Abstract
Heading date in rice is an important agronomic trait controlled by several genes. In this study, flowering time of variety Dianjingyou 1 (DJY1) was earlier than a near-isogenic line (named NIL) carried chromosome segment from African rice on chromosome 3S, when grown in both long-day (LD) and short-day (SD) conditions. By analyzing a large F2 population from NIL × DJY1, the locus DTH3 (QTL for days to heading on chromosome 3) controlling early heading date in DJY1 was fine mapped to a 64-kb segment which contained only one annotated gene, a MIKC-type MADS-box protein. We detected a 6-bp deletion and a single base substitution in the C-domain by sequencing DTH3 in DJY1 compared with dth3 in NIL, and overexpression of DTH3 caused early flowering in callus. Quantitative real-time PCR revealed that the transcript level of dth3 in NIL was lower than that DTH3 in DJY1 in both LD and SD conditions. The Early heading date 1 (Ehd1) which promotes the RFT1, was up-regulated by DTH3 in both LD and SD conditions. Based on Indel and dCAPs marker analysis, the dth3 allele was only present in African rice accessions. A phylogenetic analysis based on microsatellite genotyping suggested that African rice had a close genetic relationship to O. rufipogon and O. latifolia, and was similar to japonica cultivars. DTH3 affected flowering time and had no significant effect on the main agronomic traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
African cultivated rice (Oryza glaberrima), which originated from Oryza barthii (formerly known as Oryza breviligulata) in the Niger River delta (Jones et al. 1997; Khush 1997), has tolerances to deep water, iron toxicity and infertile soils, and resistance to bacterial leaf blight (Linares 2002). As a valuable genetic resource O. glaberrima is not fully explored. The genetic control of heading date in African rice when grown in China is not well understood. In fact, heading date is one of the most important traits for adaptation of rice (Oryza sativa L.) to cultivation areas and crop seasons. Large variations in heading date occur among cultivated rice varieties. Heading date is determined by a combination of basic vegetative growth (BVG), photoperiod-sensitivity (PS) and temperature-sensitivity (TS), which are each controlled by genetic factors and environmental conditions (Chang et al. 1969; Hosoi 1981; Sato and Takahashi 1983). Quantitative trait loci (QTLs) contributing to heading date in Oryza species have been intensively explored and mapped. The duration of BVG was controlled by seven loci, including Ef-1 on chromosome 10 (Tsai 1986), Ef-3 on chromosome 2 (Yang et al. 2005) and Ef-7 on chromosome 6 (Yuan et al. 2009). Most of the known heading date loci control PS. For example, Hd1, which is sensitive to day-length, is on chromosome 6 (Yano et al. 2000).
Regulatory mechanisms controlling flowering have been studied extensively in Arabidopsis (Arabidopsis thaliana) as a long-day (LD) model plant. In this LD species, flowering is promoted by the CONSTANS (CO) gene and regulated by the GIGANTEA (GI) gene, a circadian clock gene (Park et al. 1999; Sothern et al. 2002). CO activates expression of FLOWERING LOCUS T (FT) (Kardailsky et al. 1999; Kobayashi et al. 1999). SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) is a flowering activator downstream of CO and integrates signals from multiple flowering pathways such as photoperiod, gibberellin and vernalization (Lee et al. 2000; Onouchi et al. 2000; Moon et al. 2003). In contrast to Arabidopsis, rice is a short-day (SD) plant whereby flowering is inhibited during early developmental stages, but induced when the day-length shortens in the fall. Although the roles of genes involved in the photoperiod pathway are generally conserved between rice and Arabidopsis, some differences exist. In rice, OsGI (a rice ortholog of Arabidopsis GI) inhibits flowering in LD conditions, and promotes flowering in SD conditions (Hayama et al. 2003). Hd1, a rice ortholog of Arabidopsis CO, is regulated by OsGI and promotes flowering in SD conditions (Yano et al. 2000). Hd3a is a homolog of Arabidopsis FT, and is positively regulated by Hd1 in SD conditions (Kojima et al. 2002). Hd1 represses flowering by down-regulating Hd3a expression in LD conditions (Kojima et al. 2002). Hd3a is also regulated by Early heading date 1 (Ehd1), which encodes a B-type response regulator (Doi et al. 2004). Additionally, RID1/OsID1/Ehd2, which is homologous to maize (Zea mays) Indeterminate 1 (ID1), is necessary for the expression of Ehd1 regardless of photoperiod (Matsubara et al. 2008; Park et al. 2008; Wu et al. 2008). Ghd7, encoding a CCT domain protein, is an important regulator of heading date and yield potential in rice. It represses the expression of Ehd1 and Hd3a, thereby delaying flowering in LD conditions (Xue et al. 2008). DTH8/Ghd8, encoding a putative HAP3 subunit of the CCAAT-box-binding transcription factor, suppresses flowering and influences plant height and yield potential, and represses Ehd1 in LD conditions (Wei et al. 2010; Yan et al. 2010).
Many flowering regulators, such as RID1/OsID1/Ehd2, Ghd7, DTH8/Ghd8, OsCOL4, OsphyB and others, play important roles in influencing many agronomic traits (Takano et al. 2005; Matsubara et al. 2008; Park et al. 2008; Wu et al. 2008; Lee et al. 2010). For example, plants with the ehd2 mutation flower much later than wild type, and have more tillers and smaller spikelets (Matsubara et al. 2008). Ghd7 and DTH8/Ghd8 suppress flowering in LD conditions and influence plant height and yield (Xue et al. 2008; Wei et al. 2010; Yan et al. 2010). However, these genes are difficult to manipulate directly in rice breeding because their large effects lead to disharmony among heading date, plant height and yield.
We developed a NIL (Near-isogenic line) by MAS (molecular marker-assisted selection) and back-crossing to the japonica variety DJY1 (Dianjingyou 1), and the African rice accession IRGC102203 as the donor parent. Therefore, the objectives of this study were to elucidate the molecular basis of heading date differences between Asian and African rice, and we study the differentiation of DTH3 (days to heading on chromosome 3 in DJY1) in Asian rice and African rice.
Materials and methods
Parental materials and mapping populations
The japonica variety DJY1 (Dianjingyou 1) is from Yunan Province, China, and IRGC102203 is an O. glaberrima accession. We developed a NIL by backcross and molecular marker-assisted selection (MAS) using DJY1 as the recurrent parent, and IRGC102203 as the donor. To assess the purity of the DJY1 background of NIL, we surveyed it using 192 SSR markers that were evenly distributed across the 12 chromosomes of rice (McCouch et al. 2002). Only one foreign chromosomal fragment of about two BACs was identified on chromosome 3 (Supplementary Fig. 1a, b). NIL and DJY1 were grown in Nanjing (day-length >14 h) and Hainan (day-length <11 h). For analysis of diurnal expression patterns of flowering genes, NIL and DJY1 were grown in LD (14 h light/10 h dark) and SD (10 h light/14 h dark) conditions for 45 days. Nipponbare, Nip (hd1), ZS (ghd7), Mh (Ghd7), Asominori and CSSL61 (dth8) were grown under LD conditions for 40 days.
Mapping the heading date gene
The inserted fragment in NIL is between markers g91 and Ins11. We surveyed heading dates of the NIL × DJY1 F2 population (n = 298) in the summer of 2008 in Nanjing. In 2009, we planted a large F2 population (n = 18,000) in Nanjing, selected 2,000 individuals with the latest heading dates, and 1,500 with the earliest heading dates for fine mapping following the approach described by Zhang et al. (1994). We also planted an F2 population (n = 10,000) in Hainan (NSD) during the winter of 2009 for mapping. All materials were planted at a spacing of 16.5 cm × 16.5 cm.
DNA was extracted from fresh leaves of each plant following the method of Dellaporta et al. (1983). The PCR protocol was as described by Chen et al. (1997). PCR products were separated on an 8% non-denaturing polyacrylamide gel and detected using the silver staining method of Sanguinetti et al. (1994). To develop new SSR markers, an appropriate genomic sequence was obtained from the International Rice Genome Sequencing Project database (IRGSP; http://www.rgp.dna.affrc.go.jp/IRGSP/index.html1). Suitable microsatellite sequences were selected using on-line SSR searching software SSRIT (http://www.gramene.org/microsat/) and subjected to SSR primer design using Primer Premier 5.0 software. Additionally, insertion–deletion (InDel) markers were developed (insertions or deletions of 5 bp) by analyzing sequence differences between japonica variety Nipponbare and indica variety 93-11 in the delimited region using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/). PCR primers of products 100–300 bp in length were designed at about 10-kb intervals based on the sequence differences. We designed dCAPS markers with dCAPS Finder 2.0 on website http://helix.wustl.edu/dcaps/dcaps.html. The primers for mapping see Supplementary Table 1.
RNA extraction and QRT-PCR
RNA extraction and QRT-PCR total RNA extraction kits were used following the manufacturer’s instructions (Beijing Dingguo Biotechnology Co. Ltd., http://www.dingguo.com). First strand cDNA was reverse transcribed from DNase I-treated RNA with oligo (dT) as the primer. Gene expression was measured by QRT-PCR using the Ubiquitin gene as an internal control. QRT-PCR for Ubiquitin, DTH3, OsGI, Hd1, Hd3a, Ehd1, Ghd7, RID1, OsMADS51, DTH8, OsMADS56 and RFT1 are listed in Supplementary Table 1. QRT-PCR was carried out in total volumes of 25 μL containing 2 μL of the reverse-transcribed product above, 0.2 μM of each primer and 13 SYBR green PCR master mix (TaKaRa Co. Ltd., http://www.takara.com.cn). The PCR was performed with a Bio-Rad iCycler (http://www.bio-rad.com/) using the following program: 95°C for 30 s, then 40 cycles of 95°C for 5 s, 60°C for 34 s. Changes in gene expression were calculated via the DDCT method (Livak and Schmittgen 2001).
Vector construction and transformation
The full-length coding region of DTH3 was isolated by PCR with primer pair m1 from NIL and DJY1. The PCR product was subcloned into a pGE blunt end cloning vector. The cDNA was subcloned into pCUbi1390 binary vector using primer vector DTH3 with the restriction enzyme sites BamHI and SpeI (Supplementary Table 1). The resultant plasmid was introduced into NIL and DJY1 by means of Agrobacterium-mediated transformation (Hiei et al. 1994).
Statistical analyses
In the analysis of genetic relationships between African rice and different groups of cultivars, each polymorphism detected by 26 pairs of SSR markers was treated as a unit character by assigning a score of 1 or 0 to the presence or absence of a fragment (Supplementary Table 2). The genetic similarity coefficients among accessions assayed were estimated by genetic distance using Nei’s (1987) unbiased genetic distance coefficient. The resulting genetic distance matrix was used for cluster analysis according to the unweighted pair-group method with arithmetic averages (UPGMA), using the software program NTSYSpc version (Rohlf 1992). To assess further the genetic relationships of African rice populations and selected rice varieties, a principle component analysis (PCA) was conducted based on the SSR variation patterns converted into the 1 and 0 matrixes. The correlation matrix was selected to calculate coefficients of the first three principal components, using MINITAB version 14.13 (Minitab Inc, State College, PA, USA). We used primers: F, 5′-TGGGGCATCGCTTGGCTAT-3′, R, 5′-CACCTGATTTGCTTCCCTTGA-3′ to detect the six-base deletion, and AACATGGATGTCGAAACTGAGGAAT to detect the one-base substitution with restriction enzyme site EcoRI.
Results
Identification of flowering time in DJY1 and NIL
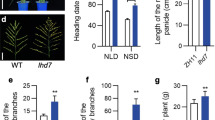
The flowering time of NIL was 7–10 days later than DJY1, whether grown in Hainan (natural short-day, NSD, day-length <11 h) or in Nanjing (natural long-day, NLD, day-length >14 h) (Fig. 1a, c). In artificial conditions (LD, 14 h light 10 h dark; and SD, 10 h light 14 h dark), NIL also flowered 7–10 days later than DJY1 (Fig. 1b). After heading, various traits, such as plant height, panicle and other agronomic traits, were not significantly different between NIL and DJY1 (Table 1; Fig. 1c, d, e).
a Comparisons of NIL and DJY1 heading dates in Nanjing (NLD) and Hainan (NSD); b days to heading for DJY1 and NIL under controlled SD (14 h light/10 h dark) and LD (10 h light/14 h dark); DAG, days after germination; c, d phenotypes of NIL and DJY1 grown under conditions at heading and maturity; e inflorescences of NIL and DJY1 grown under NSD
Fine mapping and prediction of flowering gene
The inserted fragment containing Hd9 in NIL was flanked by markers g91 and Ins11 on the short arm of chromosome 3 (Supplementary Fig. 1c). But our materials have the different phenotype, so we named DTH3 (QTL for days to heading on chromosome 3) for early flower gene in DJY1 and dth3 for late flower gene in NIL. The heading dates of the NIL × DJY1 F2 population (n = 298) was surveyed in LD conditions, and we found that early heading plants (n = 63) possessed markers mainly from DJY1 (DTH3), and late heading genotypes (n = 82) had alleles mainly from NIL (dth3) (Fig. 2). We then planted a NIL × DJY1 F2 population (n = 18,000) in Nanjing (NLD), selected 2,000 individuals with the latest heading dates, and 1,500 individuals with extremely early heading dates for fine mapping. The gene DTH3 from DJY1 controlling early flowering was located between markers g71 and RM523, the distance between the two markers was 64 kb (Fig. 3), and the F2:3 generation of recombinant individuals verified the fine mapping results (Table 2).
High-resolution mapping of DTH3. a The genetic map of DTH3 was based on recombination events among 18,000 F2 plants from NIL × DJY1. The numbers below markers indicate the numbers of recombinants. The wide horizontal lines represent BAC/PAC clones of cv. Nipponbare with the accession numbers indicated. b The wide horizontal lines represent BAC/PAC clones of O. glaberrima with the accession numbers indicated
It was reported that Hd3a promoted heading under SD conditions whereas Hd3b caused late heading under LD and natural field conditions in Japan. The genetic distance between these two genes was about 1.6 cM (Monna et al. 2002). In our materials, NIL delayed flowering both in LD and SD conditions. In order to determine if separate genes controlled flowering under LD and SD conditions, we planted a NIL × DJY1 F2 population (n = 10,000) in Hainan (NSD). The results suggested that the position mapped in both environments was the same. The genetic distances between markers g71 and RM523 in African rice (http://www.gramene.org/Oryza_glaberrima/Info/Index) and in Nipponbare (http://rgp.dna.affrc.go.jp/E/IRGSP/index.html) were 54 and 64 kb, respectively (Fig. 3). Based on candidate gene analysis by The Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/), we found a transcription factor, MIKC-type MADS-box gene, and genome length of 29.7 kb. The cDNA sequence of this MADS-box gene have two differences between DJY1 and NIL at the code region. Six bases were inserted and base T instead of C occurred in the C-domain in NIL (Fig. 4). We considered the MADS-box gene as a good candidate for DTH3.
Overexpression of DTH3 causes early flowering in callus
The transcript levels of DTH3/dth3 gene in leaves of 45 day-old seedlings were measured. The expression level of dth3 in NIL was lower than DTH3 in DJY1, both in LD and SD conditions (Fig. 5a). To verify its function, DTH3 was overexpressed in NIL and DJY1 driven by the maize ubiquitin (ubi) promoter (Fig. 6a). Overexpression of DTH3 caused early flowering in callus of both DJY1 and NIL (Fig. 6b, c, d). The transcript level in the NIL callus with Ubi::DTH3 was much higher than in NIL with the empty vector (Fig. 6e). Similar results were obtained by overexpression of the dth3 in both NIL and DJY1 (date not show), suggesting that DTH3 and dth3 have the same function in both lines, but the expression level of dth3 is weaker than DTH3. It is likely that dth3 is a weaker allele of DTH3.
Analyses of DTH3 overexpressing plants. a Schematic diagram of DTH3 construct. Ubi maize ubi promoter. b Overexpression DTH3 in NIL; note flowering in callus. s stigma, o ovary. c Enlarge the red frame in b. d Callus with empty vector. e Quantitative real-time RT-PCR analyses of DTH3 in callus, CK, NIL callus with empty vector (color figure online)
Expression patterns of DTH3 in DJY1 and NIL
To determine whether the difference of expression patterns existed between DJY1 and NIL, mRNA level of DTH3/dth3 was measured every 4 h in both LD (14L/10D) and SD (10L/14D). Transcript levels of DTH3/dth3 changed over the 24-h cycle, the highest being at dawn and lowest at dusk in both LD and SD conditions and in both lines (Fig. 7a, b). However, the transcript levels of dth3 were lower than DTH3 over the 24-h cycle in both day-length environments. The DTH3 transcript level in DJY1 gradually increased to a maximum at 9 weeks after germination, and then declined (Fig. 7c). The transcript level in NIL reached its maximum 1 week later than in DJY1. This apparently explained why the heading date of DJY1 was about 1 week earlier than NIL. Presumably, the accumulation of DTH3 at the mRNA level plays an important role in promoting flowering.
Diurnal expression of DTH3 in leaves under LD (a) and SD conditions (b) for 45 days. RNA were prepared from leaf blades at 4-h intervals over 24 h. c, Real-time RT-PCR analyses of DTH3 at various developmental stages under LD conditions. RNA samples were collected from second leaf blades from the tops of plants grown under LD: 5, 6, 7, 8, 9, 10, 11 weeks after germination
Relationship of DTH3 with other flowering time regulators
The expression of dth3 was reduced in NIL relative to DJY1 in both LD and SD conditions. To investigate the relationship of DTH3 with other flowering time regulators, the expression levels of DTH3 and ten flowering-related genes (Hd1, Hd3a, Ehd1, Ehd2, RFT1, OsMADS56, OsGI, DTH8, OsMADS51, Ghd7) were analyzed in NIL and DJY1 grown in both environments (Fig. 5). Hd1 was undetected both in LD and SD conditions over the 24-h cycle (data not shown). The mRNA levels of DTH8 (Fig. 5d), OsGI (Fig. 5e), OsMADS56 (Fig. 5f), Ehd2 (Fig. 5g), OsMADS51 (Fig. 5i) and Ghd7 (Fig. 5j) did not differ between the two lines. However, the expression of Ehd1 (Fig. 5b) and RFT1 (Fig. 5c) in NIL were significantly lower than DJY1 in both environments, and Hd3a expression was reduced in LD, but unchanged in SD (Fig. 5h). These results indicate that DTH3 accelerates rice flowering by up-regulating Ehd1 which in turn activates RFT1 both in LD and SD conditions, and up-regulates Hd3a only in LD. We further showed that the expression of DTH3 was not influenced by Hd1, Ghd7 and DTH8 by examining Hd1, Ghd7 and DTH8 NILs grown for 40 days under LD conditions (Fig. 8).
a Expression in cv. Nipponbare (Nip) and Nip (hd1) was introgressed from cv. Kasalath in a cv. Nipponbare genetic background; b expression in Mh (Ghd7) introgressed from Minghui 63 and a NIL counterpart with the non-functional ZS (ghd7) allele from Zhenshan 97; c expression of DTH8 in Asominori and a non-functional dth8 allele from IR24 in CSSL61; d Expression of DTH3 in Nip and Nip (hd1); e expression of DTH3 in Mh (Ghd7) and ZS (ghd7); f expression of DTH3 in Asominori and CSSL61. QRT-PCR was performed with total RNA from leaves of 40-day-old plants under LD conditions. Samples were collected at the initiation of the light phase (ZT 0 h). These experiments were repeated at least three times
Differentiation of DTH3 in Asian rice, African rice and wild rice: molecular classification of related varieties
Pairs of Indel and dCAPs markers were designed from the 6-bp deletion and single base substitution in NIL. Two hundred and twenty Asian rice varieties, including 127 indica and 93 japonica, 22 wild rice and 53 African rice accessions were tested with these markers. All of the Asian cultivated rice (indica and japonica) accessions and wild rice lacked the six bases, whereas all African rice accessions contained them. All the Asian rice accessions and wild rice carried T at the polymorphic position, whereas both C and T were present in African rice (Table 3; Supplementary Table 3). It seems that during domestication of Asian rice, the six-base deletion and single nucleotide became fixed in Asian rice.
To understand the genetic relationship of African cultivated rice (O. glaberrima) with other rice groups we analyzed 26 pairs of SSR markers covering the entire rice genome. The dendrogram constructed from the results showed four groups, viz. japonica, O. glaberrima, indica, and wild rice. African rice was closely related to O. rufipogon and O. latifolia, and was more similar to japonica than to indica or other wild rice groups (Supplementary Fig. 2).
Discussion
The foreign fragment insert in NIL has the same position as Hd9 in chromosome 3S (Lin et al. 2002, Supplementary Fig. 1c). The Nip (hd9) in which a small chromosomal segment of Kasalath including Hd9 was substituted into the genetic background of Nipponbare. Nip (hd9) flowered later than Nipponbare in LD condition and was no difference in SD condition (Lin et al. 2002). The cDNA sequences of DTH3 have single base substitution in Nipponbare and Kasalath (Supplementary Fig. 3). Interestingly, OsMADS50, reported by Ryu et al. (2009) as a LD-specific flowering activator was also present in our fine mapping region. It was likely that Hd9, OsMADS50, and DTH3 are multiple alleles.
Rice is a SD plant. In SD conditions, signals from light and circadian clocks are received by OsGI, which regulates the expression of Hd1 and OsMADS51 (Izawa et al. 2003; Kim et al. 2007). Hd1 up-regulates Hd3a expression (Kojima et al. 2002), while activating OsMADS51 (Doi et al. 2004; Kim et al. 2007). In our study, Hd1 was not detected in NIL and DJY1, and Ehd2, OsMADS56, OsGI, DTH8, OsMADS51 and Ghd7 were unchanged. DTH3 up-regulated Ehd1 and RFT1 in both LD and SD conditions. It had been reported that Ehd2/RID1/OsID1, OsPhyB, and OsCOL4 could up-regulate Ehd1 in both situations (Takano et al. 2005; Matsubara et al. 2008; Park et al. 2008; Wu et al. 2008; Lee et al. 2010). But mutation or deletion of these genes not only influences heading date, but also agronomic traits, such as tiller number and 1,000-grain weight (Takano et al. 2005; Matsubara et al. 2008; Park et al. 2008; Wu et al. 2008; Lee et al. 2010). Thus these genes are difficult to use in rice breeding. While the phenotype in our materials, dth3 delayed flowering both in LD and SD, and DJY1 and NIL are not very different in the main agronomic traits, so it was more useful in rice breeding and variety introduction without regard to the day-length.
Most flowering time regulators involved in photoperiod control are under circadian regulation. The mRNA level of DTH3/dth3 showed a rhythm: high at dawn and low at dusk both in LD and SD conditions, a similar pattern to COL4 and RFT1 (Komiya et al. 2009; Lee et al. 2010). The transcript level in NIL was lower than in DJY1, and the transcript level in NIL reached its maximum 1 week later than in DJY1. This result was consistent with the observation that flowering time in NIL was about 1 week later than DJY1 in both LD and SD conditions. Recent reports suggested that the SPL (SQUAMOSA BINDING FACTOR-LIKE) family of transcription factors in Arabidopsis influences a series of phase transitions from juvenile to adult, as well as from vegetative to reproductive phase transitions, involved in age-related regulation of SOC1 (Schwab et al. 2005; Wu and Poethig 2006; Wang et al. 2009). A MADS-box gene SOC1 acts as an integrator in multiple flowering pathways involving photoperiod, gibberellin, aged-dependent regulation and vernalization in Arabidopsis (Lee et al. 2000; Onouchi et al. 2000; Moon et al. 2003). It is likely that DTH3 is involved in age-dependent regulation or plays another role in the rice flowering pathway.
DTH3 encodes MIKC-type MADS-box proteins (Kaufmann et al. 2005), and has seven exons. MIKC-type MADS-box proteins contains four domains: MADS-domain (M) is required for DNA binding and dimerization (Schwarz-Sommer et al. 1990); The I domain, a less-conserved region, is also necessary for dimerization (Krizek and Meyerowitz 1996); the K domain plays an important role in the interaction between MIKC-type proteins (Fan et al. 1997; Yang and Jack 2004); and the C-domain is the least-conserved region, in some cases either possessing transactivation activity or contributing to the formation of multimeric complexes among MADS proteins (Cho et al. 1999; Honma and Goto 2001). In Arabidopsis, SOC1 interacts directly with AGL24, a MADS-box transcription factor. This direct interaction confers a positive-feedback regulation of the expression of AGL24 and SOC1 to a quantitative threshold required for the transition from vegetative to reproductive growth (Liu et al. 2008). Recent research using ChIP analysis showed that SOC1 directly binds to the modified CArG box in LFY. A missense mutation of Arg24 resulted in the loss of SOC1 binding to the LFY’s promoter suggesting that SOC1 does not work alone (Lee et al. 2008). OsMADS56 was a flower repressor under LD condition. Yeast-two hybrid was used to test the interaction between DTH3/dth3 and OsMADS56; the result showed that DTH3 can bind to OsMADS56, while dth3 can not (Supplementary Fig. 4). It was further studied, whether DTH3 and OsMADS56 form complex to regulate heading date in rice. In our research, dth3 had two mutations in the C-domain that may affect transcriptional activation. The C-terminal domain of Arabidopsis thaliana AP1 (a member of the MADS superfamily) and its homologs perform a transcriptional activation function (Cho et al. 1999).
NIL contains a single DNA fragment from African rice in the background of japonica variety Dianjingyou 1 (DJY1). African rice is more resistant than Asian cultivars to diseases and pests, and superior in tolerance to fluctuations in water depth, iron toxicity, infertile soils and severe climatic conditions. Some O. glaberrima types also mature faster than Asian rice (Linares 2002). These characteristics are potentially useful for improving Asian rice. Here, we analyzed a flowering gene derived from African rice in NIL. Compared with DTH3 in Asian rice, the dth3 sequence in the segment from African rice was unique in possessing an additional 6-bp insert and in a single base substitution. African rice clustered in a group that was little differentiated from Asian rice and wild rice. African rice had a close genetic relationship to O. rufipogon and O. latifolia, and was more similar to japonica cultivars than to the other groups. The ancestor of African rice is O. barthii, and the ancestor of Asian rice is O. rufipogon; they presumably had a common ancestor before separation of the continents (Khush 1997). Our results showed that Asian rice and African rice had a common ancestor by using molecular markers (SSR, Indel and dCAPS) for analyzing genetic relationships between Asian rice and African rice. The two cultivated species were differentiated and domesticated in parallel in their respective geographical areas. DTH3 was restricted to Asian rice and presumably differed from African rice as a result of selection. We also investigated the transcript level of DTH3 in 43 cultivars including 20 japonica and 23 indica cultivars. The results showed that DTH3 was not the main reason to affect heading date in Asian rice (Supplementary Fig. 5). To introduce the new favorable gene from other species of rice could broaden the genetic base, and it is beneficial for rice breeding.
Abbreviations
- DJY1:
-
Dianjingyou 1
- Ehd1 :
-
Early heading date 1
- LD:
-
Long-day
- MAS:
-
Marker-assisted selection
- NILs:
-
Near-isogenic lines
- SD:
-
Short-day
- SSR:
-
Simple sequence repeat
References
Chang TT, Vergara BS, Li CC (1969) Component analysis of duration from seeding to heading in rice by the basic vegetative phase and photoperiod sensitive phase. Euphytica 18:79–91
Chen X, Temnykh S, Xu Y, Cho Y, McCouch S (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Cho S, Jang S, Chae S, Chung KM, Moon YH, An G, Jang SK (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol 40(3):419–429
Dellaporta S, Wood J, Hicks J (1983) A plant DNA minipreparation: VersionII. Plant Mol Biol Rep 1:19–21
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18(8):926–936
Fan HY, Hu Y, Tudor M, Ma H (1997) Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J 12(5):999–1010
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422(6933):719–722
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6(2):271–282
Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409(6819):525–529
Hosoi N (1981) Studies on meteorological fluctuation in the growth of rice plants. V. Regional differences of thermo-sensitivity, photosensitivity, basic vegetative growth and factors determining the growth duration of Japanese varieties. Jpn J Breed 31:239–250
Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6(2):113–120
Jones MP, Mande S, Aluko K (1997) Diversity and potential of Oryza glaberrima Steud. in upland rice breeding. Jpn J Breed 47:395–398
Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286(5446):1962–1965
Kaufmann K, Melzer R, Theissen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347(2):183–198
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35(1–2):25–34
Kim SL, Lee S, Kim HJ, Nam HG, An G (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145(4):1484–1494
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286(5446):1960–1962
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43(10):1096–1105
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136(20):3443–3450
Krizek BA, Meyerowitz EM (1996) Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc Natl Acad Sci USA 93(9):4063–4070
Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14(18):2366–2376
Lee J, Oh M, Park H, Lee I (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J 55(5):832–843
Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Cho LH, Choi H, An G (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63(1):18–30
Lin H, Ashikari M, Yamanouchi U, Sasaki T, Yano M (2002) Identification and characterization of a Quantitative Trait Locus, Hd9, controlling heading date in rice. Breed Sci 52(1):35–41
Linares OF (2002) African rice (Oryza glaberrima): history and future potential. Proc Natl Acad Sci USA 99(25):16360–16365
Liu C, Chen HY, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135(8):1481–1491
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25(4):402–408
Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148(3):1425–1435
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9(6):199–207
Monna L, Lin X, Kojima S, Sasaki T, Yano M (2002) Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor Appl Genet 104(5):772–778
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35(5):613–623
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 106–107
Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12(6):885–900
Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285(5433):1579–1582
Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Xuan YH, Colasanti J, An G, Han CD (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 56(6):1018–1029
Rohlf E (1992) Numerical taxonomy and multivariate analysis system. NTSYS-pc program. Applied Biostatistics Inc., Setauket
Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, An G (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32(10):1412–1427
Sanguinetti C, Dias N, Simpson A (1994) Rapid silver staining and recover of PCR products separated on polyacrylamide gels. Biotechniques 17:915–919
Sato T, Takahashi N (1983) The effect of air and water temperature on the number of days to heading in Japonica rice cultivars. Jpn J Breed 33:111–118
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8(4):517–527
Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H (1990) Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250(4983):931–936
Sothern RB, Tseng TS, Orcutt SL, Olszewski NE, Koukkari WL (2002) GIGANTEA and SPINDLY genes linked to the clock pathway that controls circadian characteristics of transpiration in Arabidopsis. Chronobiol Int 19(6):1005–1022
Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17(12):3311–3325
Tsai K (1986) Genes controlling heading time found in a tropical Japonica variety. Rice Genet Newslett 3:71–73
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138(4):738–749
Wei XJ, Xu JF, Guo HN, Jiang L, Chen SH, Yu CY, Zhou ZL, Hu PS, Zhai HQ, Wan JM (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153(4):1747–1758
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133(18):3539–3547
Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105(35):12915–12920
Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40(6):761–767
Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF (2010) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. doi:10.1093/mp/ssq070
Yang Y, Jack T (2004) Defining subdomains of the K domain important for protein–protein interactions of plant MADS proteins. Plant Mol Biol 55(1):45–59
Yang YJ, Wang XD, Wu XJ, Zhang HY, Zhang P, Zhao HX (2005) The discovery, genetic analysis and gene mapping of earliness rice D64B (Oryza sativa L.). Yi Chuan Xue Bao 32(5):495–500
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12(12):2473–2484
Yuan Q, Saito H, Okumoto Y, Inoue H, Nishida H, Tsukiyama T, Teraishi M, Tanisaka T (2009) Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor Appl Genet 119(4):675–684
Zhang Q, Shen BZ, Dai XK, Mei MH, Saghai Maroof MA, Li ZB (1994) Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc Natl Acad Sci USA 91(18):8675–8679
Acknowledgments
We thank Professor Tao Dayun, Yunnan Academy of Agricultural Sciences, for kindly providing us with the Dianjingyou 1 (DJY1) and near-isogenic line (NIL). This research is supported by the grants from the National Natural Science Foundation of China (30871497), National Transform Science and Technology Program (2008ZX08001-06), Jiangsu Science and Technology Development Program (BE2009301-3), Doctor Foundation of Education Development of China (20090097110011), the earmarked fund for Modern Agro-industry Technology Research System and Jiangsu PAPD Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bian, X.F., Liu, X., Zhao, Z.G. et al. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1 . Plant Cell Rep 30, 2243–2254 (2011). https://doi.org/10.1007/s00299-011-1129-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1129-4