Abstract
Key message
A novel isoflavone 7- O -glucosyltransferase PlUGT1 was isolated from Pueraria lobata . PlUGT1 could convert daidzein to daidzin, genistein to genistin as well as formononetin to ononin.
Abstract
Pueraria lobata roots are traditionally consumed as a rich source of isoflavone glycosides that have various human health benefits. However, to date, the genes encoding isoflavone UDP-glycosyltransferases (UGTs) have only been isolated from the roots of soybean seedlings (GmIF7GT), soybean seeds (UGT73F2) and Glycyrrhiza echinata cell suspension cultures (GeIF7GT). To investigate the isoflavone metabolism in P. lobata, 40 types of partial UGT cDNAs were isolated from P. lobata, and seven full-length UGT candidates with preferential expression in roots were identified. Functional assays in yeast (Saccharomyces cerevisiae) revealed that one of these UGT candidates, designated PlUGT1 (official UGT designation UGT88E12), efficiently glycosylated isoflavone aglycones at the 7-hydroxy group. Recombinant PlUGT1 purified from Escherichia coli cells was characterized and shown to be relatively specific for isoflavone aglycones, while flavonoid substrates were poorly accepted. The biochemical results suggested that PlUGT1 was an isoflavone 7-O-glucosyltransferase. The deduced amino acid sequence of PlUGT1 shared only 26 % identity with GeIF7GT, 27 % with UGT73F2 and 63 % with GmIF7GT. The PlUGT1 gene was highly expressed in P. lobata roots relative to other organs and strongly induced by methyl jasmonate signal in P. lobata cell suspension culture. The transcript abundance of PlUGT1 was correlated with the accumulation pattern of isoflavone glycosides such as daidzin in P. lobata plants or in cell suspension culture. The biochemical properties and gene expression profile supported the idea that PlUGT1 could play a role in isoflavone glycosylation in P. lobata.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The glycosylation of flavonoids contributes to the chemical diversity and medicinal importance of flavonoid glycosides due to differences in glycosylation positions, various numbers of the glycosyl moiety, glycosidic structures and the nature of glycosidic bonds (carbon–carbon or oxygen–carbon). Being a flavonoid branch, isoflavonoids are predominantly biosynthesized in legumes, and over 50 kinds of glycoconjugated isoflavones have been identified (Nagashima et al. 2004). Some of the isoflavonoid malonylglycosides function as key signaling molecules in the formation of the nitrogen-fixing nodules on leguminous plant roots (Dakora et al. 1993). In addition, isoflavonoid glycosides exhibit a number of pharmacological and nutraceutical properties that are beneficial to human health (Alekel et al. 2000; Lamartiniere 2000; Merz-Demlow et al. 2000). Therefore, it is important to understand the glycosylation of isoflavonoids and to engineer these compounds for plant and human benefit.
Although the formation of the isoflavonoid skeleton has been extensively characterized (Akashi et al. 1999, 2005; Jung et al. 2000; Steele et al. 1999), the sequential modifications of isoflavonoid metabolism, such as glycosylation, O-methylation and prenylation, remain relatively unexplored. Glycosylation is mainly catalyzed by UDP-glycosyltransferases (UGTs) that transfer the glycosyl group from an activated donor sugar onto low-molecular weight acceptors. A large number of glycosyltransferase families have been reported, and the glycosyltransferases involved in secondary metabolism are categorized into family 1 (Bowles et al. 2005; Sawada et al. 2005). Complementary DNAs (cDNAs) encoding isoflavone 7-O-glucosyltransferases have so far only been identified from Glycyrrhiza echinata cell suspension cultures (Nagashima et al. 2004), the roots of soybean (Glycine max) seedlings (Noguchi et al. 2007) and soybean seeds (Dhaubhadel et al. 2008). At the protein level, relatively pure isoflavone 7-O-glucosyltransferase has been isolated from soybean seedlings (Noguchi et al. 2007) and chickpea (Cicer arietinum L.) (Koster and Barz 1981).
Pueraria lobata is a leguminous plant, and its roots have been ascribed a number of pharmacological properties, including chemoprevention of migraine, hypertension and alcoholism (Keung et al. 1996; Thiem 2003); antioxidants associated with the reduced cardiovascular disorder (Lee 2004) and other medicinal effects (Boue et al. 2003). Most pharmacological activities of P. lobata extracts are due to the presence of isoflavone C- and O-glycosides, such as daidzin (Boue et al. 2003; Keung and Vallee 1998), puerarin (Lin et al. 1996; Wu et al. 2009; Zhang et al. 2004), genistin (Jin et al. 2012; Yasuda et al. 2005; Zhao et al. 2011) and possibly ononin (Auyeung et al. 2012; Li et al. 2010).
Despite the isoflavone glycosides in P. lobata exhibiting great benefits to human health, the key genes for isoflavone glycosylations in this plant have received limited study. From P. lobata, He et al. (2011) recently used a genetic approach to identify a glycosyltransferase designated as GT04F14 that could catalyze the O-glycosylations of flavones and isoflavones. Compared to some flavone substrates, the relative activities of GT04F14 toward all the isoflavones tested in their study were much lower, suggesting that it might not be an isoflavone-specific glycosyltransferase. Although there is still no molecular evidence to elucidate the isoflavone glycosylations in P. lobata, biochemical experiments (He et al. 2011) and knowledge of C-glycosylation of flavonoids (Brazier-Hicks et al. 2009) in cereals indicated that puerarin biosynthesis might be in competition with daidzin synthesis utilizing the same substrate 2,7,4′-trihydroxyisoflavanone (Fig. 1). As part of an attempt to understand the isoflavone glycosylations at the molecular level, we investigated and report here on the molecular cloning and functional characterizations of a novel isoflavone 7-O-glucosyltransferase (denoted PlUGT1, official UGT designation UGT88E12) from the roots of P. lobata. These findings will aid understanding of the isoflavonoid metabolism in P. lobata and provide new gene resources to facilitate further metabolic engineering of isoflavone O-glycosides production in other plants and microbial hosts.
Materials and methods
Plant materials and chemicals
Seeds of P. lobata were collected from Langxi County, Anhui Province, China. The seeds were sterilized with 10 % hydrogen peroxide, and germinated on the agarized (0.8 %) Murashige and Skoog (MS) medium (Murashige and Skoog 1962) at 25 °C in darkness. After germination, the seedlings were grown at 25 °C with 14/10 h of a light (150 μmol m−2s−1)/dark cycle. The 45-day-old axenic seedlings were then used as the plant materials in the current research. The chemical standards isoliquiritigenin, apigenin and luteolin were purchased from Shanghai Forever Biotech Company (Shanghai, China) and other chemical standards were purchased from Shanghai Source Leaf Biological Technology Company (Shanghai, China). All organic solvents used for high-performance liquid chromatography (HPLC) were from Wuhan Analytical Reagent Company (Wuhan, China). Unless specified otherwise, all enzymes were from Takara Company (Takara, Dalian, China).
Partial sequence-isolation and spatial expression analysis of UGT cDNAs
Total RNA was extracted from the roots of P. lobata seedlings using an EASYspin Plant RNA isolation kit (Aidlab, Beijing, China). The first-strand cDNA was prepared from 1 μg of the total RNA using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and the primer 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTT-3′. To isolate 3′-terminus cDNA sequences of family 1 UDP-glycosyltransferases from P. lobata, a degenerate oligonucleotide 5′-TGACNCAYTGYGGNTGGAAYWC-3′ was designed based on the highly conserved Putative Secondary Plant Glycosyltransferase (PSPG) motif found at the C-termini (Hughes and Hughes 1994; Paquette et al. 2003). Some partial glycosyltransferase cDNAs were obtained by oligo (dT)-adaptor primer and degenerate primer designed based on the PSPG (Nagatoshi et al. 2011; Yu et al. 2011). The partial transcripts of glycosyltransferase cDNAs were obtained by reverse transcription PCR (RT-PCR) with the degenerate primer 5′-TGACNCAYTGYGGNTGGAAYWC-3′ and the adaptor primer 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG-3′. The PCR products were cloned into the pMD18-T vector (Takara) and sequenced.
For gene expression analysis in different organs, total RNAs were prepared from the roots, stems and leaves of P. lobata seedlings with an EASYspin Plant RNA isolation kit and treated with DNase I to remove contaminating genomic DNA. M-MLV reverse transcriptase (Invitrogen) and oligo dT primer were used for synthesis of the first-strand cDNA. Quantitative RT-PCR (qRT-PCR) was performed on an ABI StepOne Plus Cycler with FastStart Universal SYBR Green Master (ROX) (Roche, Mannheim, Germany). The thermal cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and then 55 °C for 1 min. All real-time PCRs were performed in three independent biological and three technical repeats. Expression was calculated relative to P. lobata actin gene (GenBank accession no. HO708075) to minimize variations in cDNA template levels. Two-tailed t test (confidence interval 95 %) was performed using GraphPad Prism 5 software to evaluate the difference in the gene expressions among different organs. All primers used for qRT-PCR are listed in Table S1.
Cloning and phylogenetic analysis of the full-length cDNAs of UGT candidates
PlUGT1, PlUGT13, PlUGT18, PlUGT22, PlUGT35, PlUGT39 and PlUGT40 were considered as candidates for isoflavone biosynthesis in P. lobata. The 5′-ends of PlUGT1, PlUGT13, PlUGT18 and PlUGT22 cDNAs were successfully recovered by 5′ rapid-amplification of cDNA ends (RACE) technique. Based on the RACE sequence information, the full-length cDNAs of PlUGT1, PlUGT13, PlUGT18 and PlUGT22 were then amplified by standard RT-PCR. For the other candidates, a BLAST search showed that the 3′-end transcripts of PlUGT35, PlUGT39 and PlUGT40 displayed extremely high identities with the previously identified putative glucosyltransferase genes from P. lobata, GT19J14 (GenBank accession no. HQ219051), GT18P15 (accession no. HQ219050) and GT21C20 (accession no. HQ219052), respectively. Therefore, based on the nucleotide sequences of these previously isolated putative glucosyltransferases, the full-length cDNAs of PlUGT35, PlUGT39 and PlUGT40 were simply amplified by RT-PCRs using the P. lobata root cDNA as the template. All primers for the cDNA 5′-end amplifications and full-length clones are listed in Table S2 and Table S3.
For the phylogenetic analysis, the deduced amino acid sequences of the glucosyltransferases from P. lobata were aligned with other members of the family 1 glucosyltransferases using the CLUSTAL X Version 2.0 program (Larkin et al. 2007). The phylogenetic tree was generated by the neighbor-joining method of the MEGA 6.0 program (Tamura et al. 2013), using 1,000 bootstrap replications. The GenBank accession numbers of the sequences used in the phylogenetic analysis are summarized in Table S4.
In vivo functional analysis of UGT candidates in yeast
The coding regions of PlUGT1, PlUGT13, PlUGT18, PlUGT22, PlUGT35, PlUGT39 and PlUGT40 were cloned into the pESC-HIS yeast (Saccharomyces cerevisiae) expression vector (Stratagene, La Jolla, CA) under the control of a GAL 1 promoter at the multicloning sites. The primers used for cloning are shown in Table S3. The above constructs, as well as the empty vector pESC-HIS as a control, were transformed into S. cerevisiae WAT11 strain (Pompon et al. 1996) separately using a standard lithium acetate protocol (Gietz and Woods 2002). In vivo yeast assays were performed as previously described with minor modifications (Huang et al. 2012). After growth in synthetic dropout (SD) liquid medium containing 2 % (w/v) glucose, the transgenic yeast cells were washed three times with sterile water, and then re-suspended to an OD600 of 0.8 in SD medium containing 2 % (w/v) galactose. The substrate, daidzein, genistein, formononetin, daidzin or puerarin, was then added to a final concentration of 50 μM. After 48 h of galactose induction, the cultures were extracted with an equal volume of ethyl acetate. The extracts were air dried at room temperature and re-dissolved in methanol for HPLC analysis.
To investigate the roles of the above seven glucosyltransferase candidates in puerarin biosynthesis, the primers 5′-ATGTTGCTGGAACTTGCAC-3′ and 5′-TCAAGAAAGGACTTTAGC-3′ were used to get a cDNA encoding isoflavone synthase (IFS) (GenBank accession no. KC202929) from P. lobata of Langxi County by RT-PCR based on the IFS sequences (GenBank accession no. HQ219053) reported before. The coding region of IFS was cloned into the yeast expression vector pESC-TRP (Stratagene) to give the construct pESC-TRP—IFS and the construct was transformed into S. cerevisiae WAT11. The yeast cells expressing the IFS converted liquiritigenin to daidzein, indicating that the IFS had isoflavone synthase activity (data not shown). The construct pESC-TRP–IFS was co-transferred into S. cerevisiae WAT11 cells separately with each of the seven glucosyltransferase candidates that were constructed in the yeast expression vector pESC-HIS. The yeast cells harboring the two empty vectors pESC-HIS and pESC-TRP were used as the control yeast strain. After 48 h of growth in SD liquid medium containing 2 % (w/v) glucose, all transgenic yeast cells were washed with sterile water and re-cultured in SD liquid medium containing 2 % galactose. The substrate liquiritigenin at a final concentration of 50 μM was added into the cultures. After 48 h of galactose induction, the extracts were prepared as above for subsequent HPLC analysis.
In vitro PlUGT1 and PlUGT13 enzyme activity assays
The coding regions of PlUGT1 and PlUGT13 were cloned into the E. coli expression vector pET28a (Novagen, Darmstadt, Germany) in frame with the vector sequence encoding an N-terminal HIS tag. Primers used for the cloning are listed in Table S3. Protein expressions were induced in E. coli Rosetta (DE3) cells with 1 mM isopropyl-d-thiogalactopyranoside (IPTG) at 25 °C for 7 h and purified using Ni Sepharose 6 FAST Flow resin (GE Healthcare, Piscataway, NJ, USA) following the provided protocol. The purified polypeptides were analyzed by SDS-PAGE according to Sambrook and Russell (2001). Proteins in enzyme preparations were quantified by Bio-Rad protein assay.
The standard in vitro enzyme assays were conducted according to the previous studies with some modifications (He et al. 2011; Noguchi et al. 2007). 200 μl of the reaction mixture consisted of 2 μg of the purified enzyme, 50 mM trisHCl at pH 8.0, 1 mM UDP-glucose and 100 μM acceptor substrates (daidzein, genistein, formononetin, cyanidin, naringenin, liquiritigenin, quercetin, kaempferol, apigenin and luteolin). Except for the substrate cyanidin, the reactions were carried out at 30 °C for 3 h—cyanidin was found to be unstable at 30 °C for 3 h (data not shown), and so this assay was performed for only 0.5 h. The reactions were stopped with 200 μl of methanol, and 1 μl of the reaction mixture was directly injected for HPLC analysis.
For kinetic analysis of PlUGT1, a 200 μl reaction mixture containing 50 mM trisHCl at pH 8.0, 1 mM UDP-glucose and 0–480 μM of daidzein or genistein was pre-warmed at 30 °C, and the reaction initiated by adding 1 μg of the purified PlUGT1. The mixture was then incubated for 10 min and stopped with 200 μl of methanol prior to HPLC analysis. Kinetic parameters were determined by hyperbolic regression analysis using the Prism program (GraphPad).
Isoflavone glycosides biosynthesis and gene expression elicited by methyl jasmonate (MeJA)
Callus cultures were induced from the young leaves of P. lobata seedlings with MS medium supplemented with 1 mg l−1 naphthalene acetic acid (NAA), 2 mg l−1 benzylaminopurine and 8 g l−1 agar. The suspension cell line was initiated from the callus culture on a B5 liquid medium containing 1 mg l−1 2, 4-D, 1 mg l−1 NAA, 0.5 mg l−1 kinetin and 0.2 % casein acid hydrolysates, and sub-cultured every two weeks on a rotary shaker at 120 rpm at 25 °C. After 6–8 generations of the subculture, the homogenous cell line was treated with MeJA at a final concentration of 10 μM on the 4th day after the subculture (i.e., the time point within the logarithmic growth phase of the cell line). For the control, the cell culture was treated with equal volume of ethanol. At 0, 4, 24 and 48 h after the treatments, the cells were harvested, lyophilized and stored at −80 °C for investigation of isoflavone production and gene expression analysis. For the isoflavone extraction, 20 mg of the cells was extracted with 1 ml of methanol for 24 h at room temperature. The clear methanol extracts were volatilized to dryness, re-dissolved in 2 ml of HPLC grade methanol and filtered through a 0.22-μm nylon syringe filter prior to HPLC analysis. For the gene expression analysis, total RNA was extracted from the lyophilized cells using an EASYspin Plant RNA isolation kit, and the first-strand cDNA was prepared using Superscript III reverse transcriptase (Invitrogen) and oligo dT primer. qRT-PCR was performed to investigate the effects of MeJA treatment on the expression of the PlUGT1, PlUGT13 and GT04F14 genes (He et al. 2011). All real-time PCRs were performed in three biological and three technical repeats. The real-time PCR conditions for amplifying specific gene transcripts were the same as described above. The primers used for this part are listed in Table S1.
HPLC and LC–MS/MS analysis
HPLC analyses were performed on an LC-20AT instrument equipped with a binary pump, an autosampler and a photodiode array detector (Shimadzu, Kyoto, Japan). A Shimadzu Inertsil ODS-SP reverse phase column (250 × 4.6 mm, 5 μm) was used and the column temperature was set at 25 °C. To analyze the products extracted from the yeast strains co-expressing PlUGTs and IFS, 0.1 % phosphoric acid in Milli-Q water (solvent A) and HPLC grade acetonitrile (solvent B) were used as mobile phase and the system was equilibrated at 15 % B, and samples were separated with a multi-step gradient from 15 to 85 % B (10 min), 85 % B (6 min), 85 to 15 % B (6 min) and 15 % B (4 min) at a flow rate of 0.8 ml min−1 and detected at 260 nm. Other analyses were performed as follows: the system was equilibrated at 5 % B, and samples were separated with a multi-step gradient from 5 to 85 % B (10 min), 85 % B (6 min), 85 to 5 % B (6 min) and 5 % B (4 min) at a flow rate of 0.8 ml min−1. The detection wave length was set at 260 nm for daidzein, genistein, puerarin and formononetin; 280 nm for naringenin and liquiritigenin; 350 nm for apigenin and luteolin; 370 nm for quercetin and kaempferol; and 520 nm for cyanidin. For chemical quantifications, a standard calibration curve was assembled from a range of concentrations (1–100 μg ml−1) of each chemical standard.
LC–MS/MS analysis was performed using Accela LC system coupled with TSQ Quantum Access Max mass spectrometer (Thermo Scientific, USA). The column and analysis method was same with the HPLC analyses as described above. Electrospray ionization source (ESI) was applied and operated in positive ion mode. The MS data were recorded with ranges of m/z 100–500. Other parameter settings are the same as those described previously (Chen et al. 2013).
Results
Seven full-length putative UGT cDNA candidates were isolated from P. lobata roots
A UDP-glucosyltransferase (UGT) cDNA library was constructed from the roots of P. lobata. Through sequencing 1,005 clones, the sequences representing the 3′-terminus of 40 different UGT cDNAs were identified and designated PlUGT1–PlUGT40. The nucleotide sequences of these cDNA fragments were submitted to the GenBank database (Accession nos. KC473565–KC473604).
The transcript abundances of the UGTs in different organs of P. lobata were examined by qRT-PCR. Of the 40 UGTs, 10 showed preferential expression in the roots (Fig. 2), six (PlUGT5, PlUGT12, PlUGT28, PlUGT29, PlUGT34 and PlUGT38) were preferentially expressed in the stems, five (PlUGT3, PlUGT9, PlUGT11, PlUGT20 and PlUGT33) in the leaves and others showed similar expression in the roots, stems or leaves. Given that the isoflavone glucosides were predominantly biosynthesized in the roots of P. lobata, the UGTs with higher expression in the roots were considered as candidates for further study. Of these UGTs highly expressed in the root, PlUGT16 and PlUGT23 were most homologous with a previously reported P. lobata UGT GT04F14, which showed a higher activity toward flavones than isoflavones (He et al. 2011). The nucleotide sequences of PlUGT14 showed high identities with another previously identified P. lobata GT01K01, which seemed not to be active with both isoflavones and other flavonoids (He et al. 2011) (Table S5). Taken together, the other seven UGT genes (PlUGT1, PlUGT13, PlUGT18, PlUGT22, PlUGT35, PlUGT39 and PlUGT40) preferentially transcribed in the roots relative to other organs were subjected to functional analysis in the present study.
The gene expression analysis of different glycosyltransferase genes in different organs of P. lobata. Actin was used as an internal standard. The mRNA levels of each glucosyltransferase gene in different organs were normalized to that of actin and expressed relative to the values of roots (control), which were given an arbitrary value of 1. Error bars represent the standard errors (SE) of the means calculated from three biological replicates in three technical replicates. Asterisks indicate significant differences. *P < 0.05
5′ RACE and standard RT-PCR techniques were applied to successfully amplify their respective full-length cDNAs, which were deposited in the NCBI database (www.ncbi.nlm.nih.gov) (GenBank accession nos. KC473565–KC473572). The deduced amino acid sequences of these UGT candidates showed 19–53 % identity with each other.
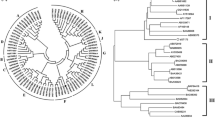
A phylogenetic tree was constructed to assess the relationships of all seven UGT candidates with other UGTs whose functions have been identified (Fig. 3). PlUGT1, PlUGT13 (official UGT designation UGT88H1) and PlUGT35 were clustered with UGT88 enzymes that included GmIF7GT from Glycine max (UGT88E3); UGT88E1 and UGT88E2 from Medicago truncatula; RhGT1 from Rosa hybrida; Am4′CGT (UGT88D3) from Antirrhinum majus and L4′CGT (UGT88D2) from Linaria vulgaris The members of the UGT 88 family have been reported to transfer the glycosyl group onto the 7-hydroxy group of (iso)flavones (Modolo et al. 2007; Noguchi et al. 2007). With respect to a special interest on isoflavonoid C-glucosyltransferase, it was informative to include a flavonoid C-glucosyltransferase OsCGT from rice (Brazier-Hicks et al. 2009) in the phylogenetic tree. Of the PlUGTs cloned in this study, OsCGT exhibits relatively higher homology to PlUGT1, PlUGT13, and PlUGT35. PlUGT1 was in the same branch as an isoflavone 7-O-glucosyltransferase GmIF7GT, which encouraged us to speculate that PlUGT1 might specifically function in isoflavone biosynthesis of P. lobata. The deduced amino acid sequence of PlUGT1 shared 63 % identity with GmIF7GT. PlUGT39 and PlUGT40 were found within a clade that included members of family 73 UGTs. A number of family 73 UGTs in this cluster are known to be stress inducible (Hirotani et al. 2000; Nagashima et al. 2004). Interestingly, PlUGT39 was in the same branch as another isoflavone-specific UGT GeIF7GT and UGT73F2. This suggested possible roles for PlUGT39 in isoflavone glycosylations in response to external stresses. PlUGT18 was in the same cluster as members of flavonoid 5-O-glucosyltransferases, while PlUGT22 showed amino acid similarities with both 5-O- and 3-O-glucosyltransferases.
Phylogenetic analysis of glucosyltransferases from P. lobata with other glucosyltransferases of known function. The deduced amino acid sequences were aligned with CLUSTAL X Version 2.0. The tree was constructed by the neighbor-joining method of MEGA 6.0. Numbers indicate the bootstrap values of 1,000 replicates. The scale bar represents 0.1 amino acid substitutions per site
Biochemical assays showed that PlUGT1 was an isoflavone 7-O-glucosyltransferase
For in vivo biochemical assays, the coding regions of all seven UGT candidates were cloned into a yeast expression vector pESC-HIS and expressed in WAT11 yeast cells. The yeast cells transformed with the empty vector pESC-HIS were used as the control. Functional assays were performed by testing the ability of yeast cells expressing the UGT candidates to glycosylate the isoflavone substrates (i.e., daidzein, genistein, formononetin, daidzin and puerarin). Initial screenings demonstrated that the yeast cells expressing PlUGT1 could efficiently convert daidzein to daidzin, genistein to genistin as well as formononetin to ononin (Fig. 4a–c). When fed with genistein, daidzein or formononetin, the yeast cells expressing PlUGT13 also produced their respective glucosides but at a low rate (Fig. 4d–f). Yeast cells expressing PlUGT1 and PlUGT13 were not able to glycosylate daidzin and puerarin. However, none of the other five UGT candidates showed any activity toward any of the isoflavone substrates used. It was especially surprising that the isoflavone substrates were not accepted by PlUGT39, which showed the closest relationship to UGT73F1 (an isoflavone-specific UGT) in the constructed phylogenetic tree (Fig. 3). The identities of daidzin, genistin or ononin produced by yeast cells were confirmed with respective authentic standards by liquid chromatography–mass spectrometry (LC–MS) (Fig. S1) and UV spectra (Fig. S2). Also, we have found that the 7-O-isoflavonoid glycosides were eluted at different retention times compared to the isoflavonoids glycosylated at other positions, e.g., the peaks corresponding to sophoricoside and genistin were observed at different times by HPLC analysis (Fig. S3). Those confirmed that both PlUGT1 and PlUGT13 functioned in transferring a glycosyl group onto the 7-hydroxy group of isoflavones.
Functional characterization of glucosyltransferases in yeast. HPLC analysis of the activities of PlUGT1 toward daidzein, genistein and formononetin is shown in a, b and c, respectively; the corresponding activities of PlUGT13 in d, e and f; the extracts from the transgenic yeast strains fed without any substrates are shown in g. PlUGT1 converted nearly all daidzein to daidzin (peak 1), genistein to genistin (peak 2) as well as formononetin to ononin (peak 3); and PlUGT13 only converted part of daidzein to daidzin (peak 4), genistein to genistin (peak 5) as well as formononetin to ononin (peak 6). WAT11 (pESC-HIS), the yeast strain carrying the empty vector pESC-His as a control; WAT11 (pESC-HIS-PlUGT1), the yeast strain expressing PlUGT1; WAT11 (pESC-HIS-PlUGT13), the yeast strain expressing PlUGT13. The comparisons LC–MS data and UV spectra of peaks 1–6 with corresponding authentic standards are shown in Fig. S1 and Fig. S2, respectively
To verify the enzyme activities in vitro, the open-reading frames of PlUGT1 and PlUGT13 were inserted into the vector pET28a for expression in E. coli cells. The recombinant enzymes were purified by immobilized metal affinity chromatography (Fig. S4) and applied to the in vitro reactions. Consistent with the in vivo assay results, both the purified PlUGT1 and PlUGT13 recognized daidzein, genistein and formononetin as acceptor substrates—PlUGT1 displayed about 4.1, 3.6 or 1.7 times activity of that of PlUGT13 when daidzein, genistein or formononetin was used as an acceptor, respectively. Given the similarity of PlUGT1 and PlUGT13 to anthocyanidin 5, 3-O-glucosyltransferase-like protein (Table S5), the purified PlUGT1 and PlUGT13 were tested for activities with anthocyanidin—no products were observed in the assays (data not shown). The acceptor specificities of PlUGT1 and PlUGT13 were determined using various types of flavonoid aglycones that are probably present in P. lobata. Of the isoflavone acceptors, 70–97 % were converted by PlUGT1 in the standard enzyme assays (Table 1). In contrast, only around 2–7 % of flavonoid substrates including liquiritigenin, naringenin and apigenin were converted by PlUGT1. Except for the 24.8 % of the chalcone substrate isoliquiritigenin converted, the other flavonoid substrates were not accepted by PlUGT1. The purified PlUGT13 exhibited relatively low activity toward both isoflavone and flavone acceptors, of which around 11–41 % of the substrates were converted. PlUGT13 showed no activities with various other substrates. Therefore, our biochemical assays indicated that PlUGT1 was an isoflavone UGT. The kinetic parameters for PlUGT1 with the isoflavones genistein and daidzein were measured using 1 mM UDP-glucose as the donor. The kinetic parameters for PlUGT1 are shown in Table 2 and the progress curve of PlUGT1 with genistein or daidzein is shown in Fig. 5. PlUGT1 had a slightly higher substrate preference for genistein than for daidzein, and the catalytic efficiency (k cat /K m) of PlUGT1 was about 1.5 times higher for genistein than for daidzein. These kinetic values are comparable to other UGTs (Achnine et al. 2005; Dhaubhadel et al. 2008).
Hyperbolic regression analysis of PlUGT1 with genistein (a) or daidzein (b) using the Prism program (GraphPad). The best-fit regression equations of PlUGT1 reacted with genistein and daidzein are V = 6.15[S]/(20.4 + [S]) and V = 6.09[S]/(29.9 + [S]), respectively. V, reaction velocity (μmol l−1min−1); [S], substrate concentration. The experiment was replicated for three times
Puerarin is a C-glycosylated isoflavone and exhibits various pharmacological activities. Puerarin might be produced via the enzymes IFS and UGT utilizing liquiritigenin as the substrate (Fig. 1). To examine the possibility of puerarin biosynthesis for our isolated seven UGT candidates, the P. lobata IFS was isolated and confirmed to be active with liquiritigenin (data not shown). The yeast WAT11 cells were transformed for the co-expression of the P. lobata IFS separately combined with each of the seven UGT candidates. The transgenic yeast cells were fed with the substrate liquiritigenin and induced. Puerarin production was not observed in any of the transgenic yeast strains. However, daidzin was detected from cells expressing IFS/PlUGT1, or IFS/PlUGT13 (Fig. S5).
PlUGT1 expression was correlated with daidzin production elicited by MeJA treatment
The P. lobata cell suspension culture was established and the effect of MeJA treatment on the isoflavone production in the cell cultures was investigated. By screening different concentrations of MeJA, our preliminary experiments showed that 10 μM MeJA was the most efficient inducer (Fig. S6), and the major isoflavone products in the cell suspension culture were daidzin and puerarin (data not shown). Compared to control cells, daidzin production was significantly elicited at 24 and 48 h after treatment with 10 μM MeJA, (Fig. 6). There was a similar increasing trend for puerarin biosynthesis (Li et al. 2014). Given the biochemical properties of PlUGT1, PlUGT13 and GT04F14 (He et al. 2011) from P. lobata in converting daidzein to daidzin, the effects on the gene expression of PlUGT1, PlUGT13 and GT04F14 were examined. Compared with controls, the gene expression of PlUGT1 was slightly elevated, while the expressions of PlUGT13 and GT04F14 were inhibited at 4 or 24 h post-treatment with 10 μM MeJA. However, compared to controls, there was a clear increase (around twofold) in PlUGT1 expression level in cells treated for 48 h (Fig. 7), whereas the transcript levels of both PlUGT13 and GT04F14 were down-regulated (Fig. 7). This observation suggested that the transcript abundance of PlUGT1 was correlated with daidzin accumulation in the cell suspension culture.
The gene expression analysis of PlUGT1, PlUGT13 and GT04F14 in P. lobata suspension cultures treated with 10 μM MeJA. Error bars represent the standard errors (SE) of the means calculated from three biological replicates. The relative expression value on the Y-axis represents the ratio of the gene expression in MeJA-treated plants to those in control plants
Discussion
Glycosylation increases the bioavailability of natural drugs by rendering them more water soluble and less toxic, and thus increasing scientific interest has been paid to molecular characterizations of glycosylation. We undertook molecular cloning of glucosyltransferase cDNAs from P. lobata roots due to their being a rich source of highly valuable C- and O-isoflavone glycosides. This led to the isolation of seven kinds of glucosyltransferase cDNAs that showed preferential expression in roots. Heterologous expression in yeast and E. coli cells showed that the PlUGT1 and PlUGT13 exhibited enzyme activities to O-glycosylate isoflavone substrates at the 7-hydroxy group. PlUGT1 accepted isoflavones as specific substrates, whereas other flavonoid substrates were poorly glycosylated. Based on the biochemical properties of the enzymes, the analysis of the gene expression profiles and the measurements of the enzyme-catalyzed products in the plant, it would be possible to estimate the in planta roles of PlUGT1. In P. lobata plants, isoflavone glycosides such as puerarin and daidzin mainly accumulate in roots (Fig. S7), which is consistent with the organ-specific expression pattern of PlUGT1 (Fig. 2). The quantity ratio of puerarin to daidzin is 2.92 in the roots, 7.02 in the stems and 0.07 in the leaves (Fig. S7) suggesting the difference in the enzyme concentration ratio of 7-O-glucosyltransferase to 8-C-glucosyltransferase between different organs. In P. lobata cell suspension cultures, compared to the controls, a significant improvement of daidzin production was observed at 24 and 48 h after the MeJA treatment (Fig. 6), while the increase of PlUGT1 transcription already has been observed at 4 h after the MeJA elicitation (Fig. 7). It is natural that the pathway gene expressions are modulated at earlier time than metabolite production in plant cells in response to signal molecules. Within the control cells treated by 95 % ethanol, an obvious increase of daidzin production was also observed at 48-h post-induction, which could be caused by the strengthened secondary metabolite biosynthesis in the culture with limited nutrients during the late-log phase of cell growth as observed in previous studies (Boonsnongcheep et al. 2010; Korsangruang et al. 2010). Based on the experiments using the plantlet and cell suspension systems of P. lobata, we concluded that the PlUGT1 expression correlated with daidzin production in P. lobata. Combining this with the substrate specificity of PlUGT1 suggests that PlUGT1 plays a role in the isoflavone 7-O-glycosylations in P. lobata. PlUGT1 was judged to be a novel isoflavone-specific 7-O-glucosyltransferase (IF7GT) based on the following observations: (1) PlUGT1 shares low amino acid sequence identity with the previously identified IF7GTs (around 26, 27, and 63 % amino acid identity to UGT73F1, UGT73F2, GmIF7GT, respectively); (2) Of the previously cloned IFGTs, PlUGT1 is most homologous to GmIF7GT (Fig. 3); however, the substrate specificities in their biochemical functions are different. For instance, GmIF7GT shows good activities toward the flavonoid substrates including quercetin and kaempferol (Noguchi et al. 2007) that were not glycosylated by PlUGT1 at all (Table 1).
The catalytic efficiency (k cat/K m) of glycosyltransferase GT04F14, previously isolated from P. lobata, for genistein is about 794 times lower than that of PlUGT1; GT04F14 also showed much higher activities to flavonols than isoflavones (He et al. 2011). The biochemical properties of GT04F14 (He et al. 2011) and its gene expression in response to MeJA treatment in the present study (Fig. 7) suggested that GT04F14 might not be an isoflavone-specific glycosyltransferase. PlUGT13, showing 50 % amino acid identity with PlUGT1, exhibited a relatively low activity toward isoflavone and other flavonoid acceptors, providing another example of the overlapping substrate specificity of these enzymes.
Similar three-dimensional structures have been identified for the UGT71G1 from M. truncatula and other glycosyltransferases despite low sequence identities between them (Shao et al. 2005). This prompted us to speculate that there might be a similar interaction between enzymes and isoflavone acceptors for PlUGT1, GmIF7GT, UGT73F1 and UGT73F2. Crystallographic study of a multifunctional triterpenoid/flavonoid glycosyltransferase UGT71G1 from M. truncatula and a flavonoid glucosyltransferase VvGT1 from Vitis vinifera demonstrated that the residues (His-22 in UGT71G1 and His-20 in VvGT1) and (Asp-121 in UGT71G1 and Asp-119 in VvGT1) played key roles in its catalysis (Shao et al. 2005; Offen et al. 2006). In contrast, mutations of the equivalent sites (His-15 and Asp-125) in GmIF7GT (an isoflavone 7-O-glucosyltransferase from Glycine max) revealed that the two residues were not critical for the GmIF7GT activity (Noguchi et al. 2007) which indicated that the substrate binding sites for the isoflavone 7-O-GTs might be different from flavonoid GTs. It would be of particular interest to determine the key residues critical for isoflavone glycosylations using structural and mutational investigations.
Puerarin is a C-glycosylated isoflavone from P. lobata and has a diverse range of health-promoting functions. The mechanism of forming an 8-C-glycosidic bond during puerarin biosynthesis is not yet clear. It was hypothesized that the 8-C-glycosidic group might be introduced at the trihydroxyisoflavanone stage by a C-glycosyltransferase, and the resultant 8-C-trihydroxyisoflavanone was then dehydrated by a trihydroxyisoflavanone dehydratase (HID), or dehydrated spontaneously in a low pH environment, to yield puerarin (Fig. 1). The formation of daidzin and puerarin diverges at the trihydroxyisoflavanone stage, and in terms of the spontaneous dehydrations in a low pH environment, the expression levels of the glucosyltransferases could be the determinants for metabolite channeling at both branches (Fig. 1). To screen the roles of our identified glucosyltransferases in puerarin biosynthesis, the IFS from P. lobata was cloned and co-expressed with the glucosyltransferase candidates in yeast cells. Using transgenic yeast cells fed with the substrate liquiritigenin, we were unable to identify any of the cloned glucosyltransferases as active in puerarin biosynthesis, but we detected the daidzin (peak1 and peak3 in Fig. 5S) and daidzein (peak2 and peak4 in Fig. 5S) from the cells expressing IFS/PlUGT1, or IFS/PlUGT13. This suggested that, in both transgenic yeast cells, 2,7,4′-trihydroxyisoflavanone was probably formed by the IFS using liquiritigenin as the substrate, spontaneously dehydrated to daidzein, and further converted to daidzin by PlUGT1 or PlUGT13. Interestingly, in the plant, the production of puerarin was higher than that of daidzin in roots (In 45-day-old seedlings of this study, the quantity ratio of puerarin to daidzin is 2.92), while in the cell suspension cultures the quantity ratio of puerarin to daidzin is varied between 0.24 and 1.05 at different stages of the cell suspension cultures (Li et al., 2014). The difference in the enzyme activities for the 7-O- and 8-C-glucosylations in different cell environments may eventually direct different carbon fluxes to the biosynthesis of puerarin and daidzin. Further investigation is necessary to discover the C-glucosyltransferase from P. lobata. Nevertheless, our data here suggested that PlUGT1 played roles in the isoflavone 7-O-glycosylations of P. lobata in vivo.
Conclusions
In conclusion, the cDNA encoding a novel isoflavone-7-O-glucosyltransferase (PlUGT1) was isolated from P. lobata. PlUGT1 is a counterpart in P. lobata responsible for UGT88E3 of soybean, and Fabaceae plant may have a common IF7GT responsible for isoflavone glucosylation. PlUGT1 shares low amino acid sequence identities with several previously identified IF7GTs. Our study supplied a new gene resource for the biosynthesis of isoflavone 7-O-glucosides in various organisms.
Abbreviations
- cDNAs:
-
Complementary DNAs
- HID:
-
Trihydroxyisoflavanone dehydratase
- HPLC:
-
High-performance liquid chromatography
- IFS:
-
Isoflavone synthase
- IPTG:
-
Isopropyl-D-thiogalactopyranoside
- LC–MS:
-
Liquid chromatography–mass spectrometry
- MeJA:
-
Methyl jasmonate
- MS:
-
Murashige and Skoog
- NAA:
-
Naphthalene acetic acid
- PSPG:
-
Putative secondary plant glycosyltransferase
- qRT-PCR:
-
Quantitative RT-PCR
- RACE:
-
Rapid-amplification of cDNA ends
- UGTs:
-
UDP-glycosyltransferases
References
Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41:875–887
Akashi T, Aoki T, Ayabe S (1999) Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 121:821–828
Akashi T, Aoki T, Ayabe S (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137:882–891
Alekel DL, Germain AS, Peterson CT, Hanson KB, Stewart JW, Toda T (2000) Isoflavone-rich soy protein isolate attenuates bone loss in the lumbar spine of perimenopausal women. Am J Clin Nutr 72:844–852
Auyeung KK, Law PC, Ko JK (2012) Novel anti-angiogenic effects of formononetin in human colon cancer cells and tumor xenograft. Oncol Rep 28:2188–2194
Boonsnongcheep P, Korsangruang S, Soonthornchareonnon N, Chintapakorn Y, Saralamp P, Prathanturarug S (2010) Growth and isoflavonoid accumulation of Pueraria candollei var. candollei and P. candollei var. mirifica cell suspension cultures. Plant Cell Tiss Organ Cult 101:119–126
Boue SM, Wiese TE, Nehls S, Burow ME, Elliott S, Carter-Wientjes CH, Shih BY, McLachlan JA, Cleveland TE (2003) Evaluation of the estrogenic effects of legume extracts containing phytoestrogens. J Agric Food Chem 51:2193–2199
Bowles D, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263
Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284:17926–17934
Chen F, Hao F, Li C, Gou J, Lu D, Gong F, Tang H, Zhang Y (2013) Identifying three ecological chemotypes of Xanthium strumarium glandular trichomes using a combined NMR and LC-MS method. PLoS One 8:e76621
Dakora FD, Joseph CM, Phillips DA (1993) Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol 101:819–824
Dhaubhadel S, Farhangkhoee M, Chapman R (2008) Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J Exp Bot 59:981–994
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
He X, Blount JW, Ge S, Tang Y, Dixon RA (2011) A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233:843–855
Hirotani M, Kuroda R, Suzuki H, Yoshikawa T (2000) Cloning and expression of UDP-glucose: flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210:1006–1013
Huang L, Li J, Ye H, Li C, Wang H, Liu B, Zhang Y (2012) Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta 236:1571–1581
Hughes J, Hughes MA (1994) Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. Mitochondrial DNA 5:41–49
Jin SE, Son YK, Min BS, Jung HA, Choi JS (2012) Anti-inflammatory and antioxidant activities of constituents isolated from Pueraria lobata roots. Arch Pharm Res 35:823–837
Jung W, Yu O, Lau SM, O’Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18:208–212
Keung WM, Vallee BL (1998) Kudzu root: an ancient Chinese source of modern antidipsotropic agents. Phytochemistry 47:499–506
Keung WM, Lazo O, Kunze L, Vallee BL (1996) Potentiation of the bioavailability of daidzin by an extract of Radix puerariae. Proc Natl Acad Sci USA 93:4284–4288
Korsangruang S, Soonthornchareonnon N, Chintapakorn Y, Saralamp P, Prathanturarug S (2010) Effects of abiotic and biotic elicitors on growth and isoflavonoid accumulation in Pueraria candollei var. candollei and P. candollei var. mirifica cell suspension cultures. Plant Cell Tiss Organ Cult 103:333–342
Koster J, Barz W (1981) UDP-glucose: isoflavone 7-O-glucosyltransferase from roots of chick pea (Cicer arietinum L.). Arch Biochem Biophys 212:98–104
Lamartiniere CA (2000) Protection against breast cancer with genistein: a component of soy. Am J Clin Nutr 71:1705S–1707S Discussion 1708S–1709S
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lee JS (2004) Supplementation of Pueraria radix water extract on changes of antioxidant enzymes and lipid profile in ethanol-treated rats. Clin Chim Acta 347:121–128
Li G, Zhang Q, Wang Y (2010) Chemical constituents from roots of Pueraria lobata. Zhongguo Zhong Yao Za Zhi 35:3156–3160
Li Z-B, Li C-F, Li J, Zhang Y-S (2014) Molecular cloning and functional characterization of two divergent 4-coumarate: coenzyme A ligases from kudzu (Pueraria lobata). Biol Pharm Bull 37:113–122
Lin RC, Guthrie S, Xie CY, Mai K, Lee DY, Lumeng L, Li TK (1996) Isoflavonoid compounds extracted from Pueraria lobata suppress alcohol preference in a pharmacogenetic rat model of alcoholism. Alcohol Clin Exp Res 20:659–663
Merz-Demlow BE, Duncan AM, Wangen KE, Xu X, Carr TP, Phipps WR, Kurzer MS (2000) Soy isoflavones improve plasma lipids in normocholesterolemic, premenopausal women. Am J Clin Nutr 71:1462–1469
Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64:499–518
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nagashima S, Inagaki R, Kubo A, Hirotani M, Yoshikawa T (2004) cDNA cloning and expression of isoflavonoid-specific glucosyltransferase from Glycyrrhiza echinata cell-suspension cultures. Planta 218:456–459
Nagatoshi M, Terasaka K, Nagatsu A, Mizukami H (2011) Iridoid-specific glucosyltransferase from Gardenia jasminoides. J Biol Chem 286:32866–32874
Noguchi A, Saito A, Homma Y, Nakao M, Sasaki N, Nishino T, Takahashi S, Nakayama T (2007) A UDP-glucose:isoflavone 7-O-glucosyltransferase from the roots of soybean (glycine max) seedlings. Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J Biol Chem 282:23581–23590
Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25:1396–1405
Paquette S, Moller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62:399–413
Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272:51–64
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, vol 1, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T (2005) UDP-glucuronic acid: anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280:899–906
Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17:3141–3154
Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367:146–150
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thiem B (2003) In vitro propagation of isoflavone-producing Pueraria lobata (Willd.) Ohwi. Plant Sci 165:1123–1128
Wu HQ, Guo HN, Wang HQ, Chang MZ, Zhang GL, Zhao YX (2009) Protective effects and mechanism of puerarin on learning-memory disorder after global cerebral ischemia-reperfusion injury in rats. Chin J Integr Med 15:54–59
Yasuda T, Endo M, Kon-no T, Kato T, Mitsuzuka M, Ohsawa K (2005) Antipyretic, analgesic and muscle relaxant activities of pueraria isoflavonoids and their metabolites from Pueraria lobata Ohwi-a traditional Chinese drug. Biol Pharm Bull 28:1224–1228
Yu H-S, Ma L-Q, Zhang J-X, Shi G-L, Hu Y-H, Wang Y-N (2011) Characterization of glycosyltransferases responsible for salidroside biosynthesis in Rhodiola sachalinensis. Phytochemistry 72:862–870
Zhang FR, Chen JZ, Zhu JH, Wang XX, Shang YP, Guo XG, Dai HM, Sun J (2004) Effects of puerarin on number and activity of endothelial progenitor cells from peripheral blood. Zhongguo Zhong Yao Za Zhi 29:777–781
Zhao C, Chan HY, Yuan D, Liang Y, Lau TY, Chau FT (2011) Rapid simultaneous determination of major isoflavones of Pueraria lobata and discriminative analysis of its geographical origins by principal component analysis. Phytochem Anal 22:503–508
Acknowledgments
This project was partially supported by the National Natural Science Foundation of China (Project No. 31170284), the Grant for One Hundred Talents Program of the Chinese Academy of Sciences, China (Project No. Y129441R01) and the Innovation Project of Chinese Academy of Science (Project No. KSCX2-EW-J-20).
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. F. Quiros.
J. Li and Z. Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Li, Z., Li, C. et al. Molecular cloning and characterization of an isoflavone 7-O-glucosyltransferase from Pueraria lobata . Plant Cell Rep 33, 1173–1185 (2014). https://doi.org/10.1007/s00299-014-1606-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1606-7