Abstract
Catharanthus roseus is an important medicinal plant and the sole commercial source of monoterpenoid indole alkaloids (MIA), anticancer compounds. Recently, triterpenoids like ursolic acid and oleanolic acid have also been found in considerable amounts in C. roseus leaf cuticular wax layer. These simple pentacyclic triterpenoids exhibit various pharmacological activities such as anti-inflammatory, anti-tumor and anti-microbial properties. Using the EST collection from C. roseus leaf epidermome (http://www.ncbi.nlm.nih.gov/dbEST), we have successfully isolated a cDNA (CrAS) encoding 2,3-oxidosqualene cyclase (OSC) and a cDNA (CrAO) encoding amyrin C-28 oxidase from the leaves of C. roseus. The functions of CrAS and CrAO were analyzed in yeast (Saccharomyces cerevisiae) systems. CrAS was characterized as a novel multifunctional OSC producing α- and β-amyrin in a ratio of 2.5:1, whereas CrAO was a multifunctional C-28 oxidase converting α-amyrin, β-amyrin and lupeol to ursolic-, oleanolic- and betulinic acids, respectively, via a successive oxidation at the C-28 position of the substrates. In yeast co-expressing CrAO and CrAS, ursolic- and oleanolic acids were detected in the yeast cell extracts, while the yeast cells co-expressing CrAO and AtLUP1 from Arabidopsis thaliana produced betulinic acid. Both CrAS and CrAO genes show a high expression level in the leaf, which was consistent with the accumulation patterns of ursolic- and oleanolic acids in C. roseus. These results suggest that CrAS and CrAO are involved in the pentacyclic triterpene biosynthesis in C. roseus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Catharanthus roseus (L.) G. Don has been one of the most extensively investigated medicinal plants in the past two decades, because it is the sole commercial source of the anticancer monoterpenoid indole alkaloids (MIA) (van der Heijden et al. 2004; O’Connor and Maresh 2006). The MIA biosynthetic pathway has been well characterized (El-Sayed and Verpoorte 2007) and, recently, triterpenoids like ursolic acid and oleanolic acid have also been found in considerable amounts in C. roseus leaf cuticular wax layer (Usia et al. 2005; Murata et al. 2008). These pentacyclic triterpenoids exhibit diverse pharmacological activities. For example, both oleanolic acid and ursolic acid are effective in protecting against chemically induced liver injury in laboratory animals (Liu 1995) and show anti-inflammatory, anti-tumor, anti-hyperlipidemic, anti-ulcer and anti-microbial properties (Liu 1995; Farina et al. 1998; Liu 2005). Betulinic acid is a lupane-type triterpene holding great promise in the treatment of human cancers (Fulda 2008). These compounds often occur mainly in the leaves and fruit cuticular wax layers of many plant species, such as apples (Malus domestica) (Bringe et al. 2006), tomatoes (Solanum lycopersicum) (Wang et al. 2010), grapes (Vitis vinifera) (Grncarevic and Radler 1971) and Catharanthus roseus (Murata et al. 2008), which corroborate the assumption of triterpenes being involved in plant defense.
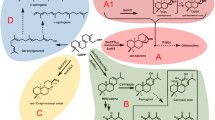
Although the pharmaceutical and physiological importance of these triterpenoids is known, understanding of their biosynthetic pathways remains limited (Augustin et al. 2011). The ursolic acid, oleanolic acid and betulinic acid are likely derived from α-amyrin, β-amyrin and lupeol, respectively, followed by a successive oxidation at the C-28 position (Fig. 1). It has been shown that the triterpene skeletons (α-amyrin, β-amyrin and lupeol) are cyclized from 2,3-oxidosqualene, a common precursor of phytosterols and triterpenoids (Abe et al. 1993). The enzymes involved in the formation of these triterpene backbones are generally named oxidosqualene cyclases (OSCs). More than 50 different OSCs have been cloned from various plant species and generate more than 100 different carbon skeletons which contribute enormously to the structural diversity of triterpenoids (Phillips et al. 2006). According to the product profiles, OSCs can be divided into two groups. One group is a monofunctional synthase yielding one specific product, such as the lupeol synthase (Herrera et al. 1998; Shibuya et al. 1999; Guhling et al. 2006), β-amyrin synthase (Kushiro et al. 1998; Kajikawa et al. 2005; Kirby et al. 2008; Shibuya et al. 2009), caloartenol synthase (Hayashi et al. 2000), isomultiflorenol synthase (Hayashi et al. 2001), cucurbitadienol synthase (Shibuya 2004), thalianol synthase (Fazio et al. 2004) and marneral synthase (Xiong et al. 2006). The other group is multifunctional synthase producing more than one product, like the AtLUP2 from Arabidopsis thaliana (Husselstein-Muller et al. 2001), PSM from Pisum sativum (Morita et al. 2000), KcMS from Kandelia candel (L.) Druce (Basyuni et al. 2006), OEA from Olea europaea (Smimaru H et al. 2007), SITTS2 from Solanum lycopersocum (Wang et al. 2010) and MdOSC1 from Malus × domestica (Brendolise et al. 2011). Interestingly, no monofunctional α-amyrin synthase has been identified and all triterpene synthases whose products including α-amyrin are classified as multifunctional OSCs.
Proposed biosynthetic pathways of ursolic-, oleanolic- and betulinic acids in plants. The ursolic- and oleanolic acid biosynthesis in C. roseus shown in the square was indicated by this work. CrAS, Catharanthus roseus mixed amyrin synthase; AtLUP1, Arabidopsis thaliana lupeol synthase; CrAO, Catharanthus roseus amyrin C-28 oxidase
Following the formation of the carbon skeletons, the triterpene alcohols are modified by various cytochrome P450s, dehydrogenases, reductases and other modification enzymes. Only a few P450s involved in triterpene biosynthesis have been identified, such as CYP93E1 responsible for the C-24 hydroxylation of β-amyrin and sophoradiol in the biosynthesis of soyasaponins (Shibuya et al. 2006), CYP88D6 for the C-11 oxygenation of β-amyrin in the glycyrrhetinic acid biosynthesis (Seki et al. 2008) and CYP51H10 (Sad2) for the avenacin biosynthesis (Qi et al. 2006). Recently, CYP716A12 from Medicago truncatula and two CYP716A12 homologs (CYP716A15 and CYP716A17) from Vitis vinifera were revealed to catalyze successive oxidation at the C-28 position of β-amyrin, α-amyrin and lupeol to yield oleanolic-, ursolic- and betulinic acids, respectively (Carelli et al. 2011; Fukushima et al. 2011). Cytochrome P450s catalyzing a three-step sequential oxidation have also been discovered in other metabolisms such as CYP71AV1 for artemisinin biosynthesis (Teoh et al. 2006), CYP701A1 for gibberellin biosynthesis (Helliwell et al. 1999) and CYP720B1 for diterpene resin acid biosynthesis (Ro et al. 2005).
To the best of our knowledge, the enyzmes involved in the triterpene biosynthesis in C. roseus have not been characterized. Here, we reported the cDNA cloning and functional characterization of a triterpene synthase (denoted as CrAS) and a triterpene C-28 oxidase (denoted as CrAO) from C. roseus. CrAS was identified to be a multifunctional OSC capable of forming α-amyrin and β-amyrin in a ratio of 2.5:1. CrAO was a multifunctional oxidase catalyzing successive oxidations at C-28 position of the pentacyclic triterpene skeletons. The expression patterns of the two genes are quite well correlated to the accumulation of the triterpenoids in C. roseus.
Materials and methods
Plant materials and chemicals
Catharanthus roseus (L.) G. Don seeds were obtained from Wuhan Botanical Garden, the Chinese Academy of Sciences. Seeds were germinated and grown in soil in a greenhouse. α-amyrin, β-amyrin, lupeol, uvaol, erythrodiol, betulin, ursolic acid, oleanolic acid and betulinic acid were purchased from Sigma-Aldrich (St Louis, MO, USA). Unless specified otherwise, all enzymes were from Takara Company (Takara, LN, China).
Full-length cDNA isolation and plasmid construction
C. roseus amyrin synthase cDNA (CrAS)
Among the ESTs in the leaf epidermomes of C. roseus in GenBank EST database (http://www.ncbi.nlm.nih.gov/dbEST) (Murata et al. 2008), 14 ESTs were selected as candidates encoding OSCs. Their dbEST Ids are FD661067, FD660779, FD660764, FD661171, FD661168, FD661154, FD661120, FD661048, FD661024, FD660932, FD660928, FD660921, FD660840 and FD661245. Except the FD661067, the other 13 ESTs could be assembled to the same contig which showed high identity to the 3′ end sequences of a putative OSC cDNA. Through the Genbank blastx search, the EST FD661067 showed a similarity to the 5′ end sequences of the same putative OSC cDNA, which led to a speculation that the above 14 ESTs might be from the same OSC cDNA. Therefore, gene-specific primers 1–2 (Table S1) were designed to amplify the putative OSC cDNA (CrAS) by RT-PCR using Phusion® Hot Start High-Fidelity DNA Polymerase (New England Biolabs, Beijing, China). CrAS was subcloned into the plasmid pESC-His (Stratagene, La Jolla, CA, USA) at the BamHI/SalI sites to give the construct pESC-His-CrAS.
C. roseus cytochrome P450 enyzme cDNAs
The C. roseus leaf epidermome ESTs in the GenBank EST database (http://www.ncbi.nlm.nih.gov/dbEST) were surveyed for sequences encoding cytochrome P450 enzymes and the selected ESTs were sorted into contigs with functional annotations (Murata et al. 2008). The contigs with unknown functions (CrContig3, CrContig4, CrContig5, CrContig8, CrContig9 and CrContig10) were selected for investigation of their potential oxidizing activities (Table S2). All the oligonucleotides used for PCRs are listed in the Supplemental Table S1. Thermal asymmetric interlaced PCR (TAIL-PCR) was performed to acquire the missing 5′ sequences of CrContig3 according to the protocol described by Liu and Whittier (Liu and Whittier 1995) using primers 3–6. The open reading frame (ORF) of the cDNA corresponding to CrContig3 was amplified by primers 7 and 8 and designated CYP71D1V1. To identify the missing 5′ sequences of CrContig4, 5′-RACE cDNA amplification was conducted using primers 9–12, following a 5′-RACE protocol (Invitrogen, Carlsbad, CA, USA). The obtained 5′ sequence indicated that CrContig4 and CrContig5 might be from a single cDNA, which was confirmed by amplifying the ORF of the cDNA using primers 13 and 14. The cDNA corresponding to CrContig4 and CrContig5 was designated CYP716AL1. 3′-RACE was performed to amplify the 3′ region of CrContig8 by primers 15–18. The ORF of the cDNA corresponding to CrContig8 was then recovered by RT-PCR with primers 19 and 20 and designated CYP71D1V2. Efforts to determine the full-length sequences of CrContig9 and CrContig10 were unsuccessful. Therefore, only cDNA clones CYP716AL1, CYP71D1V1 and CYP71D1V2 were further investigated in this study. The ORFs of CYP716AL1, CYP71D1V1 and CYP71D1V2 were digested with BamHI/KpnI, EcoRI/SacI and KpnI/NheI, respectively, and then ligated into the corresponding sites in pESC-Trp (Stratagene) to give the constructs pESC-Trp-CYP716AL1, pESC-Trp-CYP71D1V1 and pESC-Trp-CYP71D1V2.
Arabidopsis thaliana lupeol synthase cDNA (AtLUP1)
The open reading frame of AtLUP1 (Herrera et al. 1998) was amplified by RT-PCR from A. thaliana cDNA using the gene-specific primers 21 and 22 (Table S1). The resulting PCR product was sequenced and subcloned into the pESC-His vector (Stratagene) at the BamHI and SalI sites to generate a yeast expression construct pESC-His-AtLUP1.
Functional characterizations of cDNAs
In vivo functional assay of CrAS in yeast
The Sacchromyces cerevisiae WAT11 strain was transformed with the construct pESC-His-CrAS using a standard lithium acetate protocol (Gietz and Woods 2002) and an empty vector (pESC-His) was used as a control. The WAT11 strain was kindly provided by Dr. Phillippe Urban (CNRS, France) (Pompon et al. 1996). After 48 h of growth in SD dropout liquid medium containing 2 % (w/v) glucose, the transgenic yeast cells were washed three times in sterile water, resuspended to an OD600 of 0.8 in SD dropout medium containing 2 % (w/v) galactose and incubated for another 48 h. Cells were collected, refluxed with 2 mL 20 % KOH/50 %EtOH for 5 min and extracted with hexane. The hexane extracts were derivatized using Bis- N,O- (trimethylsilyl) trifluoroacetamide (BSTFA) at 80 °C for 30 min prior to GC–MS analysis.
In vitro functional screens of C. roseus P450 candidates in yeast
The constructs pESC-Trp-CYP716AL1, pESC-Trp-CYP71D1V1 and pESC-Trp-CYP71D1V2, as well as the empty vector pESC-Trp as the control, were transferred into S. cerevisiae WAT11 cells. After 48 h of galactose induction, yeast cells were collected, resuspended in 0.3 mL of 50 mM potassium phosphate buffer and broken using a Tissue Lyser (Retsch, Haan, Germany) with glass beads (0.4–0.6 mm diameter). Cell homogenates were centrifuged at 4000g for 5 min, and the supernatant was used as the total protein. The in vitro enzymatic reactions were performed in 0.5 mL of 50 mM potassium phosphate buffer (pH 7.5) containing 2 mg of the total protein, 20 mM glucose 6-phosphate, 2.5 U glucose-6-phosphate dehydrogenase, 2 mM NADPH and 80 μM substrate (α-amyrin, β-amyrin and lupeol). The reaction with each substrate was carried out independently. After incubation at 30 °C for 6 h, the reaction was stopped by adding 200 μL 2 M HCl and the reaction products were extracted with ethyl acetate three times. The ethyl acetate extracts were evaporated and derivatized using Bis- N,O-(trimethylsilyl) trifluoroacetamide (BSTFA) at 80 °C for 30 min prior to GC–MS analysis.
In vivo functional assays of CYP716AL1 in yeast
From the above in vitro experiments, CYP716AL1 was identified to be the C. roseus triterpene C-28 oxidase (CrAO). For the in vivo functional assay, CrAO was co-expressed in the WAT11 yeast strain with either CrAS or AtLUP1. Cells expressing CrAS or AtLUP1 alone were used as the controls. After 48 h of galactose induction, the transgenic yeast cells were collected and fractions from the medium, the cell surface and the cells were collected separately. The cell pellets were washed with 1 mL of alkaline buffer (pH 9.0, 50 mM Tris-HCl buffer) resulting in the cell surface fraction. The washed cells were re-suspended in 0.5 mL of neutral buffer (pH 7.0, 50 mM potassium phosphate buffer) and broken by glass beads to give the intracellular fraction. All the fractions were acidified to pH 2 using 2 M HCl, passed through a CNWBOUND LC-C18 column (CNW, Shanghai, China) and extracted with methanol. The resulting methanol extracts were air dried and derivatized with Bis-N,O-(trimethylsilyl)trifluoroacetamide (BSTFA) at 80 °C for 30 min prior to GC–MS analysis.
Analytical procedures
GC–MS analysis
GC–MS analyses were performed in an Agilent 7890A GC machine (Agilent Technologies, Waldbronn, USA) equipped with an HP-5MS column (0.25 mm ID × 30 m, 0.25 μm film thickness, Agilent) and an Agilent 5975C mass selective detector. One microliter of sample was injected into a splitless injection mode and the carrier gas was helium with a flow rate of 1.2 mL/min. The injection temperature was 250 °C. For analyzing the products from CrAO in vitro assays, the GC oven temperature was programmed from 80 °C to 310 °C at 20 °C/min and held for 15 min. For other compounds analyzed, the initial oven temperature was set at 80 °C for 2 min, followed by a 20 °C/min ramp to 290 °C, held at 290 °C for 20 min. Full mass sepctra were generated for metabolite identification by scanning within the m/z range of 50–600. Compounds were identified by comparing their retention times and mass spectra with those of authentic standards.
LC–MS analysis
LC–MS analysis was performed on an Agilent 1100 instrument (Agilent) equipped with a binary pump, an autosampler, a UV detection system and a column thermostat (thermostatted column compartment). An Agilent HC-C18 reverse phase column (250 × 4.6 mm, 5 μm) was used with acetonitrile/0.1 % formic acid (V/V = 90:10) as the mobile phase at a flow rate of 0.5 mL/min. The column temperature was set at 25 °C and the detection wave length was 214 nm. The LC/MSD trap VL mass spectrometer was equipped with an ESI source. The ionization mode was negative. Compounds were identified by comparing their retention times and mass spectra with those of authentic standards.
Determination of ursolic- and oleanolic acids in Catharanthus roseus
To investigate the accumulation of ursolic- and oleanolic acids in different tissues of C. roseus, fresh materials (roots, stems, leaves, flowers, fruits) were collected and dried at 60 °C and ground to powder. 0.2 g of dried materials was accurately weighed and extracted with 5 mL of 95 % ethanol for 2 h followed by 30 min of ultrasonic extraction. After filtration, the residues were washed three times with 95 % ethanol and all filtrates were combined and diluted to 10 mL. The extracts were filtered through a 0.22 μm microfilter prior to LC–MS analysis.
Gene expression analysis
The expression patterns of CrAS and CrAO in C. roseus were analyzed by quantitative reverse transcriptional polymerase chain reactions (QRT-PCRs). Total RNAs were isolated from roots, stems, leaves, flowers and fruits using plant total RNA isolation kit (BioTeck, Beijing, China) according to the manufacturer’s instructions. All of the total RNAs were treated with DNase I and reverse transcribed at 55 °C using the SuperScript III Reverse Transcriptase (Invitrogen) and the oligo d(T) primers. The QPCRs were performed with the Applied Biosystems StepOne Real-time PCR System using FastStart Universal SYBR Green Master (Roche, Mannheim, Germany) in three independent biological and four technical replicates. All the primers for QPCRs are shown in Supplemental Table S1. A C. roseus actin gene (GenBank accession number DQ117850) was amplified as an internal standard using the primers 23 and 24. The transcripts of CrAS were amplified with the primers 25 and 26, and the transcripts of CrAO were amplified with the primers 27 and 28. The thermal cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s.
Results
Cloning and functional characterization of a novel OSC in C. roseus
Detailed screening, assembling and blasting analysis of EST library from C. roseus leaf epidermomes (http://www.ncbi.nlm.nih.gov/dbEST) resulted in 14 putative OSC ESTs, which might belong to a single OSC cDNA. Indeed, a full-length cDNA (denoted as CrAS, GenBank Accession Number JN991165) was successfully amplified from C. roseus by RT-PCR with gene-specific primer pairs designed according to the above ESTs. The ORF of CrAS was 2289-bp long and encoded a 87.73-kDa protein (762 amino acids) with a calculated pI of 6.69. CrAS showed 86 % identity with a multifunctional OSC named OEA from Olea europaea.
The function of CrAS was investigated by overexpressing in S. cerevisiae WAT11 that synthesizes 2,3-oxidosqualene endogenously. The yeast cells overexpressing CrAS produced two new products which were not present in the control yeast cells carrying the empty vector pESC-His (Fig. 2). The two new compounds were identified as α-amyrin and β-amyrin, respectively, by comparing their retention times and mass spectra with those of authentic standards. The ratio of α-amyrin to β-amyrin was about 2.5:1. These results clearly indicated that CrAS was a multifunctional OSC producing both α- and β-amyrin.
GC–MS analysis of the products in S. cerevisiae strain expressing CrAS. β-Amyrin (peak 1) and α-amyrin (peak 2) were produced by the yeast cells expressing CrAS, while not found in the yeast cells carrying the empty vector pESC-His. Retention time and mass spectra of peak 1 and 2 are identical to the authentic β-amyrin and α-amyrin, respectively
Cloning and functional characterization of a triterpene C-28 oxidase from C. roseus
The next important step of triterpene acid biosynthetic pathway in C. roseus is the oxidation at C-28 position of the basic triterpene skeletons such as α-amyrin and β-amyrin. This C-28 oxidation has been proposed to be catalyzed by cytochrome P450 enzymes. Since cytochrome P450s occur in a diverse subfamily, it is a challenge to identify the P450 genes involved in the biosynthesis of specific compounds. When we started this study, cytochrome P450 enzymes involved in ursolic acid biosynthesis had not yet been isolated from any plants.
Ursolic acid is produced exclusively in the epidermis of many plant species including C. roseus (Pereira et al. 2005; Yu et al. 2007), which drives us to screen triterpene C-28 oxidation cytochrome P450 enzymes candidate genes from a C. roseus leaf epidermis EST database available in GenBank (Murata et al. 2008). The ESTs encoding cytochrome P450s were assembled into 15 contigs, 8 of which were annotated as indole alkaloid biosynthetic genes (Table S2). Six contigs (CrContig3, CrContig4, CrContig5, CrContig8, CrContig9 and CrContig10) with unknown functions were considered as potential candidates. None of the EST candidates were full-length cDNA; therefore, RACE and TAIL-PCR techniques were used to isolate their full-length sequences. Three full-length cDNAs were successfully amplified from C. roseus leaf cDNAs, while those of CrContig9 and CrContig10 were not able to be amplified from the same cDNAs pool.
Further sequence analysis revealed that CrContig4 and CrContig5 belonged to a single cytochrome P450 gene of CYP716A subfamily, and this full-length cDNA was designated CYP716AL1 (GenBank Accession Number-JN565975). The functions of most members of the CYP716 family are poorly understood. The CYP716AL1 ORF encodes a polypeptide of 480 amino acids showing 80 % identity with a cytochrome P450 enzyme from Panax notoginseng (AED99868), 76 % identify with CYP716A15 from Vitis vinifera (BAJ84106) and 74 % identity with CYP716A12 from Medicago truncatula (ABC59076).
The full-length cDNAs obtained corresponding to CrContig3 and CrContig8 encode cytochrome P450 enzymes belonging to the CYP71D subfamily and were named CYP71D1V1 (GenBank Accession Number JN613015) and CYP71D1V2 (GenBank Accession Number JN613016), respectively. CYP71D1V1 showed 39–54 % identity with the CYP71 enzymes of V. vinifera, Populus trichocarpa and Ricinus communis in GenBank, while CYP71D1V2 had 48–56 % identity with the sequences of CYP71D1 from C. roseus and CYP71D51 from Nicotiana tabacum.
For the functional analysis in vitro, cell-free extracts prepared from yeast overexpressing CYP71D1V1, CYP71D1V2 and CYP716AL1 were incubated with the ursane-type substrate α-amyrin, respectively, followed by GC–MS analysis of the enzymatic products. Two new compounds corresponding to uvaol and ursolic acid were produced by CYP716AL1-transformed yeast extract, but were not detected in the control reaction (Fig. 3a). No activity was found from CYP71D1V1- or CYP71D1V2-transformed yeast extracts (data not shown). The identities of uvaol and ursolic acid were confirmed by comparing their retention times and MS fragmentation patterns with those of the authentic standards (Fig. 3a, Fig. S1). The putative aldehyde intermediate was hardly detected probably due to the low amount of the product in the reaction. These results suggested that CYP716AL1 was a C. roseus α-amyrin oxidase (referred to as CrAO later) capable of converting α-amyrin into ursolic acid via the alcohol and aldehyde intermediates.
GC–MS analysis of the products in the in vitro assays using the total protein extracted from the yeast cells expressing CrAO. The total protein from the yeast cells harboring the empty vector pESC-Trp was used for the control reactions. Total ion chromatograms are shown for the reactions with the substrate α-amyrin yielding uvaol (peak 3) and ursolic acid (peak 4) (a), β-amyrin yielding erythrodiol (peak 5) and oleanolic acid (peak 6) (b), and lupeol yielding betulin (peak 7) and betulinic acid (peak 8) (c). All the products were confirmed with the corresponding authentic standards (Fig. S1)
Given the structural similarity between ursane-, oleanane-, and lupane-type substrates, β-amyrin and lupeol were included in the CrAO activity assay. The results indicated that CrAO catalyzed also similar successive oxidation at the C-28 position of both β-amyrin (Fig. 3b) and lupeol (Fig. 3c). The retention times and mass spectra of erythrodiol, oleanolic acid, betulin and betulinic acid were identical to those of their authentic standards (Fig. 3b, c, Fig. S1). Based on these results, CrAO was characterized to be a multifunctional C-28 oxidase with a triple oxidizing activity on three different types of basic triterpene skeletons (α-amyrin, β-amyrin and lupeol).
Co-expression of CrAS and CrAO in yeast
To further confirm the enzymatic activities described above, in vivo assays were conducted by co-expressing CrAS and CrAO in yeast and the de novo synthesis of ursolic- and oleanolic acids in the transgenic yeast is expected. The construct pESC-His-CrAS was co-transferred into the S. cerevisiae WAT11 strain with pESC-Trp-CrAO. The yeast cells transformed with the vector pESC-His-CrAS alone was used as the control. Compared with the control yeast strain carrying CrAS alone, the yeast cells co-expressing CrAS and CrAO synthesized four new compounds (Fig. 4a), erythrodiol, uvaol, oleanolic acid and ursolic acid, which were confirmed with the authentic standards. The ratio of ursolic- and oleanolic acid produced was 2.2:1 and the yields were 0.1 and 0.045 mg/L, respectively. To further confirm the oxidizing activity of CrAO with the substrate lupeol, the ORF of a lupeol synthase from A. thaliana (AtLUP1) was co-transferred into yeast cells together with CrAO. The yeast cells expressing AtLUP1/CrAO produced betulin and betulinic acid, which were not observed in the extracts from yeast cells expressing AtLUP1 alone (Fig. 4b). Moreover, small amount of oleanolic acid was detected in the AtLUP1/CrAO-expressing yeast strain because AtLUP1 is a multifunctional OSC which produces β-amyrin as one of the minor products (Herrera et al. 1998). Clearly, these in vivo experiments further demonstrated that CrAO catalyzes a successive three-step oxidation at the C-28 position of α-amyrin, β-amyrin and lupeol to yield the corresponding acids. In addition, nearly 90 % of the acids produced by the transgenic yeast were secreted into the media (data not shown).
In vivo functional assays of CrAO co-expressed with either CrAS (a) or AtLUP1 (b) in the yeast cells. GC–MS analysis was shown for the products from the yeast cells expressing CrAS/CrAO, and the cells expressing CrAS alone (a), and the yeast cells expressing AtLUP1/CrAO, and the cells expressing AtLUP1 alone (b). β-Amyrin (peak 1), α-amyrin (peak 2), uvaol (peak 3), ursolic acid (peak 4), erythrodiol (peak 5), oleanolic acid (peak 6), betulin (peak 7) and betulinic acid (peak 8) were confirmed with the corresponding chemical standards
CrAS and CrAO are correlated to the biosynthesis of ursolic- and oleanolic acids in C. roseus
The expression patterns of CrAS and CrAO in different organs of C. roseus were analyzed by QRT-PCR. CrAS was highly expressed in the leaves with relatively lower expression in flowers, fruits and stems, and nearly no expression in the roots. For CrAO, the highest mRNA levels were also detected in the leaves relative to the stems, flowers and fruits, and at an extremely low level in the roots (Fig. 5). The highest expressions of CrAS and CrAO in the leaves matched the distributions of ursolic- and oleanolic acids in different organs of C. roseus (Fig. 6). Moreover, in all the tissues investigated, the ratios of ursolic acid to oleanolic acid in were about 3:1. This in planta accumulation pattern of the acid products in C. roseus is quite similar to that observed in yeast cells co-expressing CrAS and CrAO. Thus, the enzymatic activities and gene expression patterns of CrAS and CrAO strongly suggest that the two enzymes are involved in the biosynthesis of pentacyclic triterpenoid acids in C. roseus.
QRT-PCR analysis of the transcripts of CrAS and CrAO in different organs of C. roseus. Primers specific for CrAS and CrAO were used to measure the transcript levels of the two genes in root, stem, leaf, flower and fruit. The expression level was normalized to that of a C. roseus actin gene. Error bars represent the standard errors (SE) of the means calculated from three independent experiments in four technical replicates
Discussion
Pentacyclic triterpenoids have attracted many researchers’ interests due to their anti-inflammatory, anti-tumor and antimicrobial activities. Ursolic acid and oleanolic acid accumulate predominantly in the leaf epidermis of C. roseus (Murata et al. 2008). To understand the biosynthetic pathway of these triterpene acids in C. roseus, molecular cloning and functional analysis of cDNAs encoding a 2,3-oxidosqualene cyclase (CrAS) and a cytochrome P450 enzyme (CrAO) from this plant species have been performed. The CrAS was identified to be a novel multifunctional OSC producing α- and β-amyrin in a ratio of 2.5:1. Monofunctional α-amyrin synthase has not been identified yet (Morita et al. 2000; Basyuni et al. 2006; 2007; Wang et al. 2010; Brendolise et al. 2011). The co-appearance of ursane- and oleanane-type triterpenoids in many plant species suggested that the monofunctional α-amyrin synthase might not exist in nature. To date, several multifunctional triterpene synthases have been reported to form α- and β-amyrin together with other triterpene products in various proportions, but only MdOSC1 from apple (Malus × domestica) exclusively produces α- and β-amyrin (Brendolise et al. 2011). CrAS is the second enzyme identified to entirely synthesize α- and β-amyrin. The key amino acid residues specifically controlling α-amyrin formation were not identified; this could be discovered by the crystal structure and site-directed mutagenesis of the enzyme.
Using the degenerate primers designed on the alignment of CrAS with other published amyrin synthases, we were not able to amplify any amyrin synthase sequences other than CrAS from C. roseus (data not shown), which suggested that CrAS might be the only amyrin synthase in C. roseus. Although the presence of other triterpene synthases in C. roseus could not be completely ruled out, the enzymatic property and gene expression pattern of CrAS strongly suggested its involvement in the triterpenoid biosynthesis in C. roseus.
Following the formation of the triterpene backbones by OSCs, these intermediates are usually modified by various cytochrome P450 enzymes, reductases and dehydrogenases. C-28 oxidized triterpenes are widely distributed in many plant species including C. roseus, but biosynthetic enzymes catalyzing the C-28 oxidations of triterpenes in C. roseus have not been characterized. CYP716A subfamily members were recently identified to catalyze the C-28 carboxylation of triterpene skeletons (α-amyrin, β-amyrin and lupeol) in M. truncatula (Carelli et al. 2011) and V. vinifera (Fukushima et al. 2011). In this work, we identified CrAO catalyzing a similar reaction from C. roseus. CrAO shared 74, 76 and 76 % identities with CYP716A12, CYP716A15 and CYP716A17 at the amino acid level, respectively, suggesting that the CYP716A subfamily is highly conserved in triterpene C-28 oxidations. Besides C-28 carboxylation, C-24 hydroxylation (Shibuya et al. 2006) and C-11 oxygenation (Seki et al. 2008) have been reported to be involved in the triterpenoid biosynthesis as well.
QRT-PCR results revealed that both CrAS and CrAO were expressed in all the aerial tissues with the highest expression levels in the leaves, but not in the roots of C. roseus. The gene expression data were consistent with the triterpenoid accumulation pattern in C. roseus, in which the highest concentrations of ursolic- and oleanolic acids were in the leaves. Moreover, the ratios of ursolic- and oleanolic acid produced in the yeast cells co-expressing CrAS and CrAO were close to that observed in all the aerial tissues of C. roseus. Therefore, it is reasonable to assume that CrAS and CrAO are involved in the productions of ursolic- and oleanolic acids in C. roseus. Although CrAO and other CYP716A members from different plants all function in C-28 oxidation of triterpene backbones, their organ-specific expressions are quite different. CYP716A12 from M. truncatula was expressed in both aerial and subterranean parts and preferentially expressed in the roots, which was consistent with the sapogenin distribution in the plant (Carelli et al. 2011). CYP716A15 and CYP716A17 from V. vinifera were coordinately expressed in the aerial parts, especially in the stems and fruit skins (Fukushima et al. 2011). Thus, the expression patterns of CYP716A subfamily members are closely associated with the distributions of triterpenoids in individual plant species.
Synthetic biological approaches have been used in the production of natural products with interesting pharmacological properties in microorganism by co-expressing genes from different plant origins; these compounds include mono-, sesqui- and diterpenoids (Reiling et al. 2004; Ro et al. 2006; Bohlmann and Keeling 2008). The identification of CrAO in this study allows potentially the production of structurally related triterpenes with C-28 carboxyl groups using synthetic biological approaches. Given that betulinic acid is a promising anti-cancer and anti-viral agent, we have shown the possibility of producing betulinic acid in yeast cells by co-expressing AtLUP1 from A. thaliana and CrAO from C. roseus. The betulinic acid yield in the transgenic yeast cells was pretty low (roughly below 0.1 mg/L) at the moment, and further optimizations to improve the yields of betulinic acid in microbial hosts will be of particular interests. Through the manipulations of the upstream isoprenoid pathway and downstream sterol pathway, a moderate increase in β-amyrin levels was obtained in S. cerevisiae (Kirby et al. 2008). With more detailed manipulation and optimization, it will be possible to engineer the microbes capable of producing a significantly high amount of triterpenoids, like the achievement in the production of sesquiterpene precursor of antimalarial drug artemisinin in engineered yeast (Ro et al. 2006).
Abbreviations
- CrAS:
-
Catharanthus roseus mixed amyrin synthase
- CrAO:
-
Catharanthus roseus amyrin oxidase
- EST:
-
Expressed sequence tag
- ORF:
-
Open reading frame
- LC–MS:
-
Liquid chromatogram mass spectrum
- P450:
-
Cytochrome P450 monoxygenase
- QRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- TAIL-PCR:
-
Thermal asymmetric interlaced polymerase chain reaction
References
Abe I, Rohmer M, Prestwich GD (1993) Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem Rev 93:2189–2206
Augustin JM, Kuzina V, Andersen SB, Bak S (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72:435–457
Basyuni M, Oku H, Inafuku M, Baba S, Iwasaki H, Oshiro K, Okabe T, Shibuya M, Ebizuka Y (2006) Molecular cloning and functional expression of a multifunctional triterpene synthase cDNA from a mangrove species Kandelia candel (L.) Druce. Phytochemistry 67:2517–2524
Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54:656–669
Brendolise C, Yauk Y-K, Eberhard ED, Wang M, Chagne D, Andre C, Greenwood DR, Beuning LL (2011) An unusual plant triterpene synthase with predominant α-amyrin producing activity identified by characterising oxidosqualene cyclases from Malus × domestica. FEBS J 278:2485–2499
Bringe K, Schumacher CFA, Schmitz-Eiberger M, Steiner U, Oerke E-C (2006) Ontogenetic variation in chemical and physical characteristics of adaxial apple leaf surfaces. Phytochemistry 67:161–170
Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, May S, Scotti C, Calderini O (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23:3070–3081
El-Sayed M, Verpoorte R (2007) Catharanthus terpenoid indole alkaloids: biosynthesis and regulation. Phytochem Rev 6:277–305
Farina C, Pinza M, Pifferi G (1998) Synthesis and anti-ulcer activity of new derivatives of glycyrrhetic, oleanolic and ursolic acids. Il Farmaco 53:22–32
Fazio GC, Xu R, Matsuda SP (2004) Genome mining to identify new plant triterpenoids. J Am Chem Soc 126:5678–5679
Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T (2011) CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol 52:2050–2061
Fulda S (2008) Betulinic acid for cancer treatment and prevention. Int J Mol Sci 9:1096–1107
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Grncarevic M, Radler F (1971) A review of the surface lipids of grapes and their importance in the drying process. AM J Enol Vitic 20:80–86
Guhling O, Hobl B, Yeats T, Jetter R (2006) Cloning and characterization of a lupeol synthase involved in the synthesis of epicuticular wax crystals on stem and hypocotyl surfaces of Ricinus communis. Arch Biochem Biophys 448:60–72
Hayashi H, Hiraoka N, Ikeshiro Y, Kushiro T, Morita M, Shibuya M, Ebizuka Y (2000) Molecular cloning and characterization of a cDNA for Glycyrrhiza glabra cycloartenol synthase. Biol Pharm Bull 23:231–234
Hayashi H, Huang P, Inoue K, Hiraoka N, Ikeshiro Y, Yazaki K, Tanaka S, Kushiro T, Shibuya M, Ebizuka Y (2001) Molecular cloning and characterization of isomultiflorenol synthase, a new triterpene synthase from Luffa cylindrica, involved in biosynthesis of bryonolic acid. Eur J Biochem 268:6311–6317
Helliwell CA, Poole A, Peacock WJ, Dennis ES (1999) Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol 119:507–510
Herrera JB, Bartel B, Wilson WK, Matsuda SP (1998) Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry 49:1905–1911
Husselstein-Muller T, Schaller H, Benveniste P (2001) Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol Biol 45:75–92
Kajikawa M, Yamato KT, Fukuzawa H, Sakai Y, Uchida H, Ohyama K (2005) Cloning and characterization of a cDNA encoding beta-amyrin synthase from petroleum plant Euphorbia tirucalli L. Phytochemistry 66:1759–1766
Kirby J, Romanini DW, Paradise EM, Keasling JD (2008) Engineering triterpene production in Saccharomyces cerevisiae-beta-amyrin synthase from Artemisia annua. FEBS J 275:1852–1859
Kushiro T, Shibuya M, Ebizuka Y (1998) β-Amyrin synthase cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur J Biochem 256:238–244
Liu J (1995) Pharmacology of oleanolic acid and ursolic acid. J Ethnopharmacol 49:57–68
Liu J (2005) Oleanolic acid and ursolic acid: research perspectives. J Ethnopharmacol 100:92–94
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Morita M, Shibuya M, Kushiro T, Masuda K, Ebizuka Y (2000) Molecular cloning and functional expression of triterpene synthases from pea. Eur J Biochem 267:3453–3460
Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20:524–542
O’Connor SE, Maresh JJ (2006) Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep 23:532–547
Pereira SI, Freire CSR, Pascoal Neto C, Silvestre AJD, Silva AMS (2005) Chemical composition of the epicuticular wax from the fruits of Eucalyptus globulus. Phytochem Anal 16:364–369
Phillips DR, Rasbery JM, Bartel B, Matsuda SPT (2006) Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol 9:305–314
Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272:51–64
Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A, Melton R, Osbourn A (2006) A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc Natl Acad Sci USA 103:18848–18853
Reiling KK, Yoshikuni Y, Martin VJJ, Newman J, Bohlmann J, Keasling JD (2004) Mono and diterpene production in Escherichia coli. Biotechnol Bioeng 87:200–212
Ro D, Arimura G, Lau S, Piers E, Bohlmann J (2005) Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102:8060–8065
Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943
Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105:14204–14209
Shibuya M (2004) Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 60:6995–7003
Shibuya M, Zhang H, Endo A, Shishikura K, Kushiro T, Ebizuka Y (1999) Two branches of the lupeol synthase gene in the molecular evolution of plant oxidosqualene cyclases. Eur J Biochem 266:302–307
Shibuya M, Hoshino M, Katsube Y, Hayashi H, Kushiro T, Ebizuka Y (2006) Identification of beta-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J 273:948–959
Shibuya M, Katsube Y, Otsuka M, Zhang H, Tansakul P, Xiang T, Ebizuka Y (2009) Identification of a product specific beta-amyrin synthase from Arabidopsis thaliana. Plant Physiol Biochem 47:26–30
Smimaru H, Orihara Y, Tansakul P, Kang YH, Shibuya MYE (2007) Production of triterpene acids by cell suspension cultures of Olea europaea. Chem Pharm Bull 55:784–788
Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS (2006) Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett 580:1411–1416
Usia T, Watabe T, Kadota S, Tezuka Y (2005) Cytochrome P450 2D6 (CYP2D6) inhibitory constituents of Catharanthus roseus. Biol Pharm Bull 28:1021–1024
van der Heijden R, Jacobs DI, Snoeijer W, Hallared D, Verpoorte R (2004) The Catharanthus alkaloids: pharmacognosy and biotechnology. Curr Med Chem 11:607–628
Wang Z, Guhling O, Yao R, Li F, Yeats TH, Rose JKC, Jetter R (2010) Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol 155:540–552
Xiong Q, Wilson WK, Matsuda SP (2006) An Arabidopsis oxidosqualene cyclase catalyzes iridal skeleton formation by Grob fragmentation. Angew Chem Int Ed Engl 45:1285–1288
Yu MML, Konorov SO, Schulze HG, Blades MW, Turner RFB, Jetter R (2007) In situ analysis by microspectroscopy reveals triterpenoid compositional patterns within leaf cuticles of Prunus laurocerasus. Planta 227:823–834
Acknowledgments
This project was partially supported by the Grant for One Hundred Talents Program of the Chinese Academy of Sciences, China (Project No. Y129441R01), the National Science and Technology Program of China during the Twelveth Five-Year Plan period and the National Natural Science Foundation of China (Project No. 91733A1001).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, L., Li, J., Ye, H. et al. Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus . Planta 236, 1571–1581 (2012). https://doi.org/10.1007/s00425-012-1712-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1712-0