays were performed with three independent biological replicates.
Measurement of physiochemical indicators and expression analysis of stress-related genes in transgenic Arabidopsis under stresses
One week after seed germination on the medium, the seedlings of three transgenic lines and Col-0 were transplanted into a substrate with a mixture of soil and vermiculite (1:1) and grown under normal management for 2 weeks. Afterward, stress treatments of 400 mM mannitol and 200 mM NaCl were applied, respectively. After 1 week, the plants were harvested for the measurement of physiochemical indicators and expression analysis of stress-related genes. The activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and content of malondialdehyde (MDA) were measured following the previous method (Sun et al. 2017). Each treatment in this experiment was collected from six individual Arabidopsis seedlings with three independent biological replicates. Meanwhile, the transcript levels of PePIP2;7 and three other stress-related genes, AtNHX1 (AT5G27150), AtLEA (AT1G02820), and AtP5CS (AT3G55610), were analyzed using qRT-PCR with AtUBQ (NM_116771) as a reference gene (Chen et al. 2015; Du et al. 2017). Each PCR assay was run in triplicate for three independent biological replicates. The primers for qRT-PCR were shown in Appendix S2. All qRT-PCR steps followed the MIQE rules (Bustin et al. 2009).
Transformation of yeast and stress tolerance analysis
The coding sequence of PePIP2;7 was amplified using PePIP2;7-yeast-F and PePIP2;7-yeast-R primers harboring the BamHI and XbaI restriction sites, respectively (Appendix S2). The PCR fragments were subcloned into the corresponding sites of pYES2 vector (Invitrogen, USA) under the control of the inducible GAL1 promoter. The vectors of pYES2-PePIP2;7 and pYES2 (Invitrogen, USA) were introduced into INVSc1 yeast cells, respectively, using the transformation kit (Fungenome, China), which were selected on SC medium devoid of uracil (Fungenome, China). YPD solid medium (1% yeast extract, 2% peptone, and 2% glucose) was used for further culture of the yeast cells. For stress analysis, transformed yeast cells were propagated in SC-U medium containing 2% galactose. After 12 h, the cell density was adjusted to 1.0 of OD600, which was diluted to a range of different concentrations and inoculated on YPD medium supplemented with PEG6000 (4%) or NaCl (200 mM). The yeast cells were incubated at 30 °C and monitored, and the digital images were captured using a SONY camera DSC-HX50 after 2 days. All tolerance analyses were performed with three independent biological replicates.
Statistical analysis
Data analyses were performed using SPSS Statistics for Windows (Version 17.0. SPSS Inc.). Figures were created with Office Excel for Windows (Microsoft Office 365. Microsoft Inc.). All data are presented as the mean ± standard error of three replications. One-way analysis of variance was used for the evaluation of the statistical significance of differences among means using the software in SPSS. Single and double asterisks indicate significant differences at the levels of p < 0.05 and p < 0.01, respectively.
Results
Expression analysis of AQP genes in different tissues in moso bamboo
The expression analysis of 26 AQP genes in different tissues of moso bamboo was performed using the transcriptome data (Peng et al. 2013). The results demonstrated that they had different expression profiles in seven tissues, among them, 12 genes (PePIP1;2, PePIP1;3, PePIP2;1, PePIP2;2, PePIP2;3, PeTIP2;1, PeTIP2;2, PeNIP1;3, PeNIP2;1, PeNIP3;1, PeSIP1;1, and PeSIP2;1) had high expression levels in roots, ten genes (PePIP1;1, PePIP2;4, PePIP2;6, PeTIP1;1, PeTIP1;2, PeNIP1;1, PeNIP1;2, PeNIP1;4, PeNIP2;2, and PeNIP3;2) showed high expression levels in panicles, and the other three ones (PePIP2;5, PePIP2;7, and PeTIP4;1) presented high expression levels in leaves (Fig. 1). Tissue-specific expression of the AQP genes suggested that they may have specific functional roles in bamboo growth and development. Moreover, the AQP genes with relatively high expression levels were mostly concentrated in the PIP subfamily, and the gene that had the highest expression level in leaves was PePIP2;7, which was 11.9-fold of the second higher level (PePIP2;5).
Tissue-specific expression analysis of PeAQPs based on the transcriptome data generated from different tissues of moso bamboo. S1: 20 cm height of shoots; S2: 50 cm height of shoots; S3: bamboo rhizomes; S4: bamboo roots; S5: bamboo leaves; S6: panicle at early flowering stage; S7: panicle at late flowering stage. Expression levels were visualized using heat maps. Green and red colors indicated gene expression levels from low to high (color figure online)
To further validate the expression profile of PePIP2;7, semi-quantitative PCR and qRT-PCR were performed with cDNA templates from roots (T1), stems (T2), mature leaves (T3), immature leaves (T4), leaf sheaths (T5), and tender shoots (T6) of moso bamboo. The result of semi-quantitative PCR showed that PePIP2;7 was specifically expressed in leaves, and the expression level was relatively higher in immature leaves than that in mature ones (Fig. 2a). The qRT-PCR result further confirmed the expression profile of PePIP2;7 (Fig. 2b). The highly leaf-specific expression of PePIP2;7 indicated that it might have a special important function in bamboo leaves.
Expression profiles of PePIP2;7 in different tissues of moso bamboo. T1–T6 represent roots, stems, mature leaves, immature leaves, leaf sheaths, and tender shoots, respectively. a The result of semi-quantitative PCR using PeActin as a reference gene. b The result of qRT-PCR using PeNTB as a reference gene. Error bars represent standard deviation of biological triplicate with three technical replicates. Asterisks indicate a significant difference at **p < 0.01
Molecular characterization of PePIP2;7 and phylogenetic analysis
Based on the nucleotide sequence of PePIP2;7 (PH01000332G0830) in BambooGDB (Zhao et al. 2014), the ORF and genomic sequences of PePIP2;7 were obtained by end-to-end PCR with primers of PePIP2;7-F and PePIP2;7-R (Appendix S2), which were 864 bp and 1268 bp, respectively, containing four exons and three introns (Appendix S3). The introns were in full compliance with the intron splicing principles of GT-AG, in which the composition of A + T and G + C were 42.06% and 57.94%, respectively. Also, the promoter region upstream of the start codon (1711 bp) was also isolated, in which light-responsive regulatory elements such as 3-AF1-motif, AE-box, G-box, SP1-box GATA-motif, and TCT-motif, as well as defense and stress-responsive elements of TC-rich repeats and MBS, were found (Appendix S4). These elements suggested that PePIP2;7 might be involved in response to stresses such as light, drought, and salinity.
PePIP2;7 encodes a predicted protein of 287 amino acids with a calculated molecular weight of 30.127 kDa and pI of 9.07. The result of BLASTX in GenBank showed that the deduced protein sequence of PePIP2;7 shared the highest identity (88%) with OsPIP2;7 (Q651D5.2) in Oryza sativa. The protein structure of PePIP2;7 was predicted (Appendix S5A) using c2w2eA as a template (Appendix S5B). Tertiary structure analysis of PePIP2;7 demonstrated that it had six transmembrane helices and five interhelical loops, in which the second and fourth loops, as well as N- and C-terminal were inside the cytoplasmic, and the other three loops were outside the cell (Appendix S5C).
Multiple sequence alignment was conducted using PePIP2;7 and other PIP2;7s from different species. The result showed that the members of PIPs were conservative, and characterized by six highly conserved transmembrane helices (TM1-TM6) and two typical NPA motifs in Loop B and Loop E, respectively. However, the N-terminus of PIPs had a large variability (Appendix S6). A phylogenetic tree was constructed using the amino acid sequence of PePIP2;7 and some known PIP2s in A. thaliana, Brachypodium distachyon, Glycine max, Hordeum vulgare, O. sativa, Populus trichocarpa, and Setaria italica. Further analysis indicated that the PIP2 members mainly clustered into two groups belonging to monocotyledons and dicotyledons respectively (Appendix S7). Moreover, PePIP2;7 and OsPIP2;7 were in the same cluster, indicating Ph. edulis and O. sativa were close in the genetic relationship, which was distant with A. thaliana, G. max and P. trichocarpa. Those finding was consistent with the stated phylogeny and classification in the previous study (Peng et al. 2013).
Expression profile of PePIP2;7 in bamboo leaves under stresses
Bamboo leaves have extensive contact with external environment. Therefore, the response-mechanism of leaves under abiotic stress is particularly important. PePIP2;7 was specifically expressed in bamboo leaves, and a large number of light and stress-responsive elements were found in PePIP2;7 promoter region (Appendix S4), indicating that PePIP2;7 might be correlated with leaf photosynthesis and stress response. Under the treatment of high light (1200 μmol m−2 s−1), the expression level of PePIP2;7 was steadily upregulated with the extension of treatment time (Fig. 3a). Meanwhile, the value of Fv/Fm in leaves had an opposite trend of decreasing continuously. The value of NPQ increased significantly at 0.5 h after treatment and then decreased slowly and reached a relatively stable state after 8-h treatment (Fig. 3b). Thermal dissipation associated with NPQ is an essential photo-protective mechanism of photosystem II (Demmig-Adams and Adams 2006). These results indicated that the expression of PePIP2;7 was induced by high light, suggesting that PePIP2;7 might be involved in photoprotection through indirect way, which required further study.
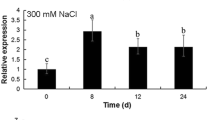
Expression profiles of PePIP2;7 and chlorophyll fluorescence parameters under stress treatment. The expression profiles of PePIP2;7 under 1200 μmol m−2 s−1 (a), drought (c), and NaCl (d) stresses based on qRT-PCR method using PeNTB as the reference gene. b The chlorophyll fluorescence parameters in moso bamboo leaves under 1200 μmol m−2 s−1 condition are also analyzed. Solid line with dots represents the Fv/Fm. Unfilled line with dots represent the NPQ kinetics. Asterisks indicate a significant difference at *p < 0.05 and **p < 0.01
Moreover, the expression profiles of PePIP2;7 in leaves under drought and NaCl treatments were investigated. The results demonstrated that the expression level of PePIP2;7 was upregulated significantly to more than 14-fold of the control during the first 2 h treated by drought stress, and then quickly declined to about 1.9-fold at 4 h of the control (Fig. 3c). Similar expression trend of PePIP2;7 was found in leaves under NaCl treatment, which increased rapidly up to more than 18-fold of the control in the first hour, then declined gradually and reached to about 2.1-fold at 4 h of the control (Fig. 3d). These results indicated that both drought and NaCl stresses could cause the upregulated expression of PePIP2;7 significantly.
Localization of PePIP2;7 transcript in leaves
To investigate the transcript of PePIP2;7 in leaves, the specific 35 nt RNA probe best matching the 5′ of PePIP2;7 mRNA sequence was synthesized. In situ hybridization was performed on sections of immature and mature leaves using the Cy3-labeled PePIP2;7 RNA probe. The result showed that the expression of PePIP2;7 was mainly detected in mesophyll cells, but not in the vascular cells and the epidermal cells of the mature leaves (Fig. 4a–f) and that in immature leaves was mainly in the blade edge cells besides part of mesophyll cells, but not in all the mesophyll cells (Fig. 4g–i). Interestingly, PePIP2;7 was strongly expressed in mesophyll cells near the bulliform cells (Fig. 4j–l), which was further confirmed by the digoxigenin-labeled PePIP2;7 RNA as a probe (Fig. 5). These results suggested that PePIP2;7 was related to water transport in the mesophyll cells of leaves and contributed to leaf expansion.
Fluorescence in situ hybridization of PePIP2;7 in mature leaves (a–f) and immature leaves (g–l) of moso bamboo. a, d, g, j Images with fluorescein using the Cy3-labeled PePIP2;7 RNA probe only; b, e, h, k images with 4′,6-diamidino-2-phenylindole (DAPI) only; c, f, i, l merged images. BC bulliform cell, BEC blade edge cell, EC epidermal cell, MC mesophyll cell, VC vascular cell. Bar = 100 μm
Subcellular localization of PePIP2;7
To examine the subcellular localization of PePIP2;7 in cells, an in silico prediction was performed on its peptide sequence followed by experimental investigation using laser scanning confocal microscopy. Based on the signal peptide presented at N-termini of the protein sequence, PePIP2;7 was predicted to be localized in the plasma membrane. To further assess this prediction, transient expression assay in onion epidermal cells with the construct of CaMV 35S-PePIP2;7::GFP (Appendix S8) and positive control of vector CaMV 35S-GFP were conducted by using gold-particle biolistics. The result showed that the fusion protein of PePIP2;7::GFP was visualized as a membrane protein (Fig. 6a–c), while the free GFP protein was visualized throughout the onion epidermal cell (Fig. 6d–f). These histological observations supported that PePIP2;7 was a plasma membrane intrinsic protein, which was consistent with the previous prediction.
Subcellular localization of PePIP2;7::GFP fusion protein and green fluorescent protein (GFP) in living onion epidermal cells. Onion epidermal cells were transformed with plasmids expressing PePIP2;7::GFP (a–c) or GFP only (d–f). a, d Fluorescence image; b, e bright-field image; c, f conformity for both. Bar = 100 μm
Transgenic Arabidopsis overexpressing PePIP2;7 and germination analysis
To examine the function of PePIP2;7, the overexpression vector of PePIP2;7 was constructed and transformed into A. thaliana (Col-0) for heterologous expression. After screened by kanamycin and verified by PCR, a total of five transgenic lines were obtained (Appendix S9). There are no obvious differences in phenotype between transgenic and Col-0 plants under normal conditions. For further analysis, three T3 generation transgenic lines (L1, L2, and L4) without resistance segregation were selected for validation.
A comparative analysis of germination rates between PePIP2;7 transgenic and Col-0 plants was conducted on 1/2MS medium supplemented with 300 mM mannitol or 150 mM NaCl, respectively (Appendix S10). The result showed that transgenic and Col-0 seeds had similar germination rates (L1: 98.0%; L2: 96.5%; L4: 97.0%; Col-0: 97.0%) on 1/2MS medium after 8 days of cultivation. Under the mannitol mimics drought stress, the germination rate of Col-0 declined to 44.6%, and those of transgenic lines (L1, L2, and L4) declined to 93.6%, 71.6%, and 83.9% respectively. Additionally, the reduced seed germination rates were also found under NaCl stress, which was 49.3%, 91.5%, 85.5%, and 89.5% for Col-0, L1, L2, and L4, respectively (Fig. 7).
The germination rate of transgenic lines and Col-0 seeds all decreased significantly under the treatments of mannitol and NaCl. However, the germination rate of transgenic lines reduced slower than those of Col-0. Besides, PePIP2;7 transgenic plants under mannitol and NaCl stresses showed longer taproot length than those of Col-0 (Appendix S11). These results indicated that overexpressing PePIP2;7 could improve the tolerance of transgenic Arabidopsis in response to drought and salt stresses.
Expression analysis of stress-related genes in PePIP2;7 transgenic Arabidopsis plants under stresses
Previous studies had proved that AtP5CS, AtLEA, and AtNHX were stress-responsive genes (Chen et al. 2015). P5CS is important in the process of proline accumulation when facing salinity or drought stress (Szabados and Savoure 2010; Zhang et al. 1995). As a ubiquitous polypeptide, LEA is always correlated with the acquisition of desiccation tolerance (Pedrosa et al. 2015). AtNHX1 is an important ion transporter involved in regulating the Na+/H+ transporting activity (Yamaguchi et al. 2003). Although the transgenic and Col-0 plants had similar phenotype under normal conditions, the rosette leaves of PePIP2;7 transgenic plants were greener than those of Col-0 plants under mannitol and NaCl treatments after 7 days (Appendix S12), which suggested that overexpressing PePIP2;7 conferred the tolerance of transgenic plants in response to drought and salt stresses. Therefore, we investigated the expression changes of three stress-response genes in transgenic plants by qRT-PCR.
The results show that the expression level of AtLEA in three transgenic lines (L1, L2, and L4) and only that of AtP5CS in L2 were higher than those in Col-0, while those of AtP5CS and AtNHX in L1 and L4 were lower than those in Col-0, and that of AtNHX in L2 was similar to that in Col-0 under mannitol treatment. However, the increased transcripts of AtP5CS, AtLEA, and AtNHX were all observed in three transgenic lines compared with those in Col-0 under NaCl treatment (Appendix S13). These results suggested that an indirect transcriptional regulatory mechanism may be responsible for the enhanced stress tolerance of PePIP2;7-overexpressing Arabidopsis plants in response to drought and salt stresses, which still need to be further validated.
Antioxidant enzyme activities and MDA content analysis
The measurement of SOD, POD, and CAT activities, as well as MDA content in transgenic and Col-0 plants, were performed. The results showed that there were some differences between transgenic and Col-0 plants even under normal conditions. For example, the values of SOD in L2 and L4 were significantly higher than those in L1 and Col-0 plants (Appendix S14). The value of POD in L1, those of CAT in three lines, and the content of MDA in L2 were similar to those in Col-0 plants, while the value of POD in L4, and that MDA in L4 was significantly higher, except that the value of POD in L2 and the content of MDA in L1 were significantly lower than those in Col-0 plants (Appendix S15). These results suggested that overexpressing PePIP2;7 could affect the enzyme activities of SOD and POD and the content of MDA in transgenic plants under normal conditions.
Furthermore, the values of SOD in three lines under both mannitol and NaCl treatments (Appendix S14) and those of POD in L1 and L4 under NaCl treatment were significantly higher than those in Col-0 plants (Appendix S15). The value of POD in L1 under mannitol treatment and that of POD in L2 under NaCl were similar to those in Col-0 plants. All the values of CAT in transgenic lines were similar to those in Col-0 plants under mannitol and NaCl treatments except that in L4 was significantly lower under NaCl treatment. The content of MDA in L1, L2 and L4 under mannitol treatment and that in L4 under NaCl treatment were lower than those in Col-0 plants, while those in L1 and L2 under NaCl treatment were higher than that in Col-0 plants (Appendix S15). The results of higher SOD activity and lower MDA content indicated that overexpressing PePIP2;7 could improve the tolerance of transgenic Arabidopsis to drought and salt stresses.
Although not all lines had higher antioxidant enzyme activities and lower MDA content under drought and salt stresses, these results still showed that overexpression of PePIP2;7 could confer abiotic stress tolerance in transgenic plants. This was supported by the phenotypes of transgenic plants with greener rosette leaves than Col-0 under drought and salt stresses (Appendix S12), suggesting that the expression of PePIP2;7 might play an important role in hydraulic conductivity balance of leaves.
Transgenic yeast expressing PePIP2;7 under drought and salt stresses
A yeast expression system was used to validate the function of PePIP2;7 in maintaining water and hydraulic conductivity balance. Transgenic yeast cells harboring pYES2-PePIP2;7 or pYES2 vector and the original INVSc1 cells could grow well on the YPD medium after 10−7 dilution (Fig. 8a). The cells containing pYES2 vector grew only until 10−5 dilution, while those harboring pYES2-PePIP2;7 grow well until 10−7 dilution on the YPD medium supplemented with 4% PEG6000 (Fig. 8b). A similar result was found on the YPD medium supplemented with 200 mM NaCl (Fig. 8c). However, none of the INVSc1 yeast cells were able to grow on the YPD medium with 4% PEG6000 and 200 mM NaCl (Fig. 8b, c). These results suggested that PePIP2;7 can increase the drought and salt tolerance by maintaining water and hydraulic conductivity balance of yeast cells.
Effect of PePIP2;7 expression in yeast under drought and salt stresses. Original yeast cells only (Yeast only), yeast cells with pYES2 vector only (Vector only), and yeast cells harboring PePIP2;7 expressing construct (PePIP2;7) were subjected to 4% PEG6000 and 200 mM NaCl. Cell density adjusted to OD600 at 1.0 and serial dilutions were made at each step. Two microliters of each dilution were inoculated on a YPD medium, b YPD medium supplemented with 4% PEG6000, and c YPD medium supplemented with 200 mM NaCl, respectively. Photographs were taken after 48 h of incubation at 30 °C
Discussion
With the development of in silico methods and sequenced genomes, more than 1200 AQP genes have been identified in 31 plant species (Deshmukh et al. 2016). A total of 63 AQP members were identified in moso bamboo genome, in which 37 members were added (Zhao et al. 2018). However, only PeTIP4;1-1 was cloned and characterized up to now (Sun et al. 2017). In the present study, a leaf-specific AQP gene PePIP2;7 was cloned and characterized based on the expression analysis of transcriptome data. PePIP2;7 shared high sequence similarity with PIP2 sequences from O. sativa, H. vulgare, S. italica, and B. distachyon. PIPs are the most abundant aquaporins in the plasma membrane of plant cells (Boursiac et al. 2008). In silico prediction showed that PePIP2;7 is a plasma membrane intrinsic protein located in the cell membrane. The transient expression assay in onion epidermal cells showed the same results (Fig. 6), which further confirmed PePIP2;7 is a plasma membrane intrinsic protein belonging to the family of PIPs.
Research on AQP delineates ever-expanding fields in plant integrative biology thereby establishing their crucial role in plants (Maurel et al. 2015). It has been reported that varying light environment adjusts water flow at the cellular level; water flow is maximized at a certain light intensity and then will be reduced again by the increased light intensity (Kim and Steudle 2009). Many light-responsive elements were found in the promoter region of PePIP2;7 suggested that it might respond to light stimulus, which was supported by the continuously upregulated expression of PePIP2;7 under high light (Fig. 3a). This result was consistent with the transcript levels of JrPIP2,1 and JrPIP2,2 of walnut under high illumination, which were positively correlated with the leaf hydraulic conductance (Cochard et al. 2007). Although no direct evidence can link PePIP2;7 with high light stress, it might be linked with other proteins that participated in the process of oxidative gating (Kim and Steudle 2009). However, details of PePIP2;7 function under high light still need further investigation.
PIPs are generally localized to the plasma membrane and also be found to co-localize to the chloroplast envelope (Beebo et al. 2013; Uehlein et al. 2008). The multi-location characteristics of PIPs may endow them with multiple functions such as participating in water regulation and photosynthesis (Groszmann et al. 2017). PePIP2;7 was a leaf-specific PIP gene, which was validated by semi-quantitative PCR, qRT-PCR (Fig. 2), and in situ hybridization (Fig. 4). It was interesting that PePIP2;7 was strongly expressed in mesophyll cells nearby the bulliform cells of immature leaves. Bulliform cells that occur in groups between vascular bundles on the adaxial epidermis of the lamina are involved in the modulation of blade curling (Itoh et al. 2005). Sufficient water is needed for bulliform cell expansion and leaf unfolding. The expression of PePIP2;7 in immature leaves suggested that it was closely related to the transport of water to bulliform cells during blade unfolding by exerting multiple indirect effects on stomatal apertures, such as ABA and pathogen pathway (Maurel et al. 2016; Rodrigues et al. 2017). We will carry out this further in next study. These results indicated that PePIP2;7 may play vital roles in leaf physiology for maintaining bamboo growth and development.
As the most sensitive tissue in bamboo, the leaf is easy to sense various environmental stimuli such as drought and salinity. PePIP2;7 was upregulated with different trends of initial increase followed by decrease under both drought and NaCl stresses. The same results reported that PIP genes presented different expression profiles under diverse stresses. For example, CsPIP1;1 and CsPIP2;4 were upregulated in citrus leaves under drought stress (Martins et al. 2015), and OsPIP2;5 was found to be significantly upregulated in roots of rice in exposure to chilling stress (Ahamed et al. 2012). The accumulation of mRNA of PePIP2;7 in a short time under drought and NaCl stresses suggested that PePIP2;7 protein was needed in resistance of those stresses. We also found that the expression peak under NaCl stress comes earlier than drought stress. It might be explained that the amount of PePIP2;7 channel is different when facing drought and NaCl stresses (Pou et al. 2016). Therefore, we speculate that PePIP2;7 might have a non-water channel function or interplay with other endogenous aquaporins and structural proteins in response to stresses.
PePIP2;7 expression in yeast certainly resulted in increased stress tolerance under high drought and salt conditions (Fig. 8). However, it was complex in PePIP2;7 transgenic Arabidopsis. Only the increased transcript of AtLEA was observed in PePIP2;7 transgenic lines, but those of AtP5CS and AtNHX were uncertain under mannitol stress. However, the transcriptional levels of AtP5CS, AtLEA, and AtNHX1 in PePIP2;7 transgenic lines were all higher than those in Col-0 under NaCl treatment (Appendix S13). The activity of SOD increased significantly in PePIP2;7 overexpressing transgenic lines (Appendix S14). No significant activity difference of CAT was observed between transgenic lines and Col-0, while that of POD and the content of MDA in transgenic lines were uncertainty of increase or decrease compared with Col-0 under mannitol and NaCl stresses (Appendix S15). These results suggested that overexpression of PePIP2;7 could confer drought and salt tolerance in transgenic plants and yeast. Previous research found that some transcription factors cooperate with aquaporins to confer drought and salt tolerance (Rae et al. 2011). Whatever, the tolerance conferred by PePIP2;7 may also be a complex process involving transcription factors. Therefore, further studies are required to reveal these mechanisms.
PIPs have been confirmed to function as the transporters of water, glycerol, H2O2, CO2, and urea, and be involved in abiotic stress response (Afzal et al. 2016). Several pieces of evidence support the idea that specific aquaporin in leaves may have specific functional roles in response to environmental stresses (Heckwolf et al. 2011). Overall, our findings demonstrated that PePIP2;7 was a functional gene specifically expressed in moso bamboo leaves, and it might play an important role in response to stresses. Besides, plant–water balance under adverse conditions is a complex regulatory network (Matsumoto et al. 2009). The highly complex and integrated roles of PIPs in response to different environmental stimuli and involved in other regulating processes are still unclear. As we all know, the response of plants to stress is very complicated, and their regulation is a complex network, involving transcription factors, structural genes and small RNAs. Co-expression of NtPIP1;1 with NtPIP2;1 significantly enhanced water transport activity in oocytes (Mahdieh et al. 2008). RhPIP2;1 interacts with a membrane-tethered MYB protein and serves as a key player in orchestrating the trade-off between growth and stress survival in Rosa (Zhang et al. 2019). A calcium-dependent protein kinase (OsCPK17) compromises cold tolerance of rice by phosphorylating OsPIP2;1/OsPIP2;6 (Almadanim et al. 2017). After all, bamboo molecular biology research is still in its infancy, and enormous experimental evidence is needed to elucidate the precise roles of PIP genes in bamboo.
Abbreviations
- AQP:
-
Aquaporin
- bp:
-
Base pair
- CAT:
-
Catalase
- cDNA:
-
DNA complementary to RNA
- CDS:
-
Coding sequence
- dNTP:
-
Deoxyribonucleoside triphosphate
- F v/F m :
-
Maximum quantum yield in photosystem II
- GIP:
-
GlpF-like intrinsic protein
- HIP:
-
Hybrid intrinsic protein
- kDa:
-
Kilodalton
- MDA:
-
Malonaldehyde
- MW:
-
Molecular weight
- NPQ:
-
Non-photochemical quenching
- PIP:
-
Plasma membrane intrinsic protein
- POD:
-
Peroxidase
- NIP:
-
Nodulin 26-like intrinsic protein
- ORF:
-
Open reading frame
- pI:
-
Isoelectric point
- qRT-PCR:
-
Quantitative real-time PCR
- SIP:
-
Small basic intrinsic protein
- SOD:
-
Superoxide dismutase
- TIP:
-
Tonoplast intrinsic protein
- TM:
-
Transmembrane
- XIP:
-
Uncategorized X intrinsic protein
References
Afzal Z, Howton TC, Sun Y, Mukhtar MS (2016) The roles of aquaporins in plant stress responses. J Dev Biol. https://doi.org/10.3390/jdb4010009
Ahamed A, Murai-Hatano M, Ishikawa-Sakurai J, Hayashi H, Kawamura Y, Uemura M (2012) Cold stress-induced acclimation in rice is mediated by root-specific aquaporins. Plant Cell Physiol 53:1445–1456. https://doi.org/10.1093/pcp/pcs089
Alavilli H, Awasthi JP, Rout GR, Sahoo L, Lee BH, Panda SK (2016) Overexpression of a barley aquaporin gene, HvPIP2;5 confers salt and osmotic stress tolerance in yeast and plants. Front Plant Sci 7:1566. https://doi.org/10.3389/fpls.2016.01566
Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484. https://doi.org/10.1007/s11103-005-0352-1
Almadanim MC, Alexandre BM, Rosa MTG, Sapeta H, Leitao AE, Ramalho JC, Lam TT, Negrao S, Abreu IA, Oliveira MM (2017) Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ 40:1197–1213. https://doi.org/10.1111/pce.12916
Beebo A, Mathai JC, Schoefs B, Spetea C (2013) Assessment of the requirement for aquaporins in the thylakoid membrane of plant chloroplasts to sustain photosynthetic water oxidation. FEBS Lett 587:2083–2089. https://doi.org/10.1016/j.febslet.2013.05.046
Boursiac Y, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Maurel C (2008) Stimulus-induced downregulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J 56:207–218. https://doi.org/10.1111/j.1365-313X.2008.03594.x
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Chen Y, Li L, Zong J, Chen J, Guo H, Guo A, Liu J (2015) Heterologous expression of the halophyte Zoysia matrella H(+)-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol Biochem 91:49–55. https://doi.org/10.1016/j.plaphy.2015.04.004
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cochard H, Venisse JS, Barigah TS, Brunel N, Herbette S, Guilliot A, Tyree MT, Sakr S (2007) Putative role of aquaporins in variable hydraulic conductance of leaves in response to light. Plant Physiol 143:122–133. https://doi.org/10.1104/pp.106.090092
Demmig-Adams B, Adams WW 3rd (2006) Photoprotection in an ecological context: the remarkable complexity of thermal energy dissipation. New Phytol 172:11–21. https://doi.org/10.1111/j.1469-8137.2006.01835.x
Deshmukh RK, Sonah H, Belanger RR (2016) Plant aquaporins: genome-wide identification, transcriptomics, proteomics, and advanced analytical tools. Front Plant Sci 7:1896. https://doi.org/10.3389/fpls.2016.01896
Du X, Jin Z, Liu D, Yang G, Pei Y (2017) Hydrogen sulfide alleviates the cold stress through MPK4 in Arabidopsis thaliana. Plant Physiol Biochem 120:112–119. https://doi.org/10.1016/j.plaphy.2017.09.028
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 8:e56573. https://doi.org/10.1371/journal.pone.0056573
Forrest KL, Bhave M (2007) Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct Integr Genomics 7:263–289. https://doi.org/10.1007/s10142-007-0049-4
Groszmann M, Osborn HL, Evans JR (2017) Carbon dioxide and water transport through plant aquaporins. Plant Cell Environ 40:938–961. https://doi.org/10.1111/pce.12844
Heckwolf M, Pater D, Hanson DT, Kaldenhoff R (2011) The Arabidopsis thaliana aquaporin AtPIP1;2 is a physiologically relevant CO2 transport facilitator. Plant J 67:795–804. https://doi.org/10.1111/j.1365-313X.2011.04634.x
Hove RM, Bhave M (2011) Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol 75:413–430. https://doi.org/10.1007/s11103-011-9737-5
Hu W, Yuan Q, Wang Y, Cai R, Deng X, Wang J, Zhou S, Chen M, Chen L, Huang C, Ma Z, Yang G, He G (2012) Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol 53:2127–2141. https://doi.org/10.1093/pcp/pcs154
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46:23–47. https://doi.org/10.1093/pcp/pci501
Kim YX, Steudle E (2009) Gating of aquaporins by light and reactive oxygen species in leaf parenchyma cells of the midrib of Zea mays. J Exp Bot 60:547–556. https://doi.org/10.1093/jxb/ern299
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Li GW, Zhang MH, Cai WM, Sun WN, Su WA (2008) Characterization of OsPIP2;7, a water channel protein in rice. Plant Cell Physiol 49:1851–1858. https://doi.org/10.1093/pcp/pcn166
Li R, Wang J, Li S, Zhang L, Qi C, Weeda S, Zhao B, Ren S, Guo YD (2016) Plasma membrane intrinsic proteins SlPIP2;1, SlPIP2;7 and SlPIP2;5 conferring enhanced drought stress tolerance in tomato. Sci Rep 6:31814. https://doi.org/10.1038/srep31814
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lopez D, Bronner G, Brunel N, Auguin D, Bourgerie S, Brignolas F, Carpin S, Tournaire-Roux C, Maurel C, Fumanal B, Martin F, Sakr S, Label P, Julien JL, Gousset-Dupont A, Venisse JS (2012) Insights into Populus XIP aquaporins: evolutionary expansion, protein functionality, and environmental regulation. J Exp Bot 63:2217–2230. https://doi.org/10.1093/jxb/err404
Mahdieh M, Mostajeran A, Horie T, Katsuhara M (2008) Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant Cell Physiol 49:801–813. https://doi.org/10.1093/pcp/pcn054
Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaitre B, Jaillais Y, Chory J, Geldner N, Vert G (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6:6151. https://doi.org/10.1038/ncomms7151
Matsumoto T, Lian HL, Su WA, Tanaka D, Liu C, Iwasaki I, Kitagawa Y (2009) Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant Cell Physiol 50:216–229. https://doi.org/10.1093/pcp/pcn190
Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59:595–624. https://doi.org/10.1146/annurev.arplant.59.032607.092734
Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L (2015) Aquaporins in plants. Physiol Rev 95:1321–1358. https://doi.org/10.1152/physrev.00008.2015
Maurel C, Verdoucq L, Rodrigues O (2016) Aquaporins and plant transpiration. Plant Cell Environ 39:2580–2587. https://doi.org/10.1111/pce.12814
Moshelion M, Halperin O, Wallach R, Oren R, Way DA (2015) Role of aquaporins in determining transpiration and photosynthesis in water-stressed plants: crop water-use efficiency, growth and yield. Plant Cell Environ 38:1785–1793. https://doi.org/10.1111/pce.12410
Park W, Scheffler BE, Bauer PJ, Campbell BT (2012) Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.). BMC Plant Biol 12:90. https://doi.org/10.1186/1471-2229-12-90
Pedrosa AM, Martins CD, Goncalves LP, Costa MG (2015) Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 10:e0145785. https://doi.org/10.1371/journal.pone.0145785
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, Li Y, Fan D, Guo Y, Li W, Lu Y, Weng Q, Zhou C, Zhang L, Huang T, Zhao Y, Zhu C, Liu X, Yang X, Wang T, Miao K, Zhuang C, Cao X, Tang W, Liu G, Liu Y, Chen J, Liu Z, Yuan L, Liu Z, Huang X, Lu T, Fei B, Ning Z, Han B, Jiang Z (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45:456–461. https://doi.org/10.1038/ng.2569 (461.e451–452)
Pou A, Jeanguenin L, Milhiet T, Batoko H, Chaumont F, Hachez C (2016) Salinity-mediated transcriptional and post-translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Mol Biol 92:731–744. https://doi.org/10.1007/s11103-016-0542-z
Rae L, Lao NT, Kavanagh TA (2011) Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234:429–444. https://doi.org/10.1007/s00425-011-1414-z
Rodrigues O, Reshetnyak G, Grondin A, Saijo Y, Leonhardt N, Maurel C, Verdoucq L (2017) Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc Natl Acad Sci USA 114:9200–9205. https://doi.org/10.1073/pnas.1704754114
Shivaraj SM, Deshmukh RK, Rai R, Belanger R, Agrawal PK, Dash PK (2017) Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci Rep 7:46137. https://doi.org/10.1038/srep46137
Sonah H, Deshmukh RK, Labbe C, Belanger RR (2017) Analysis of aquaporins in Brassicaceae species reveals high-level of conservation and dynamic role against biotic and abiotic stress in canola. Sci Rep 7:2771. https://doi.org/10.1038/s41598-017-02877-9
Sun H, Li L, Lou Y, Zhao H, Gao Z (2016a) Genome-wide identification and characterization of aquaporin gene family in moso bamboo (Phyllostachys edulis). Mol Biol Rep 43:437–450. https://doi.org/10.1007/s11033-016-3973-3
Sun H, Li L, Lou Y, Zhao H, Yang Y, Gao Z (2016b) Cloning and preliminary functional analysis of PeUGE gene from moso bamboo (Phyllostachys edulis). DNA Cell Biol 35:706–714. https://doi.org/10.1089/dna.2016.3389
Sun H, Li L, Lou Y, Zhao H, Yang Y, Wang S, Gao Z (2017) The bamboo aquaporin gene PeTIP4;1–1 confers drought and salinity tolerance in transgenic Arabidopsis. Plant Cell Rep 36:597–609. https://doi.org/10.1007/s00299-017-2106-3
Szabados L, Savoure A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97. https://doi.org/10.1016/j.tplants.2009.11.009
Uehlein N, Otto B, Hanson DT, Fischer M, McDowell N, Kaldenhoff R (2008) Function of Nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell 20:648–657. https://doi.org/10.1105/tpc.107.054023
Wang X, Gao F, Bing J, Sun W, Feng X, Ma X, Zhou Y, Zhang G (2019) Overexpression of the jojoba aquaporin gene, ScPIP1, Enhances drought and salt tolerance in transgenic Arabidopsis. Int J Mol Sci. https://doi.org/10.3390/ijms20010153
Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100:12510–12515. https://doi.org/10.1073/pnas.2034966100
Yang C, Zhou Y, Fan J, Fu Y, Shen L, Yao Y, Li R, Fu S, Duan R, Hu X, Guo J (2015) SpBADH of the halophyte Sesuvium portulacastrum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis. Plant Physiol Biochem 96:377–387. https://doi.org/10.1016/j.plaphy.2015.08.010
Zhang CS, Lu Q, Verma DP (1995) Removal of feedback inhibition of delta 1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J Biol Chem 270:20491–20496
Zhang L, Han Q, Xiong J, Zheng T, Han J, Zhou H, Lin H, Yin Y, Zhang D (2019) Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana. Plant Cell Physiol 60:2282–2292. https://doi.org/10.1093/pcp/pcz125
Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21:736–748. https://doi.org/10.1105/tpc.108.061655
Zhao H, Peng Z, Fei B, Li L, Hu T, Gao Z, Jiang Z (2014) BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database Oxford 2014:bau006. https://doi.org/10.1093/database/bau006
Zhao H, Gao Z, Wang L, Wang J, Wang S, Fei B, Chen C, Shi C, Liu X, Zhang H, Lou Y, Chen L, Sun H, Zhou X, Wang S, Zhang C, Xu H, Li L, Yang Y, Wei Y, Yang W, Gao Q, Yang H, Zhao S, Jiang Z (2018) Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience. https://doi.org/10.1093/gigascience/giy115
Zhou S, Hu W, Deng X, Ma Z, Chen L, Huang C, Wang C, Wang J, He Y, Yang G, He G (2012) Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS ONE 7:e52439. https://doi.org/10.1371/journal.pone.0052439
Acknowledgements
The work was supported by the National Science Foundation of China (Grant nos. 31971736; 31930075) and the Special Funds for Fundamental Scientific Research on Professional Work Supported by the International Center for Bamboo and Rattan (No. 1632018005).
Author information
Authors and Affiliations
Contributions
ZMG and HYS designed the experiments; HYS and SNW performed experiments; HYS, SNW, LYF, and ZMG analyzed data; YFL, CLZ, HSZ, YL, and XPL contributed reagents/materials/analysis tools; HYS and ZMG wrote the paper. All authors read and approved the final manuscript. HS and SW contributed equally to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Leandro Peña.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, H., Wang, S., Lou, Y. et al. A bamboo leaf-specific aquaporin gene PePIP2;7 is involved in abiotic stress response. Plant Cell Rep 40, 1101–1114 (2021). https://doi.org/10.1007/s00299-021-02673-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02673-w