Abstract
Ghd7, Ghd8, Ghd7.1, and Hd1 are all pleiotropic genes regulating heading date, plant height, and yield in rice. Early studies showed that Hd1 promoted heading under short-day conditions (SD) and delayed heading under long-day conditions (LD). Recent studies found that Hd1 also promoted heading in some genetic backgrounds under LD. In this study, we developed a series of near-isogenic lines for Ghd7, Ghd8, Ghd7.1, and Hd1 in the Zhenshan 97 (ZS97) and Minghui 63 (MH63) backgrounds and recorded their heading dates. In the ZS97 background, Ghd7 alone triggered the conversion of Hd1 function from promoting to suppressing heading under LD. Ghd8 alone and Ghd7.1 alone did not promote but enhanced Ghd7-mediated Hd1 function conversion. In the MH63 background, Ghd7 alone and Ghd7.1 alone did not convert Hd1 function under LD, but they jointly promoted Hd1 function conversion. Conversion of Hd1 function occurred only under LD but not SD. Transcript analysis showed that downregulation or upregulation of Ehd1 and Hd3a by Hd1 in the lines under LD was determined by genetic background. In summary, multiple gene combinations of Ghd7, Ghd7.1, and Ghd8 and unknown genes caused conversion of Hd1 function. Moreover, the different gene combinations had a big difference in photoperiod sensitivities. These findings lay a solid foundation for further unveiling the molecular mechanism of Hd1 function conversion and provide guidance for heading date improvement in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is an important cereal crop that is cultivated across the world. Environmental factors such as photoperiod, temperature, rainwater, and illuminance in rice planting regions vary temporally and geographically. To gain high yield, rice cultivars must avoid unfavorable growth conditions during its life cycle and adequately utilize light energy and temperature resources. Thus, flowering at a suitable time is essential to rice production.
Rice is a short-day plant whose flowering is promoted under short-day conditions (SD), while it is delayed under long-day conditions (LD). The photoperiod pathway is an important way to regulate heading in rice. Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1), orthologs to FLOWERING LOCUS T (FT) of Arabidopsis, are florigen genes in rice that switch on flowering (Komiya et al. 2008; Komiya et al. 2009; Taoka et al. 2011). Two independent pathways are mediated by Heading Data 1 (Hd1) and Early heading date 1 (Ehd1) to regulate Hd3a and RFT1 expression. Hd1, an ortholog of CONSTANS in Arabidopsis, delays and promotes flowering by downregulating and upregulating the expression of Hd3a and RFT1 under LD and SD, respectively (Yano et al. 2000). Ehd1, encoding a B-type response regulator, functions as a homodimer and induces heading by upregulating Hd3a and RFT1 under both LD and SD (Doi et al. 2004; Cho et al. 2016). Ghd7, Ghd8, and Ghd7.1 repress flowering by inhibiting Ehd1 expression under LD (Xue et al. 2008; Yan et al. 2011; Hori et al. 2013). However, recent studies have shown that Hd1 also regulates Ehd1 expression (Nemoto et al. 2016; Zhang et al. 2017), which indicates that the two pathways are tightly associated.
Genetic interactions between Ghd7, Ghd8, Ghd7.1, and Hd1 regulate rice flowering (Lin et al. 2000; Shibaya et al. 2011; Zhang et al. 2015). Hd1, Ghd7, and Ghd7.1 all encode CO, CO-like, and TOC1 (CCT)-domain proteins (Yano et al. 2000; Xue et al. 2008; Hori et al. 2013; Liu et al. 2013; Zhang et al. 2017), while Ghd8 encodes a CCAAT-box-binding transcription factor protein, HAP3H (Yan et al. 2011). Hd1 forms complexes with Ghd7 or Ghd8 to repress flowering by downregulating Ehd1 and Hd3a under LD (Nemoto et al. 2016; Du et al. 2017; Goretti et al. 2017; Zhu et al. 2017). Ghd8 interacts with Ghd7 and Ghd7.1 in yeast (Goretti et al. 2017). These results imply that Ghd7, Ghd8, Hd1, and Ghd7.1 interactively regulate flowering. However, the detailed genetic interactions and molecular mechanisms which they regulate flowering are not very clear.
Recent studies indicated that Hd1 constantly promoted heading date and exhibited photoperiod insensitivity under both SD and LD on a genetic background without Ghd7 or Ghd8 (Du et al. 2017; Zhang et al. 2017), which conflicted with earlier findings (Yano et al. 2000). The phenomenon that Hd1 can switch from its presumed function of promoting heading to repressing heading under LD was termed Hd1 function conversion. Why Hd1 function conversion occurs has been controversial. One study insisted that Ghd7 was required for the conversion (Zhang et al. 2017), while another study deemed that Ghd8 rather than Ghd7 was required for its conversion (Du et al. 2017).
To resolve the inconsistent results, the heading dates of a series of near-isogenic lines (NILs) of Ghd7, Ghd8, and Hd1 in the Zhenshan 97 (ZS97) background were investigated in this study. A series of NILs of Ghd7, Ghd7.1, Hd1, and Ghd8 on either the ZS97 or the Minghui 63 (MH63) background was also used. We found that Ghd8 converted Hd1 function depending on Ghd7, and Ghd7.1 also participated in Hd1 function conversion together with Ghd7 or Ghd8. By surveying the photoperiod sensitivity (PS) of all these NILs, we found that different combinations of Ghd7, Ghd7.1, Hd1, and Ghd8 had diverse PS. This study sheds some light in the molecular mechanism of Hd1 function conversion.
Materials and methods
Plant materials
Zhenshan 97 (ghd7ghd8ghd7.1Hd1) is a cultivar with nonfunctional Ghd7, Ghd8, and Ghd7.1 and functional Hd1 (Xue et al. 2008; Yan et al. 2011, 2013; Zhang et al. 2017). In a previous study, ZS-G7G8H1 (Ghd7Ghd8ghd7.1Hd1), a near-isogenic line (NIL) with introgressed functional Ghd7 and Ghd8 from MH63 and 93-11, respectively, was developed in the ZS97 background (Zhang et al. 2015). ZS-g7g8h1 (ghd7ghd8ghd7.1hd1) was a NIL introgressed with nonfunctional hd1 from Teqing (TQ) into the ZS97 background (Zhang et al. 2017). We obtained two F1 hybrid plants by crossing ZS-G7G8H1 with ZS-g7g8h1 and then developed two F2 populations with Ghd7, Hd1, and Ghd8 segregating by self-pollination. Eight homozygous combinations of Ghd7, Ghd8, and Hd1, namely, ZS-G7G8H1, ZS-G7G8h1, ZS-G7g8H1, ZS-G7g8h1, ZS-g7G8H1, ZS-g7G8h1, ZS-g7g8H1, and ZS-g7g8h1, were identified from these two populations by functional markers or closely linked markers (Supplementary Table 1) and selfed for many seeds for subsequent experiments (Supplementary Fig. 1A).
ZS-g7G7.1H1 (ghd7ghd8Ghd7.1Hd1) was the NIL introgressed with functional Ghd7.1 form TQ in the ZS97 background (Liu et al. 2013). It was crossed with ZS-g7g8h1 to generate an F2 population in the ZS97 background and ZS-g7G7.1H1, ZS-g7G7.1h1, ZS-g7g7.1H1, and ZS-g7 g7.1 h1 were identified from this population (Supplementary Fig. 1B). Then, ZS-g7G7.1H1 was crossed with ZS-G7g8h1 to obtain the ZS-G7G7.1H1, ZS-G7G7.1h1, ZS-G7g7.1H1, and ZS-G7g7.1h1 lines (Supplementary Fig. 1C). The NIL ZS-G7G8hd1 was crossed with ZS-g7G7.1H1, an F2 population was planted, and the homozygous genotypes ZS-g7G8G7.1H1, ZS-g7G8G7.1h1, ZS-G7G8G7.1H1, and ZS-G7G8G7.1h1 were identified from this population (Supplementary Fig. 1D).
MH63 (Ghd7ghd8Ghd7.1hd1) is an indica cultivar containing nonfunctional Ghd8 and Hd1 and functional Ghd7 and Ghd7.1 (Yan et al. 2013; Zhang et al. 2015; Zhang et al. 2017). MH-G7G7.1H1 (Ghd7ghd8Ghd7.1Hd1) and MH-G7g7.1h1 (Ghd7ghd8ghd7.1hd1) introgressed with functional Hd1 and nonfunctional ghd7.1 in the MH63 background, respectively, were identified from advanced chromosome segment substitution lines (CSSLs) in the MH63 background with ZS97 as the donor parent (Shen and Xing 2014; Zhang et al. 2017). They were crossed to get an F2 population in the MH63 background, and four homozygous MH-G7G7.1H1, MH-G7G7.1h1, MH-G7g7.1H1, and MH-G7g7.1h1 were identified (Supplementary Fig. 1E). MH-g7G7.1H1 (ghd7ghd8Ghd7.1Hd1) was an MH63-background line that contained nonfunctional ghd7 and functional Hd1 (Zhang et al. 2017). It was crossed with MH-G7g7.1h1 to get MH-g7G7.1H1, MH-g7G7.1h1, MH-g7g7.1H1, and MH-g7g7.1h1 homozygous plants (Supplementary Fig. 1F).
Field experiments and photoperiod treatment
Plant materials were sown in the middle of May at the experimental station of Huazhong Agriculture University, Wuhan, Hubei province, China (31°N). The day length of Wuhan was more than 13.5 h from the middle of May to the beginning of August (natural long-day conditions, NLD). Twenty-five-day old seedlings were transplanted to the field with 16.5 cm between plants in a row and 26.5 cm between rows. Plant materials were sown in the beginning of December at Lingshui, Hainan province, China (18°N). The average day length was less than 12 h from December to March in Lingshui (natural short-day condition, NSD). Thirty-day-old seedlings were transplanted to the field with the same planting density as was used in Wuhan.
For photoperiod treatment, five 20-day-old seedlings of each genotype were transplanted to the field at a density of approximately 10 cm × 10 cm in Wuhan. The plants were grown under SD with a day length of 8 h and darkness of 16 h by shading treatment until all plants headed. At the same time, another five 20-day-old seedlings of each genotype were simultaneously grown in the same field under NLD for LD.
Identification of the genotypes of Ghd7, Ghd8, Ghd7.1, and Hd1
The DNA of plant materials was extracted from leaves using the CTAB method (Stewart Jr. and Via 1993). Then, the linked markers MRG4436, Z9M, S56, and Indel37 were used to identify the genotypes at Ghd7, Ghd8, Hd1, and Ghd7.1, respectively. The primers of these markers are available in Supplementary Table 1.
Expression analysis of Ehd1 and Hd3a by quantitative real-time PCR
To detect Ehd1 and Hd3a expression in the series of NILs, the second leaves were collected from three plants of each genotype at 40 days old at 8:00 under NLD as biological replicates. A 100-mg sample ground with liquid nitrogen was suspended with TransZol (TransGen Biotech, China) reagent to extract RNA. Then, 3 μg of RNA treated with DNase I was used to synthesize cDNA by using reverse transcriptase M-MLV reagent (Invitrogen). Relative expression levels of Ehd1 and Hd3a were analyzed by quantitative real-time PCR conducted with an ABI PRISM 7900 Sequence Detection System (Applied Biosystems) according to the manufacturer’s instructions. The primers used are listed in Supplementary Table 1.
Data analysis
The Student’s t test was used for pairwise comparison. Multiple comparisons were made using Duncan’s test (Duncan 1955). The photoperiod sensitivity (PS) index was measured using the averages of the heading date under LD, subtracting the averages of the heading date under SD.
Results
Hd1 function conversion depends on Ghd7 in the ZS97 background
To study the roles of Ghd7 and Ghd8 in Hd1 function conversion, eight homozygous three-gene combinations of Ghd7, Ghd8, and Hd1 were selected from two NIL-F2 populations to survey the heading date under NLD. ZS-g7g8H1 and ZS-g7G8H1 flowered 9.7–13.9 and 2.7–8.6 days earlier compared to ZS-g7g8h1 and ZS-g7G8h1, respectively (Fig. 1; Supplementary Table 2). Thus, Hd1 promoted heading without Ghd7 and Ghd8, or only with Ghd8 in ZS97. However, ZS-G7g8H1 and ZS-G7G8H1 flowered 3.3–5.0 days and > 59.8 days later than ZS-G7g8h1 and ZS-G7G8h1, respectively (Fig. 1; Supplementary Table 2). This indicated that Hd1 suppressed heading date with Ghd7, or together with Ghd7Ghd8, and suggested that Ghd8 without Ghd7 did not convert Hd1 function from promoting to delaying heading date in the ZS97 background, which was not in agreement with the previous report that single Ghd8 could convert Hd1 function in the 93-11 background (Du et al. 2017). Unlike Ghd8, Ghd7 alone slightly converted Hd1 function for 3.3–5 days in the ZS97 background, consistent with our previous study (Zhang et al. 2017). However, Hd1 significantly delayed heading (more than 59.8 days) and caused failed heading in the normal growing season under the background of functional Ghd7Ghd8 combination. Taken together, the data showed that Ghd8 largely enhanced Ghd7-mediated Hd1 function conversion in the ZS97 background.
Photoperiod sensitivity of eight homozygous three-gene combinations of Ghd7, Ghd8, and Hd1 in the ZS97 background
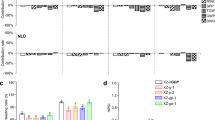
Ghd7, Ghd8, and Hd1 are photoperiod-sensitivity genes influencing rice adaptability to different ecological areas. To investigate the photoperiod sensitivity of eight homozygous three-gene combinations and evaluate whether Hd1 function conversion occurred in short days, these eight homozygous combinations of Ghd7, Ghd8, and Hd1 were grown under LD, NSD, and SD. Under SD and NSD, Hd1 always promoted heading date regardless of genetic backgrounds (Table 1), which suggested that Hd1 function conversion did not happen under SD conditions.
Estimates of photoperiod sensitivity (PS) showed that ZS-G7G8H1 had the strongest PS of more than 100.6 days, followed by ZS-G7g8H1 and ZS-g7G8H1 of 20.6 days and 16.2 days, respectively (Table 1). Hd1 was weakly sensitive to photoperiod in ZS-g7g8H1 (PS = 3 days) (Table 1). For the combinations without Hd1, their PS was weak or even absent (Table 1). For example, the PS of ZS-g7G8h1 and ZS-G7g8h1 were 5.8 days and − 1.5 days, respectively. It was surprising that ZS-G7G8h1 lost its photoperiod sensitivity and had similar heading dates of 102 and 101 days between NLD and SD, indicating its photoperiod insensitivity (Table 1).
Ghd7.1 enhances Ghd7-mediated Hd1 function conversion in the ZS97 background
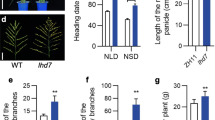
Ghd7.1 has similar functions to Ghd7 under LD conditions (Hori et al. 2013; Liu et al. 2013). We hypothesized that Ghd7.1 also triggered Hd1 function conversion. To verify our hypothesis, four homozygous combinations of Ghd7.1 and Hd1 in the ZS97 background were used to measure heading date under NLD. Both ZS-g7g7.1H1 and ZS-g7G7.1H1 flowered earlier than ZS-g7g7.1h1 and ZS-g7G7.1h1 for 16.6 and 9 days, respectively (Fig. 2b). Thus, Ghd7.1 alone did not trigger Hd1 function conversion in the ZS97 background. We also developed combinations of ZS-G7G7.1H1, ZS-G7G7.1h1, ZS-G7g7.1H1, and ZS-G7g7.1h1. Both ZS-G7G7.1H1 and ZS-G7g7.1H1 headed later than did ZS-G7G7.1h1 and ZS-G7g7.1h1 by 21 and 3.7 days, respectively (Fig. 2b), indicating that Ghd7.1 enhances Ghd7-driven Hd1 function conversion like Ghd8 in the ZS97 background.
Ghd7 alone does not promote Hd1 function conversion in the MH63 background
Accordingly, four homozygous combinations of Ghd7.1 and Hd1 in the MH63 background were used to measure heading date under NLD. MH-G7g7.1H1 headed 11.3 days earlier than MH-G7g7.1h1, indicating that Hd1 promoted heading in the MH63 background with Ghd7 and ghd7.1 (Fig. 2a). MH-G7G7.1H1 headed later than MH-G7G7.1h1 by 6.8 days (Fig. 2a), indicating that Hd1 delayed heading in the MH63 background with Ghd7.1. Thus, Ghd7.1 was also involved in Hd1 function conversion in the MH63 background. Considering that Ghd7 was functional in MH63 but nonfunctional in ZS97, it is possible that Ghd7.1-driven Hd1 function conversion is dependent on Ghd7. Thus, MH-g7g7.1H1, MH-g7G7.1H1, MH-g7g7.1h1, and MH-g7G7.1h1 were constructed to eliminate the Ghd7 effects. As expected, MH-g7g7.1H1 and MH-g7G7.1H1 headed 30.4 and 9.1 days earlier than MH-g7g7.1h1 and MH-g7G7.1h1, respectively (Fig. 2a), indicating that Hd1 promoted heading. These results indicate that both Ghd7 and Ghd7.1 were required for Hd1 function conversion in the MH63 background.
Photoperiod sensitivity of eight homozygous three-gene combinations of Ghd7, Ghd7.1, and Hd1 in the MH63 and ZS97 backgrounds
We also measured heading dates for these eight homozygous combinations of Ghd7, Ghd7.1, and Hd1 in the ZS97 and MH63 backgrounds under NLD, NSD, and SD. Hd1 always promoted heading date on either the ZS97 or MH63 background under SD and NSD (Table 2).
PS was also estimated for the eight homozygous combinations of Ghd7, Ghd7.1, and Hd1. Hd1 alone did not express photoperiod sensitivity in MH-g7g7.1H1 or ZS-g7g7.1H1 in which only Hd1 was functional, whereas Hd1 showed strong photoperiod sensitivity with the help of either Ghd7 or Ghd7.1 (Table 2). When both Ghd7 and Ghd7.1 were present, the combination of these three functional genes (ZS-G7G7.1H1 and MH-G7G7.1H1) possessed the strongest PS, at 50.6 d and 38.8 d (Table 2). When Hd1 was nonfunctional, ghd7Ghd7.1 and Ghd7Ghd7.1 still possessed a strong PS of more than 20 days in the ZS97 background, while Ghd7ghd7.1 and ghd7ghd7.1 showed a weak PS of less than 8 days (Table 2). However, Ghd7ghd7.1 and ghd7ghd7.1 were insensitive to photoperiod in the MH63 background (Table 2).
Ghd8 joinly with Ghd7.1 converts Hd1 function in the ZS97 background
As mentioned above, Ghd8 and Ghd7.1 alone did not convert Hd1 function, but Ghd8 and Ghd7.1 helped Ghd7 greatly promoted Hd1 function conversion in the ZS97 background (Figs. 1 and 2b). In addition, Ghd7 alone or Ghd7.1 alone did not convert Hd1 function in the MH63 background, but when both were present, Hd1 function conversion occurred (Fig. 2a). We hypothesized that Ghd7.1 interacted with Ghd8 to convert Hd1 function without Ghd7. To verify this hypothesis, ZS-G7G8h1 was crossed with ZS-g7G7.1H1 to construct ZS-g7G8G7.1H1, ZS-g7G8G7.1h1, ZS-G7G8G7.1H1, and ZS-G7G8G7.1h1. As expected, ZS-g7G7.1G8H1 and ZS-G7G8G7.1H1 flowered 20.7 d and more than 35.8 d later than ZS-g7G8G7.1h1 and ZS-G7G8G7.1h1, respectively, under NLD (Fig. 3a and b). These findings suggest that Ghd7.1 together with Ghd8 converted Hd1 without Ghd7.
Ghd8 cooperates with Ghd7.1 to convert Hd1 function in the ZS97 background under NLD. a and b, the difference in heading date between Hd1 and hd1 in the ZS97 background with combinations of ghd7Ghd8Ghd7.1 and Ghd7Ghd8Ghd7.1, respectively. “>165d”, the heading date was more than 165 days, beyond our survey. **, significant difference in heading date at P < 0.01 by Student’s t test
Ehd1 and Hd3a are downregulated in the lines exhibiting Hd1 function conversion under LD
In this study, Hd1 (g7g8g7.1H1) promoted heading for approximately 11.3 d and 30.4 d in the ZS97 background and MH63 background, respectively (Figs. 1 and 2a), but Hd1 function conversion was found in six genotypes. Hd1 in the genotypes of ZS-G7G8G7.1H1 (Fig. 3b) and ZS-G7G8H1 (Fig. 1; Supplementary Table 2) extremely repressed flowering (for more than 35.8 d). Hd1 strongly suppressed heading date (for about 21 d) in both ZS-G7G7.1H1 (Fig. 2b) and ZS-g7G8G7.1h1 (Fig. 3a). In the MH-G7G7.1H1 genotype, Hd1 delayed heading for 6.8 d (Fig. 2a). Hd1 slightly delayed heading date in ZS-G7g8H1 (Fig. 1; Supplementary Table 2). To unveil the transcriptional mechanism of Hd1 function conversion, we analyzed the expression levels of Ehd1 and Hd3a, the main downstream targets of Hd1, in these six genotypes under LD. Ehd1 and Hd3a expression levels in ZS-g7g8H1 and MH-g7g7.1H1 in which Hd1 promoted heading were higher than ZS-g7g8h1 and MH-g7g7.1h1, respectively (Fig. 4a and b), but Ehd1 and Hd3a expression levels in these six genotypes in which Hd1 repressed flowering were much lower than in their corresponding hd1 near-isogenic lines (Fig. 4a and b). In general, the more the flowering was delayed, the more the expression of Hd3a was inhibited (Fig. 4b). These results indicated that Hd1 in the lines without Hd1 function conversion upregulated Ehd1 and Hd3a and promoted heading under LD, but Hd1 downregulated Ehd1 and Hd3a and suppressed heading in the lines with Hd1 function conversion.
The relative expression levels of Ehd1 and Hd3a in Hd1 function conversion lines. a and b, the relative expression levels of Ehd1 and Hd3a in Hd1 function conversion lines, respectively. Ubq was used as the internal reference. * and **, significant difference level at P < 0.05 and P < 0.01 by Student’s t test, respectively
Discussion
Multiple gene combinations enhance Hd1 function conversion
Hd1 was first reported to be sensitive to photoperiod because it promoted heading date under SD but delayed heading date under LD in rice (Yano et al. 2000). Then, Hd1 was reported to constantly promote heading date in recent studies (Zhang et al. 2012; Du et al. 2017; Zhang et al. 2017). Ghd7 and Ghd8 are required to convert Hd1 function from promoting to delaying heading under LD in the ZS97 and 93-11 backgrounds, respectively (Du et al. 2017; Zhang et al. 2017). The requirement of Ghd7 for Hd1 function conversion was confirmed in this study. However, Ghd8 alone did not convert Hd1 function (Fig. 1; Supplementary Table 2) but enhanced the effect of Ghd7 on the Hd1 function conversion in the ZS97 background (Fig. 1; Supplementary Table 2). Line 93-11 carries functional Ghd7 and Ghd8 (Zhang et al. 2015; Du et al. 2017). Hd1 function conversion in 93-11 is caused by the interaction between Ghd7 and Ghd8 rather than Ghd8 alone. We uncovered that Ghd7.1 alone did not drive Hd1 conversion, but it enhanced Ghd7 to promote Hd1 function conversion (Fig. 2a and b). Moreover, Ghd7.1 together with Ghd8 promoted Hd1 function conversion without Ghd7 (Fig. 3a). MH-G7g7.1H1 headed earlier than MH-G7g7.1h1 (Fig. 2a), which was different from that in the ZS97 background (Figs. 1 and 2b). Other genes probably participate in the regulation of Ghd7-driven Hd1 function conversion, which was absent in MH63 but present in ZS97. Taken together, these findings show that Hd1 function conversion was driven by multiple gene combinations among Ghd7, Ghd8, Ghd7.1, and other unknown factors rather than a single gene.
The NF-Y-CCT complexes possibly underlie the mechanism of Hd1 function conversion in rice
Ghd7, Ghd7.1, and Hd1 all encode CCT-domain-containing proteins, while Ghd8 encodes an NF-YB protein (Yano et al. 2000; Xue et al. 2008; Yan et al. 2011; Hori et al. 2013). NF-YB family proteins form a heterotrimer complex with NF-YC and NF-YA family proteins in a wide variety of eukaryotes (Petroni et al. 2012). CO, an ortholog of Hd1 in Arabidopsis, containing a CCT domain that is highly conserved with the NF-YA binding domain, interacts with NF-YB and NF-YC family proteins physically to form an NF-Y-CO complex through its CCT domain (Wenkel et al. 2006; Hou et al. 2014). In rice, the Ghd8-Hd1, Ghd7-Hd1, and OsNF-YA-Ghd8-OsNF-YC complexes have been identified in vitro or in vivo. Hd1 interacts with OsNF-YA, OsNF-YB, and OsNF-YC in yeast, and Ghd7.1 interacts with Ghd8 in yeast (Kim et al. 2016; Nemoto et al. 2016; Du et al. 2017; Goretti et al. 2017; Zhang et al. 2017; Zhu et al. 2017). These findings suggest that the NF-YC-CCT complexes, such as Ghd7-Ghd8-Hd1-OsNF-YC, Ghd7-OsNF-YB-Hd1-OsNF-YC, Ghd7.1-Ghd8-Hd1-OsNF-YC, and Ghd7-Ghd8-Ghd7.1-OsNF-YC, might be formed in rice. Identification of these complexes would be helpful in addressing the mechanism of Hd1 function conversion mediated by Ghd7, Ghd8, and Ghd7.1.
The Ghd8-Hd1 complex increases the H3K27me3 level on the Ghd8-binding site in the Hd3a promoter and leads to inhibited Hd3a expression (Du et al. 2017). In Arabidopsis, NF-Y-CO regulates flowering time by altering the histone H3K27me3 level of the FT and SOC1 promoter (Hou et al. 2014; Liu et al. 2018). Thus, we assume that NF-Y-CCT complexes in rice are necessary to determine Hd1 function conversion by upregulating the H3K27me3 level of Ehd1 and Hd3a and covering up the effect of transcriptional activation of Hd1.
Developing varieties for global and local ecological areas according to photoperiod sensitivity of multiple gene combinations
Photoperiod sensitivity is crucial for rice adaptation to cropping areas, and photoperiod insensitivity is a prerequisite for cropping elite varieties in wide regions. A positive correlation was observed between heading date and grain yield (Hu et al. 2019). Ghd7, Ghd7.1, Ghd8, and Hd1 are sensitive to photoperiod. Nonfunctional alleles of these photoperiod-sensitive genes show photoperiod insensitivity, which leads to similar heading dates either in LD or in SD (Yano et al. 2000; Xue et al. 2008; Yan et al. 2011; Hori et al. 2013). When these functional and nonfunctional alleles combined with each other, they had varied PS. The four gene combinations of Ghd7Ghd7.1Hd1, Ghd7Ghd7.1hd1, ghd7Ghd7.1Hd1, and Ghd7ghd7.1Hd1 possessed strong PS. Cultivars carrying these gene combinations should have long life cycles, even under SD, and adapt to subtropical regions. The three combinations Ghd7ghd7.1hd1, ghd7ghd7.1Hd1, and ghd7ghd7.1hd1 were weakly sensitive or insensitive to photoperiod. They had similar heading dates between LD and SD. Of them, the combination of Ghd7ghd7.1hd1 had 80 and 90 days to heading under conditions in the ZS97 and MH63 backgrounds, respectively, which makes them promising to plant in different regions for high yield production. The combination of ghd7ghd7.1Hd1 had very short heading times of 55 and 60 days in the ZS97 and MH63 backgrounds, respectively. They are adapted to the northeast China cropping regions or as early rice in central China, where day length is long day in the rice growing season. Above all, estimation of the PS index would help breeders to develop photoperiod-insensitive varieties that could be delivered to widespread ecological areas.
References
Cho LH, Yoon J, Pasriga R, An G (2016) Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol 170:2159–2171
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1. Genes Dev 18:926–936
Du A, Tian W, Wei M et al (2017) The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol Plant 10:948–961
Duncan DB (1955) Multiple range and multiple F tests. Biometrics 11:1–42
Goretti D, Martignago D, Landini M, Brambilla V, Gómez-Ariza J, Gnesutta N, Galbiati F, Collani S, Takagi H, Terauchi R, Mantovani R, Fornara F (2017) Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet 13:e1006530
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J 76:36–46
Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5:4601
Hu Y, Li SL, Xing YZ (2019) Lessons from natural variations: artificially induced heading date variations for improvement of regional adaptation in rice. Theor Appl Genet 132:383–394
Kim SK, Park HY, Jang YH, Lee KC, Chung YS, Lee JH, Kim JK (2016) OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 243:563–576
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135:767–774
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450
Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Liu T, Liu H, Zhang H, Xing Y (2013) Validation and characterization of Ghd7.1, a major quantitative trait locus with pleiotropic effects on spikelets per panicle, plant height, and heading date in rice (Oryza sativa L.). J Integr Plant Biol 55:917–927
Liu X, Yang Y, Hu Y et al (2018) Temporal-specific interaction of NF-YC and CURLY LEAF during the floral transition regulates flowering. Plant Physiol 77:105–114
Nemoto Y, Nonoue Y, Yano M, Izawa T (2016) Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J 86:221–233
Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24:4777–4792
Shen GJ, Xing YZ (2014) Two novel QTLs for heading date are identified using a set of chromosome segment substitution lines in rice (Oryza sativa L.). J Gen Genomics 41:659–662
Shibaya T, Nonoue Y, Ono N, Yamanouchi U, Hori K, Yano M (2011) Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions. Theor Appl Genet 123:1133–1143
Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750
Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335
Wenkel S, Turck F, Singer K, Gissot L, le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4:319–330
Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, Liu T, Liu H, Zhang C, Zhang Z, Shen G, Yao W, Chen H, Yu S, Xie W, Xing Y (2013) Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Research 23(7):969–971
Yano M, Katayose Y, Ashikari M et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Zhang J, Zhou X, Yan W, Zhang Z, Lu L, Han Z, Zhao H, Liu H, Song P, Hu Y, Shen G, He Q, Guo S, Gao G, Wang G, Xing Y (2015) Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol 208:1056–1066
Zhang Z, Hu W, Shen G, Liu H, Hu Y, Zhou X, Liu T, Xing Y (2017) Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep 7:5388
Zhang ZH, Wang K, Guo L, Zhu YJ, Fan YY, Cheng SH, Zhuang JY (2012) Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice. PLoS One 7:e52538
Zhu S, Wang J, Cai M, Zhang H, Wu F, Xu Y, Li C, Cheng Z, Zhang X, Guo X, Sheng P, Wu M, Wang J, Lei C, Wang J, Zhao Z, Wu C, Wang H, Wan J (2017) The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J Exp Bot 68:553–568
Acknowledgments
We would like to appreciate Mr. Jianbo Wang for his excellent management of paddy field.
Funding
This work is supported by the National Natural Science Foundation of China (31601283, 31701054) and the Applied Basis Research Foundation of Wuhan City (2016020101010090).
Author information
Authors and Affiliations
Contributions
ZYZ and BZ are equal contributors. ZYZ and BZ performed the construction of NILs, data collection and analysis, field experiments, and gene expression detection. FXQ and HW contributed to QTLs genotyping. ZXL and YZX designed the research. ZYZ, BZ, and YZX wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 202 kb)
Rights and permissions
About this article
Cite this article
Zhang, Z., Zhang, B., Qi, F. et al. Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice. Mol Breeding 39, 92 (2019). https://doi.org/10.1007/s11032-019-1001-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1001-8