Abstract
Saccharomyces cerevisiae has been widely used as a model system for the study of basic biological processes which are usually evolutionarily conserved from yeasts to multicellular eukaryotes. These studies are very important because they shed light on mechanisms that are altered in human diseases and help the development of new biomarkers and therapies. The mitotic spindle is a conserved apparatus that governs chromosome segregation during mitosis. Given its crucial role for genome stability and, therefore, for cell viability, its structure and function are strictly regulated. Recent findings reveal new levels of regulation in mitotic spindle dynamics and link spindle pole diversification with cell fate determination, health, disease and aging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mitotic spindle organization and function is essential for faithful chromosome segregation, that is in turn fundamental for genetic stability, cell survival and health of multicellular organisms. The mitotic spindle has a conserved structure and Saccharomyces cerevisiae is a very good model system to study its dynamics. Budding yeast is a unicellular eukaryotic organism that is easy to grow in laboratory conditions and to manipulate genetically. Both direct and reverse genetics studies can be performed using yeast, and they give complementary results. In addition, yeast is suitable for microscopic analyses, cell biology studies, biochemical assays and for high-throughput screenings. The information gained by studies in this model organism are relevant to comprehend the physiology of the eukaryotic cell and to unravel the molecular mechanisms that cause human diseases (Fraschini 2019).
The mitotic spindle is formed by microtubules (MTs), cylindric structures made by protofilaments of α- and β-tubulin heterodimers assembled in a head-to-tail fashion. Each MT has a dynamic fast growing end (plus-end) and a slow growing end (minus-end). MTs are associated with several proteins that control spindle dynamics and with motor proteins that allow spindle positioning in the cell and intracellular transport. Before sister chromatids separation in anaphase, MT plus-ends bind the chromosomes via their kinetochores in a bipolar way, thus ensuring their correct migration in the daughter cells (Dhatchinamoorthy et al. 2018). In budding yeast nuclear MTs and astral MTs are nucleated from the spindle pole bodies (SPBs, the yeast MT organizing center, MTOC), that are embedded in the nuclear envelope, and the bipolar spindle is formed during S phase, concomitantly with DNA duplication. The SPBs are also important for clustering of telomeres, the ends of chromosomes; indeed they bind LINC (linker of nucleoskeleton and cytoskeleton) complexes that are essential for chromosome positioning and, consequently, for proper spindle formation and nuclear fusion during karyogamy (Katsumata et al. 2017).
Since S. cerevisiae cells divide asymmetrically and the division site is determined before mitotic spindle formation, the mitotic spindle must be properly positioned at the bud neck and correctly oriented towards the bud before cells the cell enters into mitosis (Fraschini 2017). These processes are regulated by the Kar9 pathway and the Dyn1 pathway. During S phase, Kar9 is asymmetrically recruited to the SPB that will migrate into the bud, then Kar9, associated with the MT binding protein Bim1, moves to the microtubule plus-ends where the complex interacts with the actin-associated myosin Myo2, which then pulls Kar9 and the associated microtubule into the bud (Lee et al. 2000; Fraschini et al. 2008). The Dyn1 pathway acts during anaphase and it drives the final positioning of the spindle along the cell polarity axis: the motor protein Dyn1 and dynactin form a complex, they bind MTs and pull the SPB directed into the bud through the bud neck (Yeh et al. 1995; Fraschini et al. 2008; Heil-Chapdelaine et al. 2000). Mitotic spindle dynamics are regulated also by kinesins, such as Kip3 (Carvalho et al. 2003, 2004; Pearson and Bloom 2004) and by some proteins that shuttle from the kinetochore to the spindle midzone, the so called chromosomal passenger complex (CPC), which consists of aurora kinase Ipl1, Bir1, Sli15 and Nbl1 (Nakajima et al. 2011; Makrantoni et al. 2014).
Interplay among Swe1, Mih1 and Bik1 in mitotic spindle dynamics
Cdc28 is the catalytic subunit of the cyclin-dependent kinase Cdk1, the only Cdk that governs all cell cycle transitions in S. cerevisiae (Hartwell et al. 1973). Cdc28 activity is controlled by its binding with different type of cyclins and by phosphorylation of its tyrosine 19 (Y19). The protein kinase Swe1/hWee1 directly phosphorylates Y19 thus inhibiting its activity, and this modification is reversed by the phosphatase Mih1/hCdc25 (Russel et al. 1989). Mih1 is regulated at the post-translational level by phosphorylation (Pal et al. 2008), while Swe1 is regulated at the transcriptional level and post-translational level by phosphorylation and regulation of its subcellular localization (Asano et al. 2005). During an unperturbed cell cycle, Swe1 is produced during S phase then it is phosphorylated and translocated to the bud neck where it undergoes further phosphorylation that allows its ubiquitination and degradation (Harvey et al. 2005). To entry into mitosis, dephosphorylated Cdc28 must associate with mitotic cyclins; in case of problems in with cell morphogenesis, Swe1 degradation is inhibited, it accumulates in the cells, it inactivates Cdc28 by phosphorylation and, therefore, mitotic entry is blocked (Lew 2003). When the errors are repaired, Mih1 reverts Cdc28 Y19 phosphorylation, thereby promoting its activation and progression through mitosis. Therefore, both Swe1 and Mih1 are important for the regulation of cell cycle progression in morphogenetic stressing condition, but they have a role also during unperturbed conditions. Moreover, Swe1 is also an effector of the DNA damage checkpoint kinase Mec1/ATR, thus blocking anaphase in response to different kind of DNA lesions (Palou et al. 2017).
Previous works indicate that Swe1 is involved in mitotic spindle elongation (Raspelli et al. 2015) and in spindle pole asymmetry (Lengefeld et al. 2017), a process that is strictly connected with mitotic spindle position and orientation in the cell (see next paragraph). In accord with our previous study, we recently observed that Swe1 plays a positive role in mitotic spindle positioning. Our genetic analyses show that Swe1 lack exacerbates the phenotype of some mutants involved in spindle dynamics, so we concluded that Swe1 likely acts in parallel to the chromosome passenger Ipl1 and Sli15, the kinesin Kip3, Kar9, Bim1 and Dyn1 (dynein). Conversely, the concomitant absence of Swe1 and Bik1 does not cause an additive effect, indicating that these two proteins act in concert (Raspelli et al. 2018).
Bik1 (BIlateral Karyogamy defect 1) is a microtubule-associated protein (MAP) that binds MT plus-ends (+ TIP); it is involved in MT dynamics, it targets Dyn1 to microtubule plus-ends and controls the asymmetric localization of Kar9 to the SPBs (Moore et al. 2006). In addition, it is a negative regulator of MT assembly. Bik1 is the ortholog of mammalian CLIP-170 (CLIP1), and defects in microtubule association in bik1 mutants are functionally complemented by a human CLIP-170 (Lin et al. 2001).
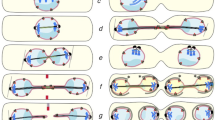
Bik1 is a 51 kDa protein and its levels are constant during the cell cycle; however, its post-translational modifications change. In particular, it is unmodified in G1 phase and gets phosphorylated (p-Bik1) when the bipolar spindle is formed; later, as cells progress through mitosis, elongate the spindle and divide, Bik1 is dephosphorylated (Raspelli et al. 2018). Bik1 has a CDK phosphorylation consensus motif, but this direct phosphorylation has never been proved in vivo. In addition, despite proteome chip analysis data, Bik1 is not directly phosphorylated by Swe1, as its slow migrating forms rise in the absence of Swe1 and are reduced, and not enhanced, in presence of high Swe1 levels. Co-immunoprecipitation and genetic data indicate that, instead, Swe1 could phosphorylate and inhibit Mih1 that, in turn, could dephosphorylate Bik1; this is also supported by the fact that p-Bik1 increases in the absence of Mih1 and that the two proteins physically interact (Raspelli et al. 2018). Interestingly, cells lacking Mih1 elongate the mitotic spindle with a delay respect to wild-type cells, and show a defect in spindle positioning (Raspelli et al. 2018). All these data fit into a model in which p-Bik1 is inactive while Bik1 plays a positive role in mitotic spindle elongation: Bik1 is likely phosphorylated by Cdc28 and by an unknown kinase, and this modification is reversed by Mih1, that allows Bik1 activation. Mih1 activity, in turn, is also finely regulated by Swe1 and by the polo-like kinase Cdc5, a key mitotic regulator (Botchkarev and Haber 2018), thus linking Bik1 activation to cell cycle progression (Fig. 1).
To perform its function, Bik1 is localized to the astral MT plus-ends, to the SPBs, and there is also a soluble pool in the cytoplasm from which it can be recruited during MT polimerization (Carvalho et al. 2004). Bik1 is recruited to MTs + TIPs by its binding with the kinesin Kip2, and moves to the periphery of the cell as the astral MTs grow. There is a very stable Bik1 pool at the SPBs, likely recruited by its binding with the MAP Stu2 and the SPB component Spc72 (Chen et al. 1998). Even if Bik1 subcellular localization has been very well described, it is not known its interplay with Bik1 activation, so it will be very interesting to investigate the connections between Bik1 subcellular localization and its phosphorylation state. To clarify the role of Bik1 phosphorylation in driving its localization, it will be necessary to identify Bik1 residues that are phosphorylated in vivo, mutagenize them to create strains that express non-phosphorylatable and phosphomimic variants, and then analyze their phenotype. Importantly, it has been shown that CLIP-170 has very similar dynamics in MT binding, so the relationship between Bik1 mutations and their effect on Bik1 activation and localization will be very helpful for understanding CLIP-170 role and function in human cells.
Dynamic interactions that are crucial for spindle positioning and alignment occur at the microtubule plus-ends. Recently, it has been described that Bim1, the human EB1, forms a complex with Bik1 and interacts with other players in spindle positioning. The Bim1–Bik1 complex is important for Bik1 localization to astral MT plus-ends and astral microtubule length (Stangier et al. 2018). However, the disruption of the complex does not significantly affect spindle pole asymmetry and spindle positioning, indicating that the previously described Bik1 role in these processes does not rely on its interaction with Bim1. Since both the proteins are conserved, it is possible to hypothesize that the CLIP-170-EB1 module is evolutionarily flexible.
CLIP-170 has been implicated in tumorigenesis and autosomal recessive intellectual disability (Tame et al. 2016; Larti et al. 2015). In particular, it has been shown that depletion of CLIP-170 induces spindle positioning defects and chromosome misalignment, both involved in cancer development. In addition, CLIP-170 has been implicated in MT-mediated transport in axons and dendrites, as the protein is absent in lymphoblastoid and skin fibroblast cell lines established from patients affected by autosomal recessive intellectual disability (ARID), thus pointing to a role for CLIP-170 in neuronal development. It is, therefore, important to gain detailed informations on Bik1 regulation and function to improve our knowledge of the molecular basis of human diseases and to help the discovery of new biomarkers and treatments.
Spindle poles maturation and asymmetric division
The polarity is an important matter for eukaryotic cells, indeed most cell types are polarized and polarity allows cells to perform specific functions. A symmetric cell division generates two identical daughter cells, while asymmetric cell division gives rise to cell diversity and this is important for both development and aging. During vertebrate development, the asymmetric division of stem cells produces a daughter cell that will differentiate and another cell that will maintain the ability to proliferate. During cell division, some molecules and structures are differentially segregated depending on their age thus influencing the aging of the cell.
Within the cell, asymmetry is built by the polarization of several cytoplasmatic factors in specific positions, and these factors are evolutionarily conserved. In metazoan cells, the PAR complex (PAR-3, PAR-6, aPKC) and Crumbs complex (Crb, PALS, PATJ, Lin7) localize asymmetrically at the cell membrane, are bound by microtubules asters with their associated proteins, and this process allows the alignment of the mitotic spindle to the polarity axis of the cell. The position of the spindle defines the division site, and, therefore, drives asymmetric division (Siller and Doe 2009).
Saccharomyces cerevisiae is a very good model organism to study polarity establishment, as budding yeast divides asymmetrically: the daughter cell is emanated from the mother cell surface as a bud that grows in a polarized way, then it becomes round shaped thanks to switch to isotropical growth. The bud is the equivalent of the stem cell that maintain pluripotency and the ability to divide, while the mother cell gets old and reflects the destiny of the cell that differentiate. In budding yeast, the localization of polarity factors generates an asymmetry of the cytoskeleton that determines the bud emergence site. Differently from higher eukaryotes cells, in yeast the bud neck defines the site of cell division and the mother-bud axis in late G1 phase, before DNA replication and mitotic spindle formation. This implies that to ensure proper chromosome partitioning, the mitotic spindle must be aligned with respect to this axis before nuclear division (Lee et al. 2000). This process is highly controlled since it is fundamental to maintain genetic integrity and cell vitality. The spindle orientation checkpoint (SPOC) inhibits mitotic exit and cytokinesis in case of spindle mispositioning (Caydasi and Pereira 2012). If the checkpoint fails, the nucleus can divide even if the spindle is not properly oriented, and cytokinesis occurence leads to the formation of aneuploid cells.
The mitotic spindle is formed thanks to the spindle pole bodies (SPBs) that are embedded in the nuclear envelope and are able to nucleate MTs. The SPB is duplicated during S phase, then the two SPBs undergo different steps of regulation (maturation) that make them different from each other: the old SPB and the new SPB. The SPB inheritance network (SPIN) and the mitotic exit network (MEN), both related to the metazoans Hippo pathway, build up SPBs asymmetry. Interestingly, the spindle poles do not segregate randomly during mitosis, but usually the old SPB migrates into the bud. This task is achieved by the action of long astral MTs that contact the bud cortex; in addition, Kar9 protein plays a major role in spindle pole asymmetry and segregation. Indeed, it has been shown that differential Kar9 recruitment to spindle poles drives the movement of the selected SPB to the bud neck and that this process helps spindle alignment (Fig. 1) (Liakopoulos et al. 2003). The MEN kinases Dbf2/20 phosphorylate Kar9 thus allowing its recruitment to astral MTs emanated from the old SPB (Hotz et al. 2012). Kar9 may also represent a crucial connection between spindle positioning and cellular metabolism, as recent data indicate that the energy sensor Snf1/AMPK is localized to the bud neck and promotes spindle orientation acting in concert with Kar9 and in parallel with Dyn1 (Tripodi et al. 2018). Interestingly, the active form of human AMPK binds the centrosomes and the spindle midzone, playing an important role during mitosis and opposing tumor progression in several cancer types; consistently, it has been indicated as a possible metabolic tumor suppressor and a potential target for cancer (Li et al. 2015).
In addition, the SPIN network controls SPB identification. The kinase Swe1 phosphorylates the SPB component Nud1 during G1 phase, then subsequent Swe1 inactivation in G2 prevents Nud1 phosphorylation on the newly formed SPB (Lengefeld et al. 2017). The downstream SPIN components Kin3 and NuA4 recognize the SPB marked by Swe1, and further phosphorylate Nud1 and the SPB component Spc72. As Nud1 is a scaffold for MEN activation at SPBs, it is, therefore, clear that SPIN action helps the localization of MEN components to the old SPB: SPIN network marks the old SPB and MEN pathway loads Kar9 on the same SPB. More recent data revealed that the asymmetry of spindle poles is due to Kar9 recruitment and to SPB positioning close to the bud neck, rather than the kinetics of SPB maturation (Lengefeld et al. 2018).
Similarly, in animal cells, the spindle poles are not identical and do not segregate randomly, instead centrosome inheritance is concord with cell fate decision: usually the old MTOC nucleates more astral MTs and is surrounded by more PCM than the new one, indicating that the old one is fully active while the new one is immature (Lerit and Rusan 2013). In mouse neural stem cells, the old centrosome migrates into the stem daughter cell (Wang et al. 2009). In the stem cells of Drosophila male germline, the old MTOC migrates in the renewing daughter cell while the new centrosome is inherited by the differentiating cell (Yamashita et al. 2007). Similar data were obtained in mouse radial glia progenitors and in Drosophila neuroblasts (Januschke et al. 2011), indicating that asymmetry of MTOCs and fate decision is a common feature of eukaryotic cells.
It is important to point out that MEN and SPIN components are evolutionarily conserved and all their orthologues localize at centrosomes. Therefore, specific MTOC segregation seems to be an ancient process that has been preserved along evolution of species.
The age-dependent MTOC segregation clearly indicates that the old spindle pole is not less functional than the newly synthesized and is not consistent with the simple hypothesis that links molecules’ age with physiological cellular aging. However, it is known that old organelles are less functional than young ones so likely the influence of aged cellular structures on aging is more complex that previously imagined.
Conclusion
In conclusion, the recent data highlight new important functions of some evolutionarily conserved proteins in mitotic spindle dynamics. The protein kinase Swe1 is involved in spindle pole asymmetry, spindle positioning and elongation, while the phosphatase Mih1 regulates spindle positioning and elongation via the MAP Bik1. Moreover, Bik1 undergoes multiple levels of regulation that involve post-translational modifications (namely phosphorylation) and modification of its subcellular localization, and the interplay between these processes is currently under investigation. Altogether these results contribute to underline the importance of these proteins for cell physiology and help the researchers in the endless battle against diseases that are incurable today but hopefully will be defeated in the near future.
References
Asano S, Park JE, Sakchaisri K, Yu LR, Song S, Supavilai P, Veenstra TD, Lee KS (2005) Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J 24(12):2194–2204
Botchkarev VV Jr, Haber JE (2018) Functions and regulation of the Polo-like kinase Cdc5 in the absence and presence of DNA damage. Curr Genet 64(1):87–96
Carvalho P, Gupta ML Jr, Hoyt MA, Pellman D (2004) Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell 6(6):815–829
Carvalho P, Tirnauer JS, Pellman D (2003) Surfing on microtubule ends. Trends Cell Biol 13(5):229–237
Caydasi AK, Pereira G (2012) SPOC alert when chromosomes get the wrong direction. Exp Cell Res 318(12):1421–1427
Chen XP, Yin H, Huffaker TC (1998) The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol 141:1169–1179
Dhatchinamoorthy K, Mattingly M, Gerton JL (2018) Regulation of kinetochore configuration during mitosis. Curr Genet 64(6):1197–1203
Fraschini R (2017) Factors that control mitotic spindle dynamics. Adv Exp Med Biol 925:89–101
Fraschini R (2019) Divide precisely and proliferate safely: lessons from budding yeast. Front Genet 9:738
Fraschini R, Venturetti M, Chiroli E, Piatti S (2008) The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans 36(Pt 3):416–420
Hartwell LH, Mortimer RK, Culotti J, Culotti M (1973) Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74(2):267–286
Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR (2005) Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 122(3):407–420
Heil-Chapdelaine RA, Oberle JR, Cooper JA (2000) The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151(6):1337–1344
Hotz M, Lengefeld J, Barral Y (2012) The MEN mediates the effects of the spindle assembly checkpoint on Kar9-dependent spindle pole body inheritance in budding yeast. Cell Cycle 11(16):3109–3116
Januschke J, Llamazares S, Reina J, Gonzalez C (2011) Drosophila neuroblasts retain the daughter centrosome. Nat Commun 2:243
Katsumata K, Nishi E, Afrin S, Narusawa K, Yamamoto A (2017) Position matters: multiple functions of LINC-dependent chromosome positioning during meiosis. Curr Genet 63(6):1037–1052
Larti F, Kahrizi K, Musante L, Hu H, Papari E, Fattahi Z, Bazazzadegan N, Liu Z, Banan M, Garshasbi M, Wienker TF, Ropers HH, Galjart N, Najmabadi H (2015) A defect in the CLIP1 gene (CLIP-170) can cause autosomal recessive intellectual disability. Eur J Hum Genet 23(3):331–336
Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D (2000) Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287(5461):2260–2262
Lengefeld J, Hotz M, Rollins M, Baetz K, Barral Y (2017) Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation. Nat Cell Biol 19(8):941–951
Lengefeld J, Yen E, Chen X, Leary A, Vogel J, Barral Y (2018) Spatial cues and not spindle pole maturation drive the asymmetry of astral microtubules between new and preexisting spindle poles. Mol Biol Cell 29(1):10–28
Lerit DA, Rusan NM (2013) PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J Cell Biol 202(7):1013–1022
Lew DJ (2003) The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol 15:648–653
Li W, Saud SM, Young MR, Chen G, Hua B (2015) Targeting AMPK for cancer prevention and treatment. Oncotarget 6(10):7365–7378
Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y (2003) Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112(4):561–574
Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D (2001) Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol 155(7):1173–1184
Makrantoni V, Corbishley SJ, Rachidi N, Morrice NA, Robinson DA, Stark MJ (2014) Phosphorylation of Sli15 by Ipl1 is important for proper CPC localization and chromosome stability in Saccharomyces cerevisiae. PLoS One 9(2):e89399
Moore JK, D’Silva S, Miller RK (2006) The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell 17(1):178–191
Nakajima Y, Cormier A, Tyers RG, Pigula A, Peng Y, Drubin DG, Barnes G (2011) Ipl1/aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics. J Cell Biol 194(1):137–153
Pal G, Paraz MT, Kellogg DR (2008) Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J Cell Biol 180:931–945
Palou R, Palou G, Quintana DG (2017) A role for the spindle assembly checkpoint in the DNA damage response. Curr Genet 63(2):275–280
Pearson CG, Bloom K (2004) Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol 5(6):481–492
Raspelli E, Cassani C, Chiroli E, Fraschini R (2015) Budding yeast Swe1 is involved in the control of mitotic spindle elongation and is regulated by Cdc14 phosphatase during mitosis. J Biol Chem 290(1):1–12
Raspelli E, Facchinetti S, Fraschini R (2018) Novel insights into Swe1 and Mih1 role in the regulation of mitotic spindle dynamics. J Cell Sci 131(17):1–13
Russell P, Moreno S, Reed SI (1989) Conservation of mitotic controls in fission and budding yeasts. Cell 57(2):295–303
Siller KH, Doe CQ (2009) Spindle orientation during asymmetric cell division. Nat Cell Biol 11(4):365–374
Stangier MM, Kumar A, Chen X, Farcas AM, Barral Y, Steinmetz MO (2018) Structure–function relationship of the Bik1–Bim1 complex. Structure 26(4):607–618
Tame MA, Raaijmakers JA, Afanasyev P, Medema RH (2016) Chromosome misalignments induce spindle-positioning defects. EMBO Rep 17(3):317–325
Tripodi F, Fraschini R, Zocchi M, Reghellin V, Coccetti P (2018) Snf1/AMPK is involved in the mitotic spindle alignment in Saccharomyces cerevisiae. Sci Rep 8(1):5853
Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461(7266):947–955
Yamashita YM, Mahowald AP, Perlin JR, Fuller MT (2007) Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315(5811):518–521
Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K (1995) Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130(3):687–700
Acknowledgements
R. F. researches are supported by Grants from PRIN (Progetti di Ricerca di Interesse Nazionale) and from the University of Milano Bicocca (FA). E. R. is supported by a fellowship from the Associazione Italiana per Ricerca sul Cancro (AIRC), Love Design Rif: 18196.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Raspelli, E., Fraschini, R. Spindle pole power in health and disease. Curr Genet 65, 851–855 (2019). https://doi.org/10.1007/s00294-019-00941-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-019-00941-7