Abstract
Key message
CYP716A179, a cytochrome P450 monooxygenase expressed predominantly in tissue-cultured stolons of licorice ( Glycyrrhiza uralensis ), functions as a triterpene C-28 oxidase in the biosynthesis of oleanolic acid and betulinic acid.
Abstract
Cytochrome P450 monooxygenases (P450s) play key roles in the structural diversification of plant triterpenoids. Among these, the CYP716A subfamily, which functions mainly as a triterpene C-28 oxidase, is common in plants. Licorice (Glycyrrhiza uralensis) produces bioactive triterpenoids, such as glycyrrhizin and soyasaponins, and relevant P450s (CYP88D6, CYP72A154, and CYP93E3) have been identified; however, no CYP716A subfamily P450 has been isolated. Here, we identify CYP716A179, which functions as a triterpene C-28 oxidase, by RNA sequencing analysis of tissue-cultured stolons of G. uralensis. Heterologous expression of CYP716A179 in engineered yeast strains confirmed the production of oleanolic acid, ursolic acid, and betulinic acid from β-amyrin, α-amyrin, and lupeol, respectively. The transcript level of CYP716A179 was about 500 times higher in tissue-cultured stolons than in intact roots. Oleanolic acid and betulinic acid were consistently detected only in tissue-cultured stolons. The discovery of CYP716A179 helps increase our understanding of the mechanisms of tissue-type-dependent triterpenoid metabolism in licorice and provides an additional target gene for pathway engineering to increase the production of glycyrrhizin in licorice tissue cultures by disrupting competing pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glycyrrhiza uralensis is a Glycyrrhiza (licorice) species belonging to the Fabaceae. Roots and stolons of this species, G. glabla, and G. inflata, are widely used as medicines and natural sweeteners because they contain substantial amounts of glycyrrhizin, a sweet-tasting oleanane-type pentacyclic triterpenoid saponin with various pharmacological activities (Hayashi and Sudo 2009). In addition to glycyrrhizin, licorice produces other pentacyclic triterpenoids, including soyasaponins, betulinic acid, and oleanolic acid. Several studies have examined the distribution of these triterpenoids in tissue cultures and in different organs in licorice (Hayashi et al. 1988, 1993; Kojoma et al. 2010). Glycyrrhizin is accumulated mainly in thickened roots and was not detected in leaves or stems of G. glabra plants (Hayashi et al. 1993). Soyasaponins and betulinic acid are accumulated in callus and suspension cultures of G. glabla instead of glycyrrhizin (Hayashi et al. 1988). Similarly, tissue-cultured stolons of G. uralensis accumulate higher levels of oleanolic acid and betulinic acid, while glycyrrhizin production is much lower than in intact stolons (Kojoma et al. 2010). Although several attempts have been made to produce glycyrrhizin in licorice tissue cultures, the production of glycyrrhizin in these cultures remains challenging (Kojoma et al. 2010).
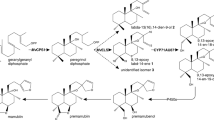
Metabolic engineering is a powerful tool for altering metabolic flow in an organism to produce desired metabolites by manipulating biosynthetic genes or regulatory genes (Oksman-Caldentey and Inzé 2004). Generally, triterpenoids are derived from 2,3-oxidosqualene, a linear isoprenoid consisting of 30 carbon atoms, then serial cyclization reactions catalyzed by oxidosqualene cyclases (OSCs) generate more than 100 different triterpene skeletons (Thimmappa et al. 2014). These triterpene skeletons are further modified through oxidation, glycosylation, and/or acetylation reactions to produce structural diversity in triterpenoids (Thimmappa et al. 2014). Cytochrome P450 monooxygenases (P450s) constitute a large gene family in plant genomes and are involved in plant metabolism and the biosynthesis of specialized metabolites (Nelson and Werck-Reichhart 2011). Recent studies have revealed that P450s play a key role in the diversification of triterpenoids through site-specific oxidation of triterpene skeletons (Seki et al. 2015). In G. uralensis, three P450s involved in triterpenoid biosynthesis have been identified (Fig. 1). CYP88D6 (β-amyrin 11-oxidase) and CYP72A154 (11-oxo-β-amyrin 30-oxidase) are key enzymes in the conversion of β-amyrin to glycyrrhetinic acid, an aglycone of glycyrrhizin (Seki et al. 2008, 2011). CYP93E3 (β-amyrin 24-hydroxylase) is a key initial enzyme in the biosynthesis of soyasaponins (Seki et al. 2008).
Proposed triterpenoid biosynthetic pathways in G. uralensis. Gray solid arrows indicate a cyclization or a single oxidation reaction catalyzed by the OSC or P450 enzymes identified thus far. A black solid arrow indicates a single oxidation reaction catalyzed by the CYP716A179 enzyme identified in this study
Among triterpenoids, oleanolic acid and betulinic acid are widely distributed in various plant species (Jäger et al. 2009). These two compounds are derived from β-amyrin and lupeol, respectively, and both have a carboxyl group at the C-28 position. The first report of a triterpene C-28 oxidase is CYP716A12, which was isolated from Medicago truncatula (barrel medic, Fabaceae) and catalyzes a three-step oxidation reaction at the C-28 position of β-amyrin to yield oleanolic acid (Carelli et al. 2011; Fukushima et al. 2011). CYP716A12 also accepts α-amyrin and lupeol as substrates to produce ursolic acid and betulinic acid, respectively (Fukushima et al. 2011). Since the discovery of CYP716A12, a number of CYP716A enzymes with similar catalytic activities were reported in various plant species (Seki et al. 2015). Although both licorice and M. truncatula are members of the Fabaceae, no CYP716A subfamily P450 in licorice has been reported to date.
Here, we identify CYP716A179 by RNA sequencing (RNA-seq) analysis of tissue-cultured stolons of G. uralensis. Heterologous expression of CYP716A179 in engineered yeast strains showed that this P450 catalyzes a three-step oxidation reaction at the C-28 position of β-amyrin, α-amyrin, and lupeol to produce oleanolic acid, ursolic acid, and betulinic acid, respectively. Transcript levels of CYP716A179 and accumulation levels of oleanolic acid and betulinic acid were highly correlated in intact roots and tissue-cultured stolons. These results may be applicable for future pathway engineering aimed at increasing the production of glycyrrhizin in licorice tissue cultures.
Materials and methods
Plant materials
The origin of the tissue-cultured stolons of G. uralensis was described previously (Kojoma et al. 2010). Tissue-cultured stolons of G. uralensis were maintained in Murashige and Skoog Medium including vitamins (Duchefa Biochemie, Haarlem, the Netherlands) supplemented with 6% sucrose and 0.01 μM 1-naphthaleneacetic acid (Sigma-Aldrich, St. Louis, MO, USA). Intact roots and leaves were collected from G. uralensis plants of a high-glycyrrhizin-producing strain (308-19) harvested in June 2011 in Takeda Garden for Medicinal Plant Conservation, Kyoto, Japan (Ramilowski et al. 2013).
Chemicals
β-Amyrin, erythrodiol, oleanolic acid, α-amyrin, uvaol, ursolic acid, lupeol, glycyrrhetinic acid (18β-glycyrrhetinic acid), echinocystic acid, and hederagenin were purchased from Extrasynthese (Genay, France). Betulin was purchased from Sigma-Aldrich. Betulinic acid was purchased from TCI (Tokyo, Japan). Soyasapogenol B was purchased from Tokiwa Phytochemical (Chiba, Japan). 22α-hydroxy-α-amyrin was extracted and purified from transgenic yeast co-expressing aAS and CYP716A2 (Yasumoto et al. 2016). Standards of oleanolic aldehyde, ursolic aldehyde, and betulinic aldehyde were synthesized chemically by Dr. Kiyoshi Ohyama (Fukushima, Ohyama et al., manuscript in preparation).
RNA extraction
Total RNA was extracted using PureLink® Plant RNA Reagent (Thermo Fisher Scientific, Waltham, MA, USA) from frozen plant tissues and treated with recombinant DNaseI (RNase-free) (Takara Bio, Shiga, Japan), then purified using the RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) following the RNA clean-up protocol.
Library construction, Illumina sequencing, and de novo assembly
A 10-μg aliquot of total RNA was used to construct a cDNA library using an Illumina® TruSeq® RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. The resulting cDNA library was sequenced using HiSeq™ 2000 (Illumina) with 100 bp paired-end (PE) reads in high-output mode. Total reads were assembled using Trinity (Grabherr et al. 2011) after adaptor sequences and low-quality reads were removed by Trimmomatic (v. 0.33) (Bolger et al. 2014).
Cloning of CYP716A179
First-strand cDNA was synthesized using the SMARTer™ RACE cDNA Amplification Kit (Clontech/Takara Bio). The full-length coding sequence (CDS) of CYP716A179 was amplified by polymerase chain reaction (PCR) using primers 1 and 2 (Supplementary Table S1) from the obtained cDNA library and PrimeSTAR® Max DNA Polymerase (Takara Bio) and cloned into pENTR™/D-TOPO® (Thermo Fisher Scientific) to produce an entry clone. The CYP716A179 cDNA was transferred into a Gateway-compatible version of the pELC vector (Seki et al. 2008) using the Gateway™ LR Clonase™ II Enzyme mix (Thermo Fisher Scientific) to generate pELC-CYP716A179, a construct for galactose-inducible dual expression of Lotus japonicus CPR1 and CYP716A179.
Yeast transformation and in vivo enzymatic assay
Saccharomyces cerevisiae INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52; Thermo Fisher Scientific) harboring pYES3-ADH-aAS, pYES3-ADH-OSC1 (bAS), or pYES3-ADH-LUS (Fukushima et al. 2011) was transformed with pELC-CYP716A179 or pELC using the Frozen-EZ Yeast Transformation II Kit™ (Zymo Research, Irvine, CA, USA). The recombinant cells were cultured as described previously (Yasumoto et al. 2016). Ten mL of yeast culture was extracted three times with ethyl acetate, concentrated in vacuo, and resuspended in 500 μL of methanol/chloroform (1:1, v/v). Then, 100 μL of the solution was evaporated and trimethylsilylated with 100 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) at 80 °C for 20 min before gas chromatography–mass spectrometry (GC–MS) analysis. Hederagenin was added as an internal standard prior to extraction.
Analysis of triterpenoid aglycone in plant tissues
Freeze-dried plant tissues (40 mg) were mixed with 200 μL of internal standard (0.1 mg/mL of echinocystic acid) and extracted twice with methanol/chloroform (1:1, v/v). After the solvents were removed, 1 mL of MeOH and 1 mL of 4 M HCl were added to the residue and hydrolyzed at 80 °C for 1 h. The hydrolyzed products were extracted twice with hexane/EtOAc (1:1, v/v), dried in vacuo, and resuspended in 400 μL of methanol/chloroform (1:1, v/v). Then, 100 μL of the solution was evaporated, resuspended in 50 μL of N,N-dimethylformamide, and trimethylsilylated with 50 μL of N,O-bis(trimethylsilyl)trifluoroacetamide + trimethylchlorosilane, 99:1 (Sigma-Aldrich), at 80 °C for 30 min before GC–MS analysis.
GC–MS analysis
GC–MS analysis was performed using a 5977A MSD (Agilent Technologies, Santa Clara, CA, USA) coupled with a 7890B GC system (Agilent Technologies) and DB-1 ms (30 m × 0.25 mm internal diameter, 0.25 μm film thickness; Agilent Technologies) capillary column. The injection component and the MSD transfer line were set at 250 °C and the oven temperature was programmed as follows: 80 °C for 1 min, followed by a rise to 300 °C at a rate of 20 °C/min, and a hold at 300 °C for 15 min (extracts from yeasts) or 28 min (extracts from plants). The carrier gas was He and the flow rate was 1 mL/min. Mass spectra were recorded in the range of 50–750 m/z (extracts from yeasts) or 50–850 m/z (extracts from plants). Peaks were identified through comparison of their retention times and mass spectra with those of the authentic standards (Supplementary Fig. S1).
Quantitative real-time PCR
First-strand cDNA was synthesized using the PrimeScript™ RT Master Mix (Perfect Real Time; Takara Bio) from 2.5 μg of total RNA in a 50-μL reaction. Then, 1× FastStart Essential DNA Green Master (Roche, Basel, Switzerland), 500 nM of primers, and 0.5 μL of cDNA were mixed together and the reaction volume was brought to 10 μL with PCR-grade water. Reactions were performed using a LightCycler® Nano (Roche) with the following conditions: 95 °C for 10 min, 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s. Data were analyzed using LightCycler® Nano Software version 1.1.0 (Roche). Relative transcript levels of each target gene were calculated using β-tubulin (Seki et al. 2008) as a reference gene. The amplification of each sample was performed three times. The primers used for quantitative real-time PCR are listed in Supplementary Table S2.
Phylogenetic analysis of P450s involved in triterpenoid biosynthesis
Full-length amino acid sequences were collected from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). A phylogenetic tree was generated using the neighbor-joining method with 1000 bootstrap replicates in the ClustalX 2.1 software (Larkin et al. 2007) and visualized with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) software.
Results
Triterpenoid profiles of intact roots and tissue-cultured stolons
We compared the triterpenoid profiles of intact roots and tissue-cultured stolons of G. uralensis. Quantification of triterpenoid aglycones in these two plant tissues was performed after the removal of sugar moieties by acid hydrolysis (Table 1, Supplementary Fig. S2). Glycyrrhetinic acid, an aglycone of glycyrrhizin, content in intact roots was almost 1000 times higher than in tissue-cultured stolons. In contrast, oleanolic acid and betulinic acid were detected only in tissue-cultured stolons. We also quantified the accumulation of soyasapogenol B, a common aglycone of soyasaponin I and soyasaponin II. Our analysis revealed that accumulation of soyasapogenol B in tissue-cultured stolons was more than 20 times higher than in intact roots.
Identification and cloning of CYP716A179
To identify P450s involved in the biosynthesis of oleanolic acid and betulinic acid in G. uralensis, we searched for a putative ortholog of CYP716A12 in our previous transcriptome data of intact roots and leaves (Ramilowski et al. 2013) or intact stolons (Sudo et al. 2009); however, no contigs or unigenes were found in either transcriptome data set. Based on the results of triterpenoid profiles (Table 1), we expected that a putative ortholog of CYP716A12 would be expressed in tissue-cultured stolons. Hence, we isolated total RNA from tissue-cultured stolons of G. uralensis (Kojoma et al. 2010) and performed RNA-seq analysis. We obtained 28,925,182 reads and assembled them into 71,673 contigs (Supplementary Table S3). The amino acid sequence of CYP716A12 was used as a query and a contig-encoding protein with 86% amino acid identity to CYP716A12 was found in the RNA-seq data from tissue-cultured stolons of G. uralensis by the TBLASTN algorithm (Altschul et al. 1997). The full-length cDNA of the corresponding contig was isolated and designated as CYP716A179 according to P450 nomenclature (DDBJ/EMBL/GenBank accession number LC157867) (Supplementary Fig. S3). The raw RNA-seq reads obtained in this study were deposited in the DDBJ Sequence Read Archive (DRA) under the accession number DRA004898.
Enzymatic assay of CYP716A179 in engineered yeast
To elucidate whether CYP716A179 catalyzes oxidation at the C-28 positions of β-amyrin, α-amyrin, and lupeol as reported for CYP716A12 (Fukushima et al. 2011), CYP716A179 was expressed together with cytochrome P450 reductase (CPR) in engineered yeast strains harboring β-amyrin synthase (bAS), α-amyrin synthase (aAS), or lupeol synthase (LUS) that endogenously produce β-amyrin, α-amyrin, or lupeol, respectively. Following the culture of each yeast strain, their in vivo metabolites were extracted and analyzed by GC–MS.
In bAS/CPR/CYP716A179-expressing yeast (Fig. 2a), erythrodiol (2), oleanolic aldehyde (3), and oleanolic acid (4) were detected. In aAS/CPR/CYP716A179-expressing yeast (Fig. 2b), uvaol (6), ursolic aldehyde (7), and urosolic acid (8) were detected. In addition to these metabolites oxidized at the C-28 position of α-amyrin, a trace amount of 22α-hydroxy-α-amyrin (9) was detected. 22α-hydroxy-α-amyrin (9) is a major product of CYP716A2 from Arabidopsis thaliana when heterologously expressed in α-amyrin-producing yeast (Yasumoto et al. 2016). Note that since aAS is a multiproduct OSC that produces β-amyrin, as well as α-amyrin, β-amyrin-derived metabolites, erythrodiol (2), oleanolic aldehyde (3), and oleanolic acid (4), were also detected in aAS/CPR/CYP716A179-expressing yeast. In LUS/CPR/CYP716A179-expressing yeast (Fig. 2c), betulin (11), betulinic aldehyde (12), and betulinic acid (13) were detected. These results were almost identical to the enzymatic activities of CYP716A12 (Fukushima et al. 2011), except for the production of 22α-hydroxy-α-amyrin (9) in aAS/CPR/CYP716A179-expressing yeast. The catalytic activities of CYP716A179 confirmed in this study are summarized in Fig. 3.
GC–MS analysis of engineered yeast co-expressing OSC, CPR, and CYP716A179. CYP716A179 was expressed in a β-amyrin-, b α-amyrin-, and c lupeol-producing yeasts. Numbers indicated in the chromatograms correspond to the compounds shown in Fig. 3
Transcript levels of triterpenoid biosynthetic genes in intact roots and tissue-cultured stolons
To address the differences between triterpenoid biosynthesis in intact roots and tissue-cultured stolons, transcript levels of triterpenoid biosynthetic genes including CYP716A179 were examined by quantitative real-time PCR analysis. As predicted from RNA-seq analysis, the transcript level of CYP716A179 in tissue-cultured stolons was about 500 times higher than in intact roots (Fig. 4). The transcript level of CYP716A179 in leaves was lower than that in intact roots (data not shown). Similarly, the transcript level of LUS in tissue-cultured stolons was about 90 times higher than in intact roots, although it was much lower than the transcript level of bAS (Fig. 4). In contrast, transcript levels of CYP88D6 and CYP72A154, two key P450 genes for glycyrrhizin biosynthesis, were much higher in intact roots; particularly, the transcript level of CYP88D6 in intact roots was about 700 times higher than that in tissue-cultured stolons (Fig. 4). The transcript levels of bAS, CYP93E3, and cycloartenol synthase (CAS) were comparable between these two samples (Fig. 4).
Discussion
P450s are key elements in the biosynthesis of various plant triterpenoids. In G. uralensis, three P450s involved in the biosynthesis of glycyrrhizin (CYP88D6 and CYP72A154) and soyasaponins (CYP93E3) have been reported thus far. Here, we successfully identified CYP716A179, which is likely to be involved in the biosynthesis of oleanolic acid and betulinic acid in licorice.
CYP716A179 shares 86% amino acid identity with CYP716A12 from M. truncatula (Supplementary Fig. S3) and shows almost identical catalytic activities against β-amyrin, α-amyrin, and lupeol as CYP716A12 (Fig. 2a–c). The CYP716A subfamily is the most abundant of the P450s involved in triterpenoid biosynthesis (Seki et al. 2015). Most of the CYP716A subfamily enzymes currently identified catalyze oxidation at the C-28 position of pentacyclic triterpene skeletons, such as β-amyrin, α-amyrin, and/or lupeol, except for CYP716A14v2 from Artemisia annua, which catalyzes oxidation at the C-3 position of α-amyrin, β-amyrin, and δ-amyrin (Moses et al. 2015). The phylogenetic tree of CYP716 family enzymes involved in triterpenoid biosynthesis indicates that CYP716A179 is an ortholog of CYP716A12 (Supplementary Fig. S4).
Identification of CYP716A179 was accomplished by RNA-seq analysis of tissue-cultured stolons of G. uralensis. Our initial attempt to identify genes similar to CYP716A12 in previous transcriptome or EST analyses of intact roots, leaves, or stolons of G. uralensis (Ramilowski et al. 2013; Sudo et al. 2009) was unsuccessful due to extremely low transcript levels of CYP716A179 in intact plants (Fig. 4). We also note that no CYP716A subfamily gene was annotated in previous EST analyses of vegetative organs (roots, stems, and leaves) of G. uralensis performed by another research group (Li et al. 2010).
Our analysis of triterpenoid profiles of intact roots and tissue-cultured stolons of G. uralensis indicates that glycyrrhizin is the most prominent triterpenoid in intact roots, while tissue-cultured stolons accumulate soyasaponins, betulinic acid, and oleanolic acid at relatively high levels, with very low glycyrrhizin production (Table 1). The transcript level of CYP716A179 is about 500 times higher in tissue-cultured stolons than in intact roots (Fig. 4). Taken together, the expression of CYP716A179 probably determines the production of oleanolic acid and betulinic acid in licorice. Plant tissue culture is an alternative method for the production of valuable specialized metabolites without harvesting wild resources (Ramachandra Rao and Ravishankar 2002). Suppression of competitive biosynthetic pathways is an effective approach to increase the production of a desired metabolite (Oksman-Caldentey and Inzé 2004). In addition to CYP93E3, CYP716A179 would be an attractive target for gene silencing or knockout aimed at increasing the production of glycyrrhizin in licorice tissue cultures (Fig. 1).
By comparing intact roots and tissue-cultured stolons of G. uralensis, we have shown that, despite a comparable bAS transcript level between these two samples, subsequent P450s directed to glycyrrhizin biosynthesis and oleanolic acid biosynthesis had opposite patterns (Fig. 4). The molecular mechanisms responsible for the differential expression patterns of these P450s in G. uralensis remain unknown. To elucidate the mechanisms of tissue-type-dependent differential regulation of these triterpenoid biosynthetic pathways in G. uralensis, identification of the transcription factors regulating each triterpenoid biosynthetic pathway is required.
Author contribution statement
HS, KS, and TM conceived and supervised the study; KT designed the experiments; KT, HS, and MK performed the experiments; KT wrote the manuscript; HS and TM made manuscript revisions.
Abbreviations
- aAS:
-
α-Amyrin synthase
- bAS:
-
β-Amyrin synthase
- CAS:
-
Cycloartenol synthase
- CDS:
-
Coding sequence
- CPR:
-
Cytochrome P450 reductase
- EST:
-
Expressed sequence tag
- LUS:
-
Lupeol synthase
- OSC:
-
Oxidosqualene cyclase
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, May S, Scotti C, Calderini O (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23:3070–3081
Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T (2011) CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol 52:2050–2061
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hayashi H, Sudo H (2009) Economic importance of licorice. Plant Biotechnol 26:101–104
Hayashi H, Fukui H, Tabata M (1988) Examination of triterpenoids produced by callus and cell suspension cultures of Glycyrrhiza glabra. Plant Cell Rep 7:508–511
Hayashi H, Fukui H, Tabata M (1993) Distribution pattern of saponins in different organs of Glycyrrhiza glabra. Planta Med 59:351–353
Jäger S, Trojan H, Kopp T, Laszczyk MN, Scheffler A (2009) Pentacyclic triterpene distribution in various plants—rich sources for a new group of multi-potent plant extracts. Molecules 14:2016–2031
Kojoma M, Ohyama K, Seki H, Hiraoka Y, Asazu SN, Sawa S, Sekizaki H, Yoshida S, Muranaka T (2010) In vitro proliferation and triterpenoid characteristics of licorice (Glycyrrhiza uralensis Fischer, Leguminosae) stolons. Plant Biotechnol 27:59–66
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li Y, Luo H-M, Sun C, Song J-Y, Sun Y-Z, Wu Q, Wang N, Yao H, Steinmetz A, Chen S-L (2010) EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genom 11:268
Moses T, Pollier J, Shen Q, Soetaert S, Reed J, Erffelinck M-L, Van Nieuwerburgh FCW, Vanden Bossche R, Osbourn A, Thevelein JM, Deforce D, Tang K, Goossens A (2015) OSC2 and CYP716A14v2 catalyze the biosynthesis of triterpenoids for the cuticle of aerial organs of Artemisia annua. Plant Cell 27:286–301
Nelson D, Werck-Reichhart D (2011) A P450-centric view of plant evolution. Plant J 66:194–211
Oksman-Caldentey K-M, Inzé D (2004) Plant cell factories in the post-genomic era: new ways to produce designer secondary metabolites. Trends Plant Sci 9:433–440
Ramachandra Rao S, Ravishankar GA (2002) Plant cell cultures: chemical factories of secondary metabolites. Biotechnol Adv 20:101–153
Ramilowski JA, Sawai S, Seki H, Mochida K, Yoshida T, Sakurai T, Muranaka T, Saito K, Daub CO (2013) Glycyrrhiza uralensis transcriptome landscape and study of phytochemicals. Plant Cell Physiol 54:697–710
Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105:14204–14209
Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K, Muranaka T (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23:4112–4123
Seki H, Tamura K, Muranaka T (2015) P450s and UGTs: key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol 56:1463–1471
Sudo H, Seki H, Sakurai N, Suzuki H, Shibata D, Toyoda A, Totoki Y, Sakaki Y, Iida O, Shibata T, Kojoma M, Muranaka T, Saito K (2009) Expressed sequence tags from rhizomes of Glycyrrhiza uralensis. Plant Biotechnol 26:105–107
Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A (2014) Triterpene biosynthesis in plants. Annu Rev Plant Biol 65:225–257
Yasumoto S, Fukushima EO, Seki H, Muranaka T (2016) Novel triterpene oxidizing activity of Arabidopsis thaliana CYP716A subfamily enzymes. FEBS Lett 590:533–540
Acknowledgements
We are very grateful to the Takeda Garden for Medicinal Plant Conservation, Kyoto, Japan (Takeda Pharmaceutical Company Limited) for the supply of licorice plants. We thank Dr. David R. Nelson (University of Tennessee) for the naming of CYP716A179 according to the P450 nomenclature. We also thank Mr. Tsutomu Hosouchi and Ms. Sayaka Shinpo (Kazusa DNA Research Institute) for technical support with the Illumina sequencing, Dr. Kazuto Mannen (Kazusa DNA Research Institute) and Dr. Ryosuke Sano (Nara Institute of Science and Technology) for technical advice with bioinformatics tools, Dr. Kiyoshi Ohyama (Tokyo Institute of Technology) for providing oleanolic aldehyde, ursolic aldehyde and betulinic aldehyde standards, and Dr. Ery Odette Fukushima (Osaka University) for helpful discussion on phylogenetic analysis of CYP716A subfamily enzymes. This work was supported by the “Health and Labour Sciences Research Grant” on the enhancement of “Comprehensive Medicinal Plant Database”, JSPS KAKENHI Grant Number JP26450123 and research funding from the Yamada Science Foundation to H. Seki, JSPS KAKENHI Grant Number JP15H04485 to TM, and the Doctor 21 scholarship from the Yoshida Scholarship Foundation to KT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by F. Sato.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tamura, K., Seki, H., Suzuki, H. et al. CYP716A179 functions as a triterpene C-28 oxidase in tissue-cultured stolons of Glycyrrhiza uralensis . Plant Cell Rep 36, 437–445 (2017). https://doi.org/10.1007/s00299-016-2092-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2092-x