Abstract
Key message
A homologous gene of MPEC from Phyllostachys edulis was isolated and characterized. Its overexpression in Arabidopsis thaliana increased chlorophyll concentration and photosynthesis efficiency, indicating it is involved in chlorophyll biosynthesis.
Abstract
Magnesium-protoporphyrin IX monomethyl ester cyclase (MPEC) is an essential enzyme in the biosynthesis of chlorophyll, which plays an important role in photosynthesis. However, limited information is available on the roles of MPEC gene in bamboo. A homologous gene, PeMPEC was identified from Phyllostachys edulis, which comprised 1474 bp and contained an open reading frame encoding 415 amino acids. PeMPEC was transcribed abundantly in leaf blade where photosynthesis predominantly occurs, which agreed with the protein accumulation pattern confirmed by Western blotting. The PeMPEC transcription was promoted by continuous darkness for 24 h, and was suppressed by increasing light intensity (100–1500 µmo1 m−2 s−1) and high temperature (42 °C). However, transcription was induced within 0.5 h and thereafter declined with prolonged treatment (up to 12 h) under low temperature (4 °C). Although PeMPEC expressed weakly in etiolated leaves, transcript levels increased gradually with subsequent light treatment (200 µmol m−2 s−1). Overexpression of PeMPEC in Arabidopsis thaliana resulted in increased chlorophyll concentration and photosynthesis efficiency in sense transgenic plants compared with a reduction in antisense transgenic plants. These changes were consistent with the transcript levels of PeMPEC. These results indicated that PeMPEC might be involved in chlorophyll biosynthesis and play important roles in maintaining the stability of photosystems, and provide a basis for the study of chlorophyll biosynthesis and dissection of photosynthesis in bamboo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorophyll is a primary photosynthetic pigment that plays an important role in the capture, transmission, and transformation of light energy. The biosynthesis of chlorophyll has been demonstrated to require an extensive set of enzymes in more than 20 enzymatic reactions, for which the properties of most key enzymes involved in the pathway are well understood (Porra 1997; Suzuki et al. 1997; Papenbrock and Grimm 2001; Kovacevic et al. 2007). However, one of the least understood reactions is the formation of the isocyclic ring. In the chlorophyll biosynthetic pathway, after a series of reactions starting from alutamate to insertion of a magnesium ion (Mg2+) into protoporphyrin IX, assembly of the isocyclic ring begins by forming Mg-protoporphyrin IX monomethyl ester (MgPME) through esterification, which is subsequently converted to protochlorophyllide by Mg-protoporphyrin IX monomethyl ester cyclase (MPEC) (Fuesler et al. 1984; Wong et al. 1985; Porra et al. 1995; Bollivar and Beale 1996; Liu and Zheng 2008). MPEC is an essential enzyme in chlorophyll biosynthesis, and knockout of the gene encoding MPEC can be lethal to plants (Pinta et al. 2002). The Arabidopsis (A. thaliana) chl27-t knock-down mutant containing a T-DNA insertion within the CHL27 promoter exhibits retarded growth and chloroplast developmental defects, which are caused by damage to PSII reaction centers (Bang et al. 2008).
The MPEC gene was first identified and isolated by differential hybridization screening of a cDNA library from Pharbitis nil cotyledons exposed to darkness (Zheng et al. 1998). Currently, functional analysis of MPEC is mainly performed by mutant analysis. Two homologous genes of MPEC identified in Chlamydomonas reinhardtii are Crd1 (Copper response defect 1) and Cth1 (Copper target homolog 1) (Moseley et al. 2002). Crd1 and Cth1 are required for maintenance of photosystem I (PSI) by interacting with light-harvesting complexes. Crd1 is exclusively expressed in copper- and oxygen-deficient conditions, whereas Cth1 is accumulated in copper- and oxygen-sufficient conditions. However, previous studies are unable to determine whether Crd1 and Cth1 are involved with MPEC in the chlorophyll biosynthetic pathway (Moseley et al. 2000, 2002). Studies on acsF (aerobic cyclization system Fe-containing subunit) in Rubrivivax gelatinosus are the earliest reports of MPEC function. The acsF mutant is unable to synthesize chlorophyll under aerobic conditions, whereas it shows a phenotype similar to that of the wild-type under low-oxygen conditions. Subsequently, the function of the bchE gene is investigated using the acsF mutant, which synthesizes chlorophyll under high oxygen conditions (Pinta et al. 2002). Based on this study, two models of MgPME conversion in R. gelatinosus have been proposed (Ouchane et al. 2004). Homologous genes of MPEC were also isolated from higher plants, such as A. thaliana and Hordeum vulgare. The phenotype and pigment concentration of transformed antisense CHL27 plants proved that CHL27 is required for MgPME conversion (Tottey et al. 2003). In H. vulgare, MPEC is composed of three gene products, consisting of a soluble protein, and two membrane-bound components encoded by Xantha-l, and Viridis-k, respectively. The acsF homologous gene has been cloned and sequenced from the H. vulgare mutant Xantha-l rather than the Viridis-k, so Xantha-l is a homologue of acsF (Rzeznicka et al. 2005).
Bamboos are ecologically and economically important forest resources. Compared with other trees, one of bamboos’ characteristics is a fast-growth rate. To some extent, it indicates that bamboos have a strong assimilative capacity, which is closely associated with photosynthesis. In order to explain this specific characteristic, the spectroscopic features of LHCIIb in photosystem II (PSII) of bamboo have been investigated, which revealed that chlorophyll a and chlorophyll b were the main pigments of the LHCIIb complexes for photosynthesis in Phyllostachys edulis (Jiang et al. 2012). However, chlorophyll biosynthesis in bamboos remains to be elucidated.
It is difficult to obtain bamboo mutants because of the infrequency and unpredictability of flowering events, resulting in severe limitations in genetic investigations on bamboo, especially for the transfer of genetic information. Therefore, model plant Arabidopsis was employed to confirm the function of a MPEC homologue from bamboo. In this study, we isolated PeMPEC from moso bamboo (P. edulis) and characterized the expression patterns in moso bamboo seedlings in response to different environmental factors. To investigate the function of PeMPEC, we constructed expression vectors with sense and antisense PeMPEC and transformed the constructs into wild-type Arabidopsis, respectively. A complementation test was performed in the Arabidopsis mutant Crd1 with sense PeMPEC. The chlorophyll concentration and chlorophyll fluorescence parameters of the transgenic plants were measured and analyzed. The results demonstrated that PeMPEC plays an important role in chlorophyll synthesis and photosystem stability.
Materials and methods
Plant materials and growth conditions
Moso bamboo (P. edulis) seedlings, Arabidopsis wild-type (ecotype Columbia-0; Col-0) and mutant Crd1 (ABRC Stocks, Columbus, OH; SALK-024716C) plants were potted in nutritional bowls (basal diameter/mouth diameter/height = 6.5/10/8 cm) containing vermiculite. The vermiculite was presoaked with plant nutrient solution (1/3 B5 medium) and grown under a 16/8 h (light/dark) photoperiod with light intensity of 200 µmol m−2 s−1 at 23 ± 1 °C. One-year-old bamboo seedlings and 2-month-old etiolated bamboo seedlings were used for further study.
Gene cloning
A search for the MPEC homologue sequence in the BambooGDB database (http://www.bamboogdb.org) constructed by our laboratory (Zhao et al. 2014) was undertaken using the OsZIP1 sequence (FJ940751) (http://www.ncbi.nlm.nih.gov/nuccore) as a reference. Primers (PM-1: 5′-ATGGCCTCCGCCATGAAGCT-3′; PM-2: 5′-TCAGTAAACAAGCCGGGGCT-3′) were designed according to the open reading frame (ORF) of the MPEC sequence in P. edulis.
Total RNA was isolated from leaves of P. edulis using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacture’s protocol. First-strand of cDNA was synthesized with 1 µg total RNA using the Reverse Transcription System (Promega Biotech Co., Ltd., Beijing, China). Genomic DNA was extracted using the CTAB method from leaves of P. edulis (Gao et al. 2006). The cDNA and genomic DNA were used as templates for PCR, which was performed with the primers PM-1 and PM-2 using Prime STAR DNA polymerase (Takara Biotech Co., Ltd., Dalian, China). The PCR amplification products were inserted into the pGEM-T easy vector (Promega) and subsequently confirmed by sequencing with an ABI 3730 DNA Analyzer (Applied Biosystems, USA).
Sequence analysis
The cDNA and genomic DNA sequences of PeMPEC were analyzed with Vector NTI software. Homology searches were performed using Blastn and Blastp with default parameters (http://www.ncbi.nlm.nih.gov/blast/). The web-based MOTIF SCAN tool (http://myhits.isb-sib.ch/cgi-bin/motif_scan) was used to identify known motifs in the sequences (Sigrist et al. 2010). A neighbor-joining tree was constructed from nucleotide sequences using the MEGA4.0 software package (Tamura et al. 2007) and the CLUSTAL algorithm in conjunction with the known DNA sequences encoding MPECs, which were downloaded from GenBank. Introns in the genomic sequence were identified by the GeneWise online tool (http://www.ebi.ac.uk/Tools/psa/genewise/), and the gene structure of PeMPEC was determined. The cis-regulatory elements of introns were analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al. 1999). The presence of a transit peptide in PeMPEC was predicted using the ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/) (Emanuelsson et al. 1999).
Tissue-specific gene expression analysis
For analysis gene expression in different tissues, total RNA was isolated from the root, stem leaf blade, and sheath of 1-year-old bamboo seedlings and cDNAs were synthesized. PCR was performed with the primers PQ-1(5′-AATCCCTCACCCCTCACTCCG-3′) and PQ-2(5′-ATGCGGAGGCGCGAATGG-3′). The final reaction volume was 20 μL and included 2 μL 10 × Buffer (Mg2+ Plus), 1.6 μL dNTPs (2.5 mM each of dATP, dTTP, dCTP and dGTP), 2 μL PQ-1 and PQ-2 (5 μM, each), 1 μL cDNA, 11.2 μL ddH2O, and 0.2 μL rTaq DNA polymerase (Takara). The PCR program included an initial denaturation step at 94 °C for 5 min, followed by 28 cycles at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s; after the last cycle, there was a final extension period at 72 °C for 7 min. A cDNA fragment of Actin, as described previously, was amplified as a positive control under the same PCR conditions (Gao et al. 2012).
Real-time quantitative PCR analysis
The cDNA templates were synthesized using total RNA isolated from leaves of one-year-old bamboo seedlings treated with different light intensities (100, 300, 500, 1000, and 1500 µmol m−2 s−1) for 2 h, low temperature (4 °C) or high temperature (42 °C) for 0, 0.5, 1, 2, 6, and 12 h. The 2-month-old etiolated bamboo seedlings were treated with laboratory light (200 µmol m−2 s−1) and total RNA was isolated after 0, 0.5, 1, 4, and 8 h, followed by cDNA synthesis. Real-time quantitative PCR (qPCR) was performed with a qTOWER2.2 Real-Time PCR System (Analytik Jena, Germany) using the Roche LightCycler®480 SYBR Green I Master Kit (Roche). The qPCR procedure consisted of 95 °C for 10 min and 50 cycles of 95 °C for 10 s, 60 °C for 10 s. The reaction volume was 10 µL and contained 5.0 µL 2× SYBR Green 1 Master Mix, 0.8 µL cDNA, 0.2 µL PQ-1 and PQ-2 each (5 μM), and 3.8 µL ddH2O. All reactions were repeated three times. For each condition, the qRT-PCR experiments were performed as biological triplicates. The relative gene expression level was calculated with the 2−∆∆ct method (Livak and Schmittgen 2001) using NTB as the reference gene (Fan et al. 2013).
Prokaryotic expression of PeMPEC and Western blotting
The mature protein of PeMPEC was predicted with the ChloroP 1.1 Server. The nucleotide sequence encoding the mature protein was amplified by PCR with primers introducing the BamHI (forward primer) and HindIII (reverse primer) restriction sites. The primers used to generate the fragment were PeMPEC-F (5′-aagcttGCCGCGAGCAAGCCGGGGC-3′) and PeMPEC-R (5′-ctcgagGTAAACAAGCCGGGGCTCAA-3′) based on the nucleotide sequence of PeMPEC. After sequencing, the fragment encoding the mature protein was introduced into the multiple cloning sites of the pET-32b vector. The recombinant plasmid pET-32b-PeMPECm was transformed into competent cells of Escherichia coli strain Rosetta-gamin B (DE3) for protein expression. The cells were cultured in medium supplemented with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 28 °C and 37 °C for 4 h (Gao et al. 2012). The recombinant protein was analyzed using SDS-PAGE (5 % stacking gel and 12 % separating gel) in accordance with the Molecular Clone procedure (Sambrook et al. 1989). The polyclonal antibody was prepared with the purified PeMPEC protein by Beijing Protein Innovation Co. Ltd. Proteins from P. edulis were extracted and Western blotting was performed in accordance with the method described previously (Gao et al. 2013).
Construction of plant expression vectors and transformation
The ORF sequence of PeMPEC was amplified by PCR with primers introducing the BamHI and XbaI sites. The primers used to generate the sense fragment were SF (5′-ggatccATGGCCTCCGCCATGAAGCTC-3′) and SR (5′-tctagaTCAGTAAACAAGCCGGGGCTC-3′), and those for the antisense fragment were AF (5′-tctagaATGGCCTCCGCCATGAAGCTC-3′) and AR (5′-ggatccGTAAACAAGCCGGGGCTCAAA-3′) based on the nucleotide sequence of PeMPEC.
The fragment was subcloned into the pCAMBIA1301 vector driven by the CaMV 35S promoter. The constructions were designated pC1301-PeMPEC-S (sense) and pC1301-PeMPEC-A (antisense), and were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. Arabidopsis plants were transformed using the floral dip method (Clough and Bent 1998). Putative transgenic plants were selected on medium supplemented with 50 mg L−1 hygromycin, and further verified by semi-quantitative PCR with the primers PQ-1 and PQ-2. AtUbiquitin (NM180850) was selected for use as an internal control under the same PCR conditions using the primers AtUbiquitin-F (5′-ATGGCTGAAGAGGATATCCAGC-3′) and AtUbiquitin-R (5′-GAAACACTTCATATGGACGATGG-3′). The phenotype of transgenic plants was observed and photographed simultaneously.
Measurement of chlorophyll concentration and chlorophyll fluorescence parameters
The chlorophyll concentration of transgenic plants, Col-0 and the Crd1 mutant of Arabidopsis was quantified by spectrophotometry in accordance with the method of Benedetti and Arruda (2002). Chlorophyll fluorescence parameters were measured using an IMAGING-PAM fluorometer (Waltz, Germany). The following parameters were calculated: the maximum quantum yield of PSII F v/F m = (F m − F o)/F m (Demmig-Adams and Adams 1996) and the actual photochemical efficiency of PSII Y(II) = (F m′ − F s)/F m′ (Bilger and Björkman 1990), where F o is the minimum fluorescence in the dark-adapted state, F s is the steady-state fluorescence after light adaptation, and F m and F m′ are the dark-adapted and light-adapted maximum fluorescence upon illumination of pulse (0.6 s) of saturating light, respectively. F o and F m were determined after 20 min dark adaptation. The maximal photo-oxidizable P700 (P m), which is a suitable parameter for representing the quantity of efficient PSI complex (Huang et al. 2010), was determined through application of a saturation pulse after far-red pre-illumination for 10 s using the Dual PAM-100 (Walz). Each parameter was measured with ten replicates per treatment. All data were statistically analyzed using SPSS software. If the results of One-way ANOVA showed the significant differences at the 0.05 significance level, we used least significance difference for multiple comparisons among the different plants.
Results
Sequence isolation and molecular characterization of PeMPEC
Based on sequence alignment, a homologous gene (FP092998) of OsMPEC was isolated from P. edulis and named as PeMPEC. The full-length gene was 1474 bp including a 23 bp 5′ untranslated region (UTR), a 203 bp 3′ UTR and an open reading frame (ORF) of 1248 bp. The ORF was determined by end-to-end PCR with the primers PM-1 and PM-2 and sequenced, which was completely consistent with the sequence of PeMPEC. The ORF encoded a putative protein of 415 amino acids containing one conserved leucine zipper domain and two copies of the EXnDEXRH motif (S1). The theoretic isoelectric point and calculated molecular mass of PeMPEC were 9.1 and 47.9 kDa, respectively. Based on the prediction with the ChloroP 1.1 Server, the deduced amino acid sequence of PeMPEC consisted of a transit peptide of 50 amino acids at the N-terminal and a mature protein (position 51-415) with a calculated molecular mass was 42.7 kDa.
The genomic sequence corresponding to the ORF of PeMPEC was isolated using the primers PM-1 and PM-2, which comprised 1854 bp (GenBank accession No. KR817911) and contained five exons and four introns (S2). The introns were in full compliance with the intron splicing principles of GT-AG, in which the composition of A+T and G+C were 59 % and 41 %, respectively. The light-responsive regulatory elements consisting of the GAG-motif, ACE, and AE-box, as well as the promoter elements TATA-box and CAAT-box, were located in the introns (S3). Although the introns are spliced out during transcription, the presence of elements in introns similar to those in the promoter indicated that the introns may play roles in the regulation of gene expression. The functions of these elements in response to light need further investigation.
Alignment and phylogenetic analysis
Sequence alignment demonstrated that the predicted protein encoded by PeMPEC shared 73.6 % homology with that of Arabidopsis, and more than 85.0 % homology with those of gramineous plants such as Oryza sativa, Brachypodium distachyon, and Triticum aestivum, which indicated that MPEC was highly conserved during evolution.
A neighbor-joining tree was constructed based on the nucleotide sequences of MPEC homologues. The genes were mainly clustered into four groups, comprising photosynthetic bacteria, algae, monocots, and dicots, respectively (Fig. 1). PeMPEC and OsMPEC were placed in the same cluster, indicating that the MPEC homologues of P. edulis and O. sativa showed a close phylogenetic relationship. In contrast, the homologues of R. gelatinosus, C. reinhardtii, and Nicotiana tabacum were indicated to be phylogenetically distant. Thus, the suggested phylogenetic relationships were consistent with the taxonomic affinities.
Neighbor-joining tree representing phylogenetic relationships among MPEC gene sequences constructed using MEGA 4.0 Numbers above major branches indicated bootstrap value estimates for 500 replicates. MPEC genes used in phylogenetic tree are OsZIP1 (FJ940751) from Oryza sativa, PeMPEC (FP092998) from Phyllostachys edulis (in rectangle), LOC101773118 (XM004967622) from Setaria italica, LOC100828682 (XM003567228) from Brachypodium distachyon, Xantha-l AY887063) from Hordeum vulgare, homologous gene (AK331887) from Triticum aestivum, Crd1 (AF236101) from Arabidopsis thaliana, PNIL34 (U37437) from Ipomoea nil, NTZIP (AY221168) from Nicotiana tabacum, Crd1 (AF237671) and CTH1 (AF337037) from Chlamydomonas reinhardtii, and acsf (AY234384) from Rubrivivax gelatinosus
Tissue-specific expression of PeMPEC
Given that the leaf is a primary photosynthetic organ, we investigated whether PeMPEC showed leaf-specific expression. Semi-quantitative PCR were performed with cDNA templates from root, stem, sheath, and leaf blade of P. edulis. PeMPEC was transcribed abundantly in the leaf blade and weakly in the sheath. Transcripts were not detected in the root and stem (Fig. 2a).
Expression analysis of PeMPEC in different tissues of P. edulis. 1 root, 2 stem, 3 sheath, 4 leaf blade. a PeMPEC transcription analyzed by semi-quantitative PCR using Actin as an internal control. The presence of transcripts was checked after 28 cycles by PCR. b PeMPEC accumulation analyzed by Western blotting. A total of 15 µg proteins were loaded on each lane. The endogenous protein was around 42 kDa marked by arrow. M protein molecular marker
Expression of the recombinant protein and Western blotting
SDS-PAGE analysis demonstrated that the recombinant PeMPEC protein was induced by IPTG at 28 and 37 °C, respectively. The protein molecular mass was about 62 kDa (S4), which agreed with the predicted mass of PeMPEC (42.7 kDa) combined with tags of pET32b (~20 kDa). Although the recombinant PeMPEC protein was abundant at 37 °C, it was in the form of inclusion bodies and the soluble protein was much less abundant. The optimal condition for induction of soluble protein expression was 0.4 mM IPTG at 28 °C for 4 h. The recombinant PeMPEC protein was purified and used to prepare a polyclonal antibody. The total proteins of the root, stem, sheath, and leaf blade from 1-year-old bamboo seedlings were extracted and separated with SDS-PAGE. The polyclonal antibody of PeMPEC was used as a probe for Western blotting. PeMPEC was detected only in leaf blade (Fig. 2b), which supported the conclusion that transcription of PeMPEC occurs mainly in the leaf.
Expression pattern of PeMPEC under abiotic stresses
To test whether the expression of PeMPEC is light-intensity dependent, we analyzed PeMPEC transcript levels in leaves exposed to different light conditions. The qPCR analysis demonstrated that PeMPEC transcription was promoted by continuous darkness for 24 h, with transcript level 2.5 times higher than those of the control (200 µmo1 m−2 s−1). However, with increasing light intensity, PeMPEC transcription gradually decreased from 2.2 times (100 µmo1 m−2 s−1) to only 55 % (1500 µmo1 m−2 s−1) that of the control (Fig. 3a). Although transcription of PeMPEC was weak in the etiolated seedlings, it increased gradually after the seedlings were exposed to the laboratory light environment (200 µmol m−2 s−1), and after 8 h treatment the level reached to 80 % of the control (Fig. 3b). These results implied that PeMPEC transcription was induced by light of low intensity.
Expression analysis of PeMPEC under different light intensities by qPCR. a PeMPEC expression in leaves treated with different light intensities. 1 control (200 µmol m−2 s−1), 2 darkness 24 h, 3 100 µmol m−2 s−1, 4 300 µmol m−2 s−1, 5 500 µmol m−2 s−1, 6 1000 µmol m−2 s−1, 7 1500 µmol m−2 s−1. b PeMPEC expression in leaves of etiolated seedlings treated with light of 200 µmol m−2 s−1 for different durations. 1 control (green leaves under continuous light of 200 µmol m−2 s−1); 2 0 h, 3 0.5 h, 4 1 h, 5 4 h, 6 8 h. Error bars represent standard errors of three independent biological replicates
Temperature was also an important factor influencing gene transcription. High temperature (42 °C) suppressed the transcription of PeMPEC but with fluctuations, transcript levels initially decreased to ~70 % of the control after 0.5 h treatment, and subsequently increased to ~92 % of the control after 1–2 h treatment, followed by rapid reduction to only 5 % of the control after 12 h treatment (Fig. 4a). However, PeMPEC transcription was initially induced by low temperature (4 °C) during the first 0.5 h treatment, but thereafter declined with prolonged treatment (1–12 h) (Fig. 4b). These results indicated that temperature had a temporally sensitive and complex effect on PeMPEC transcription, which requires further study.
Ectopic expression of PeMPEC in Arabidopsis
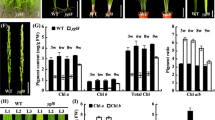
To verify whether PeMPEC was involved in chlorophyll synthesis, expression vectors containing the sense and antisense PeMPEC were constructed and transformed into Arabidopsis mediated by A. tumefaciens. A total of 39 transgenic lines, including 24 sense lines and 15 antisense lines, were obtained and confirmed by PCR. Phenotype analysis showed there were no obvious differences between the sense transgenic plants and Col-0. However, yellowish green leaves exhibiting chlorophyll deficiency were observed in the antisense transgenic plants, which also grew more slowly (Fig. 5a).
Ectopic expression of PeMPEC in A. thaliana. a Phenotype analysis of leaves from transgenic lines (bar 1 cm). b Expression analysis of PeMPEC in transgenic plants by semi-quantitative PCR. c Chlorophyll content analysis of transgenic plants. Error bars represent standard errors of three independent biological replicates. WT wild-type Col-0, S-3, S-5 sense lines, A-1, A-2, A-4 antisense lines
The transcript level of PeMPEC, the chlorophyll concentration and chlorophyll fluorescence parameters were evaluated among transgenic lines (sense lines: S-3 and S-5; antisense lines: A-1, A-2, and A-4) and Col-0. Semi-quantitative PCR analysis confirmed that PeMPEC was transcribed in transgenic plants, among which the highest abundance of transcripts was in S-5 and the lowest abundance was in A-4 (Fig. 5b). The chlorophyll concentration in sense plants was 20 % higher than that of Col-0, whereas it was reduced in all antisense plants, especially in A-2, which contained only 58 % of the chlorophyll concentration of Col-0 (Fig. 5c). The values of F v/F m, Y(II), and P m in A-1 and A-2 were maintained at a significantly lower level than those of Col-0 (p < 0.05). However, no significant difference in the values between sense lines and Col-0 was observed (Table 1). The chlorophyll concentration and chlorophyll fluorescence parameters were in agreement with the leaf phenotype. These findings indicate that the chlorophyll concentration of transgenic plants is influenced by transcription of PeMPEC, which is involved in the regulation of chlorophyll biosynthesis.
Functional complementation analysis of PeMPEC in the Crd1 mutant
Mutants play an important role in exploring gene function (Qin et al. 2007; Xu et al. 2014). PeMPEC driven by the CAMV 35S promoter was transferred into the Crd1 mutant. A total of 19 hygromycin-resistant transgenic lines were obtained. Four T3 transgenic lines (M-1, M-3, M-4, and M-5) without segregation were selected for further analysis. The phenotype of the transgenic plants was similar to that of Col-0 (Fig. 6a). PCR analysis confirmed that PeMPEC was expressed in the transgenic plants, with the highest abundance of transcripts observed in line M-4 and the lowest abundance in line M-1 (Fig. 6b).
Functional complementation analysis of PeMPEC in Crd1 mutant. a Phenotype analysis of PeMPEC transgenic plants compared with Crd1 mutant and wild-type Col-0 (bar 1 cm). b Expression analysis of PeMPEC in transgenic plants by semi-quantitative PCR. c Chlorophyll content analysis of PeMPEC transgenic plants. Error bars represent standard errors of three independent biological replicates. WT wild-type Col-0, M Crd1 mutant, M-1, M-3, M-4, M-5 transgenic plants
Chlorophyll concentration, as a physiological index, was employed as a measure of chlorophyll synthesis in the leaf. The chlorophyll concentration of sense plants was 22–50 % higher than that of the Crd1 mutants, and 7–20 % higher than that of Col-0 (Fig. 6c), indicating that PeMPEC promoted chlorophyll synthesis. No significant difference was observed between transgenic plants and Col-0 in terms of the values of F v/F m, Y(II), and P m. However, significant differences (p < 0.05) were observed between transgenic plants and the Crd1 mutants. Compared with Crd1, the values of F v/F m and Y(II) increased by 27.1–34.2 % and the P m value increased by 28.6–47.6 % in transgenic plants, which was close to those of Col-0 (Table 2). These results indicated that overexpression of PeMPEC rescued the phenotype of Crd1 by increasing chlorophyll concentration in the leaf, which played an important role in maintaining the stability of PSI and PSII.
Discussion
In higher plants, the attainment of a high yield potential depends on rapid rates of photosynthesis. Given that chlorophyll is an essential pigment involved in the absorption of radiant energy, the rate of photosynthesis is affected by the chlorophyll concentration. MPEC is a key enzyme that catalyzes conversion of MgPME into protochlorophyllide in the chlorophyll biosynthesis pathway. To investigate the function of MPEC in bamboo, we isolated the homologue PeMPEC. Gene structure analysis indicated that the ORF of PeMPEC was divided by four introns belonging to the GT-AG type. Introns have increasingly been shown to serve important biological functions, especially in transgenic plants in which introns can improve the expression of exogenous genes (Callis et al. 1987; Brinster et al. 1988). The introns in the genomic sequence of PeMPEC contain regulatory elements similar to eukaryotic promoter elements, light-response elements, and plant hormone-response elements, all of which might enhance gene transcription in different ways. Therefore, further analysis of the function of introns in PeMPEC is needed.
The analysis of protein structure showed that PeMPEC contained two copies of the EXnDEXRH motif and a leucine zipper structure. The fifth leucine was replaced by glycine, which has been observed in many leucine zipper motifs (Nantel and Quatrano 1996). This suggested that PeMPEC had a similar function to that of other ZIP proteins. As a common domain of transcription factors, the leucine zipper structure is involved in the formation of homologous and heterologous dimers (Subramaniam et al. 2001; Kuhlmann et al. 2003) and plays an important role in plant growth and development (Pysh and Schmidt 1996). However, the specific function of PeMPEC remains to be determined. Computer analysis predicted the presence of a chloroplast transit sequence of 50 amino acids in PeMPEC. This result is consistent with the finding that all chlorophyll synthesis enzymes have a transit peptide in the sequence and function in the chloroplast (Nagata et al. 2005).
Light and temperature are the most critical factors that affect chlorophyll synthesis. Light is needed in the transition of protochlorophyllide into chlorophyllide, but light of sufficiently high intensity may cause a decline in enzyme activity and accumulation of protochlorophyllide. Activity of MPEC is reduced as a result of feedback inhibition of chlorophyll synthesis. This phenomenon is widespread in the chlorophyll synthesis pathway between 5-aminolevulinic acid and protochlorophyllide (Terry and Kendrick 1999). The first plant MPEC gene to be isolated was from a dark-induced cDNA library from Pharbitis nil cotyledons (Zheng et al. 1998), which indicated that MPEC may be induced by darkness. The expression of PeMPEC under continuous darkness agreed with this hypothesis. However, PeMPEC was suppressed gradually with increasing light intensity. The expression of PeMPEC in bamboo leaves showed a fluctuating trend under temperature stress, transcription was suppressed at 42 °C, but was induced at 4 °C within 0.5 h. The up-regulation of PeMPEC might promote chlorophyll synthesis to remedy the shortage of chlorophyll, which is a compensation phenomenon of plants under abiotic stress (Feierabend and Mikus 1977).
To study the function of PeMPEC in chlorophyll synthesis, a complementation experiment was carried out using the Arabidopsis Crd1 mutant. Overexpression of PeMPEC in the Crd1 mutant resulted in recovery phenotype with increased chlorophyll concentration. However, diverse phenotypes were observed in antisense PeMPEC Arabidopsis plants, which demonstrated similar phenotypes to those of CHL27 antisense Arabidopsis plants and could be divided into four classes referred by Tottey et al. (2003). These results demonstrated that the efficiency of using an antisense approach with a heterologous gene, as shown previously using an endogenous homologous gene (Tottey et al. 2003). The antisense expression of a heterologous gene results in reduced expression level of the endogenous homologous gene, which has been confirmed by the expression of antisense FBP1 (from Phaseolus vulgaris) in Arabidopsis (Bindschedler et al. 2006), and antisense MsCOMT (from Miscanthus sinensis) and antisense GMPase (from tomato) in tobacco plants (Wang et al. 2012; Seong et al. 2013). Feedback inhibition of chlorophyll synthesis is observed. Overexpression of MPEC enhances enzyme activity, leading to accumulation of protochlorophyllide, but accumulation of chlorophylls, in turn, inhibited Mg-dechelatase activity. Thus, chlorophyll synthesis always remains at a certain level for the above reasons (Meskauskiene et al. 2001). Therefore, the chlorophyll concentration in transgenic plants expressing sense PeMPEC is unable to increase indefinitely.
Chlorophyll functions as the carrier to absorb light energy in photosynthesis. Chlorophyll fluorescence, as a reliable, rapid, noninvasive, and indirect probe, is closely associated with the photosynthesis rate (Schreiber et al. 1995). The value of Y(II) is a relative index of photosynthesis electron transport rate, and is dependent on development of the PSII reaction center and capture efficiency of PSII excitation energy. In terms of the Y(II) value, no significant difference was observed between Col-0 and sense PeMPEC transgenic plants of Col-0 and Crd1. By contrast, the Y(II) values of antisense PeMPEC transgenic plants and the Crd1 mutant were significantly lower than those of Col-0 (p < 0.05), suggesting that PSII in those plants was unstable. The P m values of A-1, A-2, and Crd1 mutant plants were also lower than those of Col-0, indicating that PSI of those plants were affected. These results were similar to those of the mutant Crd1 in R. gelatinosus, in which the PSI and light-harvesting proteins were not compounded and the corresponding proteins of PSII also showed slight degradation (Moseley et al. 2000), and in antisense transgenic plants of Arabidopsis CHL27, both PSI and light-harvesting proteins were degraded (Tottey et al. 2003).
Considering the present and previous findings, we speculate that PeMPEC might affect the biosynthesis of chlorophyll and stability of the photosystem in bamboo, which are closely associated with the rapid growth rate characteristic of bamboo.
Author contribution statement
Conceived and designed the experiments: ZHP, LY. Performed the experiments: LY, YFL, HYS. Analyzed the data: LY, YFL, ZMG. Contributed reagents/materials/analysis tools: LY, HSZ. Contributed to the writing of the manuscript: LY, ZMG.
Abbreviations
- cDNA:
-
Complementary DNA
- F o :
-
Minimal fluorescence
- F m :
-
Maximal fluorescence after darkness adaptation
- F m′:
-
Maximum fluorescence after light adaptation
- F s :
-
Steady-state fluorescence after light adaptation
- F v :
-
Variable fluorescence
- F v/F m :
-
Maximal quantum yield of PSII
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- kDa:
-
Kilodaltons
- Mg:
-
Magnesium
- MPEC:
-
Mg-protoporphyrin IX monomethyl ester cyclase
- ORF:
-
Open reading frame
- P m :
-
Maximum photo-oxidizable P700
- PSI:
-
Photosystem I
- PSII:
-
Photosystem II
- qPCR:
-
Real-time quantitative PCR
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Y(II):
-
Maximal actual quantum yield of PSII
References
Bang WY, Jeong IS, Kim DW, Im CH, Ji C, Hwang SM, Kim SW, Son YS, Jeong J, Shiina T, Bahk JD (2008) Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol 49(9):1350–1363
Benedetti CE, Arruda P (2002) Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol 128(4):1255–1263
Bilger W, Björkman O (1990) Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res 25(3):173–185
Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, Ausubel FM, Bolwell GP (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47(6):851–863
Bollivar DW, Beale SI (1996) The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase (Characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803). Plant Physiol 112(1):105–114
Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD (1988) Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA 85(3):836–840
Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1(10):1183–1200
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Demmig-Adams B, Adams WWI (1996) Chlorophyll and carotenoid composition in leaves of Euonymus kiautschovicus acclimated to different degrees of light stress in the field. Aust J Plant Physiol 23(5):649–659
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8(5):978–984
Fan C, Ma J, Guo Q, Li X, Wang H, Lu M (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8(2):e56573
Feierabend J, Mikus M (1977) Occurrence of a high temperature sensitivity of chloroplast ribosome formation in several higher plants. Plant Physiol 59(5):863–867
Fuesler TP, Wong YS, Castelfranco PA (1984) Localization of Mg-chelatase and Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase activities within isolated, developing cucumber chloroplasts. Plant Physiol 75(3):662–664
Gao ZM, Fan SH, Gao J, Li XP, Cai CJ, Peng ZH (2006) Extract genomic DNA from Phyllostachys edulis by CTAB-based method. Forest Res 19(6):725–728
Gao ZM, Wang XC, Peng ZH, Zheng B, Liu Q (2012) Characterization and primary functional analysis of phenylalanine ammonia-lyase gene from Phyllostachys edulis. Plant Cell Rep 31(7):1345–1356
Gao ZM, Liu Q, Zheng B, Chen Y (2013) Molecular characterization and primary functional analysis of PeVDE, a violaxanthin de-epoxidase gene from bamboo (Phyllostachys edulis). Plant Cell Rep 32(9):1381–1391
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Acids Res 27(1):297–300
Huang W, Zhang SB, Cao KF (2010) The different effects of chilling stress under moderate light intensity on photosystem II compared with photosystem I and subsequent recovery in tropical tree species. Photosynth Res 103(3):175–182
Jiang ZH, Peng ZH, Gao ZM, Liu C, Yang CH (2012) Characterization of different isoforms of the light-harvesting chlorophyll a/b complexes of photosystem II in bamboo. Photosynthetica 50(1):129–138
Kovacevic D, Dewez D, Popovic R (2007) Irradiation-induced in vivo re-localization of NADPH-protochlorophyllide oxidoreductase from prolamellar body to stroma of barley etioplast. Photosynthetica 45(1):105–109
Kuhlmann M, Horvay K, Strathmann A, Heinekamp T, Fischer U, Böttner S, Dröge-Laser W (2003) The alphahelical D1 domain of the tobacco bZIP transcription factor BZI-1 interacts with the ankyrin-repeat protein ANK1 and is important for BZI-1 function, both in auxin signaling and pathogen response. J Biol Chem 278(10):8786–8794
Liu HH, Zheng CC (2008) MPEC: an important gene in the chlorophyll biosynthesis pathway in photosynthetic organisms. Photosynthetica 46(3):321–328
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408
Meskauskiene R, Nater M, Goslings D, Kesster F, Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98(22):12826–12831
Moseley J, Quinn J, Eriksson M, Merchant S (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J 19(10):2139–2151
Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14(3):673–688
Nagata N, Tanaka R, Satoh S, Tanaka A (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17(1):233–240
Nantel A, Quatrano RS (1996) Characterization of three rice basic leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biology Chem 271(49):31296–31305
Ouchane S, Steunou AS, Picaud M, Astier C (2004) Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: a strategy adopted to bypass the repressive oxygen control system. J Biology Chem 279(8):6385–6394
Papenbrock J, Grimm B (2001) Regulatory network of tetrapyrrole biosynthesis-studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 213(5):667–681
Pinta V, Picaud M, Reiss-Husson F, Astier C (2002) Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J Bacteriol 184(3):746–753
Porra RJ (1997) Recent progress in porphyrin and chlorophyll biosynthesis. Photochem Photobiol 65(3):492–516
Porra RJ, Schafer W, Katheder I, Scheer H (1995) The derivation of the oxygen atoms of the 13(1)-oxo and 3-acetyl groups of bacteriochlorophyll a from water in Rhodobacter sphaeroides cells adapting from respiratory to photosynthetic conditions: evidence for an anaerobic pathway for the formation of isocyclic ring E. FEBS Lett 371(1):21–24
Pysh LD, Schmidt RJ (1996) Characterization of themaize OHP1 gene evidence of gene copy variability among inbreds. Gene 177(1–2):203–208
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17(5):471–482
Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, von Wettstein D, Merchant S, Gough SP, Hansson M (2005) Xantha-l encodes a membrane subunit of the aerobic Mgprotoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA 102(16):5886–5891
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour Lab Press, New York
Schreiber U, Bilger W, Neubauer C (1995) Chlorophyll fluorescence as a nonintrusive indicator for rapid assessment of in vivo photosynthesis. Ecophysiol Photosynthesis 100:49–70
Seong ES, Yoo JH, Lee JG, Kim HY, Hwang IS, Heo K, Kim JK, Lim JD, Sacks EJ, Yu CY (2013) Antisense-overexpression of the MsCOMT gene induces changes in lignin and total phenol contents in transgenic tobacco plants. Mol Biol Rep 40(2):1979–1986
Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucl Acids Res 38(Database issue):D161–D166
Subramaniam R, Desveaux D, Spickler C, Michnick SW, Brisson N (2001) Direct visualization of protein interactions in plant cells. Nat Biotechnol 19(8):769–772
Suzuki JY, Bollivar DW, Bauer CE (1997) Genetic analysis of chlorophyll biosynthesis. Annu Rev Genet 31:61–89
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol 119(1):143–152
Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100(26):16119–16124
Wang HS, Zhu ZJ, Feng Z, Zhang SG, Yu C (2012) Antisense-mediated depletion of GMPase gene expression in tobacco decreases plant tolerance to temperature stresses and alters plant development. Mol Biol Rep 39(12):10413–10420
Wong YS, Castelfranco PA, Goff DA, Smith KM (1985) Intermediates in the formation of the chlorophyll isocyclic ring. Plant Physiol 79(3):725–729
Xu J, Deng Y, Li Q, Zhu X, He Z (2014) STRIPE2 encodes a putative dCMP deaminase that plays an important role in chloroplast development in rice. J Genet Genomics 41(10):539–548
Zhao HS, Peng ZH, Fei BH, Li LB, Hu T, Gao ZM, Jiang ZH (2014) BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database (Oxford):bau006
Zheng CC, Porat R, Lu P, O’Neill SD (1998) PNZIP is a novel mesophyll-specific cDNA that is regulated by photochrome and a circadian rhythm and encodes a protein with a leucine zipper motif. Plant Physiol 116(1):27–35
Acknowledgments
This work received financial support from the Special Fund for Forest Scientific Research in the Public Welfare from State Forestry Administration of China (No. 201504106) and the National Science Foundation of China (No. 31370588 and No. 31400557).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by B. Li.
L. Yang and Y. Lou contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, L., Lou, Y., Peng, Z. et al. Molecular characterization and primary functional analysis of PeMPEC, a magnesium-protoporphyrin IX monomethyl ester cyclase gene of bamboo (Phyllostachys edulis). Plant Cell Rep 34, 2001–2011 (2015). https://doi.org/10.1007/s00299-015-1846-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1846-1