Abstract
DNA replication is essential to cellular proliferation. The cellular-scale organization of the replication machinery (replisome) and the replicating chromosome has remained controversial. Two competing models describe the replication process: In the track model, the replisomes translocate along the DNA like a train on a track. Alternately, in the factory model, the replisomes form a stationary complex through which the DNA is pulled. We summarize the evidence for each model and discuss a number of confounding aspects that complicate interpretation of the observations. We advocate a factory-like model for bacterial replication where the replisomes form a relatively stationary and weakly associated complex that can transiently separate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The essential process of DNA replication underlies cellular proliferation. In all organisms, multi-protein replication complexes assemble at specific DNA sequences (origins) and proceed to replicate the genome by a semi-conservative process (Meselson and Stahl 1958). Although functional replication complexes have been reconstituted in vitro from purified bacterial proteins (Zechner et al. 1992; Sanders et al. 2010), many interesting questions remain about the replication process and its regulation in the context of the cell (Mangiameli et al. 2017; Frimodt-Møller et al. 2017). In particular, the in vivo cellular-scale organization of the replication machinery (replisome) and the replicating chromosome has remained controversial in bacteria. Do replisomes translocate along the DNA molecule, or is the DNA molecule pulled through the replisomes? We refer to the first model as a track model and the latter as a factory model. Cell-biology experiments have provided evidence for both models in the bacterial cell.
Eukaryotic and bacterial replication factories. a A eukaryotic replication factory (gray) resulting from simultaneous activation of five origin sequences (black dots). Replication complexes (blue) duplicate the DNA bi-directionally from the origins. Red arrows indicate that the direction DNA is pulled into the complex. Adapted from Frouin et al. (2003). b A bacterial replication factory would involve only two replication complexes working bi-directionally from a single origin. Red arrows indicate the direction DNA is pulled into the complex
The factory model was first established in eukaryotic cells (Dingman 1974), where DNA synthesis is localized to a number of puncta, called replication factories, distributed throughout the nucleus (Newport and Yan 1996; Frouin et al. 2003). The number of puncta is much smaller than the number of origins, implying that many replisomes co-localize to each factory. Each replication factory consists of roughly five clustered and synchronously activated origins (Ma et al. 1998; Jackson and Pombo 1998). Early evidence for replication factories included the visualization of punctate foci formed by the co-localization of newly synthesized DNA with replication proteins by immunofluorescence (P. Hozák et al. 1993). More recent imaging studies have demonstrated the spatiotemporal stability of replication factories in living cells based on the sub-nuclear positioning of a fluorescent fusion of the processivity clamp (PCNA), and have hypothesized the existence of a physical linker anchoring the factory in the nucleus (Leonhardt et al. 2000). However, the analysis of replication in eukaryotic cells is confounded by the large number of replication origins distributed throughout the genome, making it difficult to study the dynamics of individual replisomes (Fig. 1).
Bacterial DNA replication is comparatively simple: Most bacteria possess only a single circular chromosome that is replicated bi-directionally from a single origin of replication. Two replisomes initiate at the origin and process DNA in opposite directions, each replicating one arm of the chromosome before meeting in the terminus region. Because replication proteins are highly conserved across all organisms (Baker and Bell 1998), sequence conservation might be expected to result in a common organization of the replication process between bacteria and eukaryotes. However, it has remained unclear whether factory-like organization is conserved in bacteria.
Although published evidence generally supports the factory model in the model organisms Caulobacter crescentus (Jensen et al. 2001) and Bacillus subtilis (Lemon and Grossman 1998, 2000; Berkmen and Grossman 2006; Mangiameli et al. 2017), a number of recent reports suggest prolonged separation of replication fork pairs Escherichia coli (Bates and Kleckner 2005; Reyes-Lamothe et al. 2008; Hiraga et al. 2000; Kongsuwan et al. 2002). We have recently published a comparative study of the organization of replication process in E. coli and B. subtilis (Mangiameli et al. 2017) where we observed unexpected similarities between the organization in these two highly divergent species (Mangiameli et al. 2017). The study, which used fluorescence microscopy to track replisome components over complete cell cycles, provided a natural explanation for the contradictory reports: There are two different scenarios that can result in two optically resolvable replication foci: (1) track-like organization consisting of individually resolvable replisomes acting on a single chromosome and (2) factory-like organization consisting unresolved replisome pairs at the quarter-cell positions after replication re-initiation (i.e., re-initiation at the origins of two separate chromosomes). As a result, analysis dependent on counting the number of observed replication foci using single images cannot distinguish between these two populations and, therefore, is not a reliable method for determining replisome organization (Mangiameli et al. 2017).
Models for the organization of replication
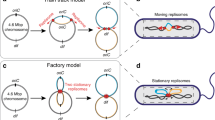
To discuss the evidence for the competing models, it is necessary to describe these models precisely. In all models, replisome pairs are coincident during and shortly after initiation. Subsequently, in the track model, the replisomes translocate in opposite directions along a (relatively) stationary DNA track, leading to sustained separation of replisome pairs as they follow chromosomal structure (Fig. 2). Since the replisomes translocate along the chromosome, the specific replisome localization predicted by the track model is dependent on nucleoid structure. For example, in C. crescentus, the chromosome is organized, such that oriC and ter are positioned at opposite cell poles (Jensen et al. 2001) (see Fig. 2b); therefore, the track model would predict end-to-end displacement of the replisomes along the DNA molecule. Due to the circular topology of the chromosome, replisome pairs may co-localize again at the terminus, but this is not necessary. If one member of a replisome pair arrives in the terminus region earlier, it may release the template DNA before the other arrives.
In the factory model, DNA is pulled or spooled through the replication complex. From a mechanistic perspective, this model may include a factor that anchors the replisomes to the cell, or to one another, but it may not. Generically speaking, the sub-cellular localization of the replisomes may change, as long as the movement is driven by chromosomal re-arrangements rather than motion of the replisomes along the DNA.
Comparison of chromosome structure in model organisms. a Schematic diagrams for the factory and track models. Most bacteria have a single circular chromosome (left), with a single origin (red dot), positioned roughly opposite the terminus (green dot). The left and right arms of the chromosome are colored pink and green, respectively. In the factory model (center), DNA is pulled through the replisomes (black dots) in the direction indicated by the red arrows. In the track model (right), replisomes translocate along the template DNA. Red arrows indicate the direction of replisome motion. b Factory model is shown for the chromosomal organizations of three model organisms. Decondensed DNA is represented by lines. Note that the E. coli chromosomal structure reflects the experimental observations in Cass et al. (2016)
The visualization of replisome dynamics
The most direct approach to characterizing the cellular organization of the replication process is by time-lapse imaging throughout the cell cycle. We have characterized the full-cell-cycle dynamics of the replisome in both B. subtilis and E. coli cells (Mangiameli et al. 2017). The first challenge in these experiments is finding a replisome-associated protein with sufficiently high stoichiometry to observe throughout the cell cycle. In both organisms, we imaged a fluorescent fusion to the processivity clamp (DnaN), expressed from the endogenous locus, as a proxy for the replisome localization. These fusions do not lead to a detectable defect in replication or growth. Furthermore, cells were propagated using a slow growth rate to avoid the complications of multi-fork replication.
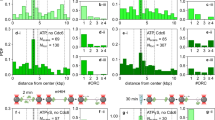
Contrary to the previous reports, E. coli and B. subtilis showed virtually indistinguishable replisome dynamics, summarized as follows: (1) A single midcell focus appears shortly before or after the beginning of the cell cycle. Cells born with a focus initiated replication before the beginning of the cell cycle. We infer that this single focus represents a pair of replisomes that cannot be optically resolved. (2) The midcell focus subsequently exhibits confined random motion (as observed previously Migocki et al. 2004), and is occasionally observed to reversibly separate, such that individual replisomes are optically resolvable (>250 nm separation). Importantly, the focus separation is both small (a fifth of a cell length on average) and transient. (3) The midcell focus disappears before the end of the cell cycle. We infer that this replisome disassembly corresponds to the termination of replication. (4) In roughly 45% of cells, foci re-appear at the quarter-cell positions. We infer that these quarter-cell foci result from re-initiation of replication prior to cell division as they persist through cell division and are unable to form in an initiation-deficient conditional mutant. Therefore, these observed quarter-cell loci represent replisome pairs. In agreement with one of the oldest studies of replisome localization in B. subtilis (Lemon and Grossman 1998), we find that replisome pair separation is diffraction-limited roughly 80% of the time (Mangiameli et al. 2017) (see Fig. 3).
Replisome dynamics in B. subtilis and E. coli based on time-lapse imaging. Schematic diagram showing full replication cycles in two individual example cells. Green spots represent diffraction-limited replisome foci, while black dots indicate the inferred number of constituent replisomes. Gray vertical lines indicate mid- and quarter-cell positions
Throughout the majority of the replication cycle, a factory-like organization is observed in both B. subtilis and E. coli. Consistent with the factory model, replisome pairs operate in close proximity for the majority (\(\approx 80\%\)) of the replication cycle. Although the two replisomes are usually not optically resolvable, they do not appear to be strongly associated since transient separation events do occur. Although these transient separation events could be consistent with the track model, this is unlikely the case. The previous reports have also noted this dynamic (Migocki et al. 2004; Berkmen and Grossman 2006), and demonstrated that these splitting and merging events continue even when the replisomes are chemically stalled (and are, therefore, not a consequence of replisome movement along the DNA) (Berkmen and Grossman 2006).
Even the time-lapse approach to studying replisome organization suffers from a number of potential shortcomings: time-lapse imaging is especially challenging in the current context due to the low stoichiometry of many replisome components (Reyes-Lamothe et al. 2010), which severely limits the number of images that can be captured before protein bleaching. We have, therefore, focused on characterizing DnaN as a proxy for the replisome, rather than characterizing each replisome protein in time-lapse image analysis. Snap-shot images of other components are consistent with our model, but the replisome has only been extensively visualized one component at a time. Studies generally find self-consistency when using different markers for the replisome; however, from the existing data, it is hard to exclude the possibility that replisome components may not be co-localized throughout the replication process. Finally, fluorescent proteins are known to form aggregates which could lead to anomalous protein localization (Swulius and Jensen 2012; Landgraf et al. 2012).
The interpretation of snap-shot images
Although the time-lapse data from DnaN seem to strongly support a stationary factory model, there are reports supporting both the factory and track models in E. coli (Koppes et al. 1999; Molina and Skarstad 2004; Hiraga et al. 2000; Kongsuwan et al. 2002; Adachi et al. 2005; Bates and Kleckner 2005; Den Blaauwen et al. 2006; Reyes-Lamothe et al. 2008; Mangiameli et al. 2017). Many of these investigations have analyzed the organization of the replisome or the newly replicated DNA (by fluorescence microscopy) using snap-shot imaging (i.e., single images). To reconstruct replisome dynamics from the snap-shot data, asynchronous populations of cells are analyzed by length, a proxy for cell age. Alternatively, a sub-culture of a synchronous population can be fixed and visualized at many time points throughout the cell cycle (Bates and Kleckner 2005). Changes in the sub-cellular focus localization organization are analyzed relative to cell length.
In sufficiently slow growth conditions, such that only a single ongoing round of replication is expected, younger cells have a single focus, while older cells often have pairs of foci localized to the quarter-cell positions. Two narrowly separated foci are also reported to occur in intermediate aged cells. In retrospect, it is clear that these data are consistent with both the factory and track model, since this approach cannot differentiate replisome splitting from re-initiation, as observed in the time-lapse data. We note that, in some cases, flow cytometry has been used as an elegant method to infer the presence of overlapping replication cycles (e.g. Molina and Skarstad 2004). This has particular importance in fast growth conditions where multiple ongoing replication cycles further complicate the interpretation of replication foci.
A shortcoming of snap-shot imaging is that populations of genetically identical cells have a wide distribution of cell cycle lengths, even under the same growth conditions (Wallden et al. 2016). Furthermore, there is a significant variation in cell length at birth and division. As a result, it is impossible to use the cell length as a precise measure of cell age (Cass et al. 2017), further complicating the snap-shot analysis. Although it is more challenging to interpret, we have demonstrated that the factory-like localization pattern can be inferred directly from snap-shot data (Mangiameli et al. 2017), providing an additional support to the factory model from protein foci that are too dim to track throughout the cell cycle. In addition to inherent cell-to-cell variation, the studies conducted to date have used a variety of growth conditions and background strains, making it difficult to make any direct comparisons across studies.
A translocating factory model
A number of more complicated models have been proposed for replisome localization. For instance, in the translocating factory model, replisome pairs are localized to midcell for roughly half the replication cycle, before abruptly transitioning to the quarter-cell positions where they remain for the rest of the cell cycle (Hiraga et al. 2000; Yamazoe et al. 2004; Onogi et al. 2002; Sunako et al. 2002). We note that studies citing results consistent with the translocating factory model argue that quarter-cell foci represent single replication forks; however, our time-lapse imaging and experiments strongly argue that these foci correspond to re-initiated pairs of replisomes (Mangiameli et al. 2017). Specifically, when replication initiation is blocked using a temperature-sensitive version of the helicase-loader protein (dnaC2 allele), quarter-cell foci do not appear.
Coupling or anchoring of the replisomes?
The observed transient separation of replisome pairs suggests that the forks are only weakly associated. If a direct link between replication fork pairs existed, it would need to frequently disassemble, or be of sufficient length to accommodate significant separations. Although it has not been excluded that the replisomes are directly linked (to each other or the cell), emerging evidence suggests that no functional dependence between the replisomes exists (Breier et al. 2005; Reyes-Lamothe et al. 2008).
One attractive feature of an anchoring mechanism is that it would immobilize the replisome, preventing it from spiraling along the DNA helix and intertwining the newly replicated DNA strands (precatenane formation). Furthermore, extrusion of the newly replicated DNA towards opposite poles of the cell could prevent mixing, further facilitating segregation (Lemon and Grossman 2001; Sawitzke and Austin 2001). However, examination of replication intermediates by two-dimensional gel electrophoresis suggests that precatenane formation does occur (J. Cebrián et al. 2015). This intermixing of the newly replicated DNA strands is consistent with the many reports citing a cohesion period before the segregation of chromosomal loci (e.g., Bates and Kleckner 2005; Bermejo et al. 2008; Joshi et al. 2013; Lesterlin et al. 2012; Wang et al. 2008). Taken together, these studies argue against an anchoring mechanism that would prevent rotation of the replisome, and imply that segregation occurs separately from replication, following demixing of the sister DNA strands by the action of TopoIV.
Mechanisms of indirect coupling
The weak association between replisome pairs is, perhaps, best explained by an indirect mechanism. One possibility is that protein complexes bridge the nascent DNA strands behind replication fork pairs, causing the replisomes to appear loosely tethered. For example, in E. coli, SeqA forms large complexes hundreds of nanometers behind the replisome that function to organize sister DNA strands (Fossum-Raunehaug et al. 2014; Helgesen et al. 2015). Fluorescence microscopy indicates that SeqA complexes trailing behind opposing replication forks generally co-localize to a single complex (Molina and Skarstad 2004). This interaction between SeqA complexes could, in-turn, influence the localization of the replisome. It has also been suggested CrfC, a dynamin homolog in E. coli, helps to maintain the proximity of the replisomes through its interaction with the processivity clamp (Ozaki et al. 2013).
Recent work on the mechanism of chromosome structure and segregation in B. subtilis is also difficult to reconcile with the track model. It has been proposed that ring-shaped assemblies of SMC are loaded at the origin and slide down the left and right arms of the chromosome to ter, drawing the arms together (Wang et al. 2017). After replication initiation, this model would act to pull the replisomes together as a consequence of the close proximity between chromosome arms. Since this mechanism of colocalizing the replisome is indirect, it could give rise to the weak association observed in experiments visualizing the replisome.
More generally, the proteins and processes that dynamically remodel the chromosome during replication and segregation could play a central role in replisome positioning. For example, nucleoid-associated proteins serve to structure and organize the chromosome. E. coli cells lacking the structural protein H-NS show anomalous replisome localization, indicating that replisome positioning is affected by changes in chromosome architecture (Helgesen et al. 2016). In addition to nucleoid-associated proteins, topoisomerases also play a central role in chromosome compaction and structure (Wang et al. 2013) and likely also affect replisome positioning.
Chromosome dynamics and the factory model
Recent studies of chromosome structure and dynamics also have important implications for the organization of the replication process. We recently analyzed the cell cycle of dynamics of loci from different regions of the E. coli chromosome (Cass et al. 2016). The factory model makes two closely related predictions about the locus dynamics: (1) loci should move towards midcell (replisome location) before replication and (2) loci should begin segregation from midcell. In the track model, loci are stationary and then segregate from their pre-replication positions. Both factory model predictions (1) and (2) are strongly supported by a statistical analysis of locus trajectories from hundreds of cells. On the other hand, the predicted locus translocation is not obvious from the inspection of a single cell due to fluctuations in the nucleoid structure (Cass et al. 2016). Under rapid growth conditions, the structure of the newly replicated nucleoids also is consistent with a centrally located factory complex (Youngren et al. 2014).
Although C. crescentus replication is also factory-like, the replisome dynamics are altered due to differences in nucleoid structure. Particularly, the origin of replication is proximal to the pole, rather than midcell, at the start of replication (Jensen et al. 2001). The replication factory is displaced towards midcell during replication, consistent with the motion being driven by the buildup of newly replicated DNA at the origin-proximal pole.
Concluding remarks
Although analysis of the replisome dynamics is complicated by differences in the chromosome structure between organisms, almost all observations appear to be consistent with DNA loci moving to, and splitting from, a pair of co-localized replisomes, as described by the replication factory model. Although replisome pairs are typically co-localized to the diffraction limit, they appear to be weakly associated, since they can transiently separate. These transient separation events are inconsistent with the most rigorous interpretation of the factory model which requires that the replisomes remain strictly immobile. Furthermore, we note that the resolution limit of fluorescence microscopy (roughly 250 nm) is large in comparison to the size scale of individual proteins or nucleotides. Although the existing evidence cannot exclude the possibility that the replisomes (at least in part) translocate along the DNA, we conclude that the replisomes are confined within a small volume in comparison to the size of the nucleoid, consistent with a factory-like model.
We propose that the conflicting reports on replication organization are the result of the misinterpretation of pairs of replisomes that re-initiate at the quarter-cell position. The inconsistencies between reports speak to the generic importance of both performing time-lapse imaging and the analysis of a large number of cells over complete cell cycles.
Although much has been learned about in vivo replisome structure, many important questions remain. For instance, it is unclear whether there is any direct interaction between replisomes when they appear co-localized to the diffraction limit. Super-resolution imaging could provide interesting insights into this question. Furthermore, although it has long been assumed that core replisome components remain stably bound and co-localized throughout the replication process, this model is being challenged experimentally. Recent work indicates that many replisome components turnover on the time scale of seconds (Liao et al. 2016; Beattie et al. 2017; Lewis et al. 2017), and that replisome disassembly and restart occurs multiple times per cell cycle due to encounters with the transcription machinery (Mangiameli et al. 2017). There is great potential for future work to offer new and fundamental insights into replication and related process (Redder 2016) in the bacterial cell.
References
Meselson M, Stahl FW (1958) The replication of DNA in Escherichia coli, Proceedings of the National Academy of Sciences of the United States of America, vol 44, pp 671–682. http://www.ncbi.nlm.nih.gov/pubmed/16590258, http://www.pubmedcentral.nih.gov/articlerender.fcgi? artid=PMC528642
Zechner EL, Wu CA, Marians KJ (1992) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem 267:4045–4053. http://www.ncbi.nlm.nih.gov/pubmed/1740452
Sanders GM, Dallmann HG, McHenry CS (2010) Reconstitution of the B. subtilis Replisome with 13 proteins including two distinct replicases. Mol Cell 37:273–281. http://www.ncbi.nlm.nih.gov/pubmed/20122408, http://linkinghub.elsevier.com/retrieve/pii/S1097276509009563
Mangiameli SM, Merrikh CN, Wiggins PA, Merrikh H (2017) Transcription leads to pervasive replisome instability in bacteria. eLife. https://doi.org/10.7554/eLife.19848. http://www.ncbi.nlm.nih.gov/pubmed/28092263, http://www.pubmedcentral.nih.gov/articlerender.fcgi? artid=PMC5305214
Frimodt-Møller J, Charbon G, Løbner-Olesen A (2017) Control of bacterial chromosome replication by non-coding regions outside the origin. Curr Genet 63:607–611. https://doi.org/10.1007/s00294-016-0671-6. http://springerlink.bibliotecabuap.elogim.com/ 10.1007/s00294-016-0671-6
Frouin I, Montecucco A, Spadari S, Maga G (2003) DNA replication: a complex matter. EMBO Rep 4:666–670. https://doi.org/10.1038/sj.embor.embor886. http://embor.embopress.org/content/ 4/7/666
Dingman CW (1974) Bidirectional chromosome replication: some topological considerations. J Theor Biol 43:187–195
Newport J, Yan H (1996) Organization of DNA into foci during replication. Curr Opinion Cell Biol 8:365–368. https://doi.org/10.1016/S0955-0674(96)80011-1
Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng P-C, Meng C, Berezney R (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143:1415–1425. https://doi.org/10.1083/jcb.143.6.1415
Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of s phase in human cells. J Cell Biol 140:1285–1295. https://doi.org/10.1083/jcb.140.6.1285. http://jcb.rupress.org/content/140/6/1285
Hozák P, Hassan AB, Jackson DA, Cook PR (1993) Visualization of replication factories attached to a nucleoskeleton. Cell. https://doi.org/10.1016/0092-8674(93)90235-I
Leonhardt H, Rahn H-P, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC (2000) Dynamics of DNA replication factories in living cells. J Cell Biol 149:271–280. https://doi.org/10.1083/jcb.149.2.271. http://jcb.rupress.org/content/149/2/271
Baker TA, Bell SP (1998) Polymerases and the replisome: machines within machines. Cell 92:295–305. https://doi.org/10.1016/S0092-8674(00)80923-X
Jensen RB, Wang SC, Shapiro L (2001) A moving dna replication factory in Caulobacter crescentus. EMBO J 20:4952–4963. http://emboj.embopress.org/content/20/17/4952
Lemon KP, Grossman AD (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516–1519
Lemon KP, Grossman AD (2000) Movement of replicating DNA through a stationary replisome. Mol Cell 6:1321–1330
Berkmen MB, Grossman AD (2006) Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol 62:57–71. https://doi.org/10.1111/j.1365-2958.2006.05356.x
Mangiameli SM, Veit BT, Merrikh H, Wiggins PA (2017) The replisomes remain spatially proximal throughout the cell cycle in bacteria. PLoS Genet 13:1–17. https://doi.org/10.1371/journal.pgen.1006582
Bates D, Kleckner N (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121:899–911
Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ (2008) Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90–102
Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M (2000) Bidirectional migration of seqa-bound hemimethylated dna clusters and pairing of oric copies in Escherichia coli. Genes Cells 5:327–341. https://doi.org/10.1046/j.1365-2443.2000.00334.x
Kongsuwan K, Dalrymple BP, Wijffels G, Jennings PA (2002) Cellular localisation of the clamp protein during DNA replication. FEMS Microbiol Lett 216:255. https://doi.org/10.1111/j.1574-6968.2002.tb11444.x
Cass JA, Kuwada NJ, Traxler B, Wiggins PA (2016) Escherichia coli chromosomal loci segregate from midcell with universal dynamics. Biophys J 110:2597–2609
Migocki MD, Lewis PJ, Wake RG, Harry EJ (2004) The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol Microbiol 54:452–463. https://doi.org/10.1111/j.1365-2958.2004.04267.x
Reyes-Lamothe R, Sherratt DJ, Leake MC (2010) Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328:498–501. https://doi.org/10.1126/science.1185757. http://science.sciencemag.org/content/328/ 5977/498
Swulius MT, Jensen GJ (2012) The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol 194:6382–6386. https://doi.org/10.1128/JB.00505-12. http://www.ncbi.nlm.nih.gov/pubmed/22904287, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3497537
Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J (2012) Segregation of molecules at cell division reveals native protein localization. Nat Methods 9:480–482. https://doi.org/10.1038/nmeth.1955. http://www.ncbi.nlm.nih.gov/pubmed/22484850www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3779060, http://www.nature.com/doifinder/10.1038/nmeth.1955
Koppes LJ, Woldringh CL, Nanninga N (1999) Escherichia coli contains a DNA replication compartment in the cell center. Biochimie 81:803–810. https://doi.org/10.1016/S0300-9084(99)00217-5
Molina F, Skarstad K (2004) Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol Microbiol 52:1597–1612. https://doi.org/10.1111/j.1365-2958.2004.04097.x
Adachi S, Kohiyama M, Onogi T, Hiraga S (2005) Localization of replication forks in wild-type and mukB mutant cells of Escherichia coli. Mol Genet Genomics 274:264–271
Den Blaauwen T, Aarsman MEG, Wheeler LJ, Nanninga N (2006) Pre-replication assembly of E. coli replisome components. Mol Microbiol 62:695–708
Wallden M, Fange D, Lundius EG, Baltekin O, Elf J (2016) The synchronization of replication and division cycles in individual E. coli cells. Cell 166:729–739. https://doi.org/10.1016/j.cell.2016.06.052
Cass JA, Stylianidou S, Kuwada NJ, Traxler B, Wiggins PA (2017) Probing bacterial cell biology using image cytometry. Mol Microbiol 103:818–828. https://doi.org/10.1111/mmi.13591
Yamazoe M, Adachi S, Kanaya S, Ohsumi K, Hiraga S (2004). https://doi.org/10.1111/j.1365-2958.2004.04389.x
Onogi T, Ohsumi K, Katayama T, Hiraga S (2002) Replication-dependent recruitment of the beta-subunit of DNA polymerase III from cytosolic spaces to replication forks in Escherichia coli. J Bacteriol 184:867–870. https://doi.org/10.1128/jb.184.3.867-870.2002. http://www.ncbi.nlm.nih.gov/pubmed/11790763, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC139520
Sunako Y, Onogi T, Hiraga S (2002) Sister chromosome cohesion of Escherichia coli. Mol Microbiol 42:1233–1241. https://doi.org/10.1046/j.1365-2958.2001.02680.x
Breier AM, Weier H-UG, Cozzarelli NR (2005) Independence of replisomes in Escherichia coli chromosomal replication. Proc Nat Acad Sci 102:3942–3947. https://doi.org/10.1073/pnas.0500812102. http://www.ncbi.nlm.nih.gov/pubmed/15738384, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC552787, http://www.pnas.org/cgi/doi/10. 1073/pnas.0500812102
Lemon KP, Grossman AD (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev 15:2031–2041. https://doi.org/10.1101/gad.913301. http://www.ncbi.nlm.nih.gov/pubmed/11511534
Sawitzke J, Austin S (2001) An analysis of the factory model for chromosome replication and segregation in bacteria. Mol Microbiol 40:786–794. http://www.ncbi.nlm.nih.gov/pubmed/11401686
Cebrián J, Castán A, Martínez V, Kadomatsu-Hermosa MJ, Parra C, Fernández-Nestosa MJ, Schaerer C, Hernández P, Krimer DB, Schvartzman JB (2015) Direct Evidence for the Formation of Precatenanes during DNA Replication. J Biol Chem 290:13725–13735. https://doi.org/10.1074/jbc.M115.642272. http://www.ncbi.nlm.nih.gov/pubmed/25829493, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4447951, http://www.jbc.org/lookup/doi/10.1074/jbc.M115.642272
Bermejo R, Branzei D, Foiani M (2008) Cohesion by topology: sister chromatids interlocked by DNA. Genes Dev 22:2297–2301. https://doi.org/10.1101/gad.1719308. http://www.ncbi.nlm.nih.gov/pubmed/18765785, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2749673
Joshi MC, Magnan D, Montminy TP, Lies M, Stepankiw N, Bates D (2013) Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet 9:e1003673. https://doi.org/10.1371/journal.pgen.1003673. http://www.ncbi.nlm.nih.gov/pubmed/23990792, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3749930
Lesterlin C, Gigant E, Boccard F, Espéli O (2012) Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBO J 31:3468–3479. https://doi.org/10.1038/emboj.2012.194. http://www.ncbi.nlm.nih.gov/pubmed/22820946, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3419930, http://emboj.embopress.org/cgi/doi/10.1038/emboj.2012.194
Wang X, Reyes-Lamothe R, Sherratt DJ (2008) Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev 22:2426–2433. https://doi.org/10.1101/gad.487508. http://www.ncbi.nlm.nih.gov/pubmed/18765793, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2532930
Fossum-Raunehaug S, Helgesen E, Stokke C, Skarstad K (2014) Escherichia coli SeqA structures relocalize abruptly upon termination of origin sequestration during multifork DNA replication. PLoS One 9:e110575. https://doi.org/10.1371/journal.pone.0110575. http://www.ncbi.nlm.nih.gov/pubmed/25333813, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4204900
Helgesen E, Fossum-Raunehaug S, Sætre F, Schink KO, Skarstad K (2015) Dynamic Escherichia coli SeqA complexes organize the newly replicated DNA at a considerable distance from the replisome. Nucleic Acids Res 43:2730–2743. https://doi.org/10.1093/nar/gkv146. http://academic.oup.com/nar/article/43/5/2730/2453293/Dynamic-Escherichia-coli-SeqA-complexes-organize, http://www.ncbi.nlm.nih.gov/pubmed/25722374, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4357733
Ozaki S, Matsuda Y, Keyamura K, Kawakami H, Noguchi Y, Kasho K, Nagata K, Masuda T, Sakiyama Y, Katayama T (2013) A replicase clamp-binding dynamin-like protein promotes colocalization of nascent DNA strands and equipartitioning of chromosomes in E. coli. Cell Rep 4:985–995. https://doi.org/10.1016/j.celrep.2013.07.040. http://linkinghub.elsevier.com/retrieve/pii/S2211124713004026, http://www.ncbi.nlm.nih.gov/pubmed/23994470
Wang X, Brando HB, Le TBK, Laub MT, Rudner DZ (2017) Bacillus subtilis smc complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355:524–527
Helgesen E, Fossum-Raunehaug S, Skarstad K (2016) Lack of the H-NS protein results in extended and aberrantly positioned DNA during chromosome replication and segregation in Escherichia coli. J Bacteriol 198:1305–16. https://doi.org/10.1128/JB.00919-15. http://jb.asm.org/lookup/doi/10.1128/JB.00919-15, http://www.ncbi.nlm.nih.gov/pubmed/26858102, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4859577
Wang X, Montero Llopis P, Rudner DZ (2013) Organization and segregation of bacterial chromosomes. Nat Rev Genet 14:191–203. https://doi.org/10.1038/nrg3375. http://www.ncbi.nlm.nih.gov/pubmed/23400100, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3869393
Youngren B, Nielsen HJ, Jun S, Austin S (2014) The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev 28:71–84. https://doi.org/10.1101/gad.231050.113. http://www.ncbi.nlm.nih.gov/pubmed/24395248, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3894414, http://genesdev.cshlp.org/cgi/doi/10.1101/gad.231050.113
Liao Y, Li Y, Schroeder JW, Simmons LA, Biteen JS (2016) Single-Molecule DNA Polymerase Dynamics at a Bacterial Replisome in Live Cells. Biophys J 111:2562–2569. https://doi.org/10.1016/j.bpj.2016.11.006. http://www.ncbi.nlm.nih.gov/pubmed/28002733, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5192695linkinghub.elsevier.com/retrieve/pii/S0006349516310335
Beattie TR, Kapadia N, Nicolas E, Uphoff S, Wollman AJ, Leake MC, Reyes-Lamothe R (2017) Frequent exchange of the DNA polymerase during bacterial chromosome replication. eLife. https://doi.org/10.7554/eLife.21763. http://www.ncbi.nlm.nih.gov/pubmed/28362256, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5403216, http://elifesciences.org/lookup/doi/10.7554/eLife.21763
Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, Horan NP, Duderstadt KE, Cox MM, Robinson A, Dixon NE, van Oijen AM (2017) Single-molecule visualization of fast polymerase turnover in the bacterial replisome. eLife. https://doi.org/10.7554/eLife.23932. http://www.ncbi.nlm.nih.gov/pubmed/28432790, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5419744
Redder P (2016) How does sub-cellular localization affect the fate of bacterial mRNA? Curr Genet 62:687–690. https://doi.org/10.1007/s00294-016-0587-1. http://www.ncbi.nlm.nih.gov/pubmed/26972734, http://springerlink.bibliotecabuap.elogim.com/10.1007/s00294-016-0587-1
Acknowledgements
The authors are grateful to Colin LaMont for the helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
The authors are supported by NSF Grant MCB1243492. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Rights and permissions
About this article
Cite this article
Mangiameli, S.M., Cass, J.A., Merrikh, H. et al. The bacterial replisome has factory-like localization. Curr Genet 64, 1029–1036 (2018). https://doi.org/10.1007/s00294-018-0830-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0830-z