Abstract
The denitrifying bacterium Acinetobacter johnsonii strain DBP-3 which was capable of removing phosphate, nitrate, and ammoniacal salt is psychrotolerant, whereas, the cold shock response mechanisms or the cold shock proteins (Csps) was unclear. In this article, the optimal growth temperature (25 °C) and cold shock temperature (7.5 °C) were determined firstly by an Arrhenius plot of the growth of the strain DBP-3. Then, among the seven cold shock-like protein genes which were cloned and identified referenced by A. johnsonii SH046 genome, qRT-PCR and shotgun-LTQ mass spectrometry showed that Csp3 and Csp4 were overexpressed under cold shock condition. Furthermore, Western blotting confirmed the result with the antibodies against Csp3 and Csp4 prepared by ourselves. Finally, the phylogenetic analysis showed that the similarity percent between Csp3 and Csp4 was 76.85 %, and Csp3 and Csp4 belonged to CspE family. The results indicated that CspE is overproduced by temperature downshift and may play an important role in the psychrotolerant process of strain DBP-3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms are adapted for optimum functioning in their normal physiological environments. Any extreme change in the environmental conditions from the optimum inflicts a stress on an organism [20, 21]. Most bacteria are able to tolerate small changes in an environmental parameter and can adapt over the time scale of minutes, hours, or days [7]. Morita [17], Bower and Daeschel [1] indicated that psychrotolerant organisms are able to grow at low temperatures with much lower growth rates, although they have growth optima in the range of mesophilic organisms (20–40 °C). Previously, A. johnsonii strain DBP-3, isolated by Li [14] from a eutrophic water body through low temperature-oriented enrichment culture, is able to grow and perform denitrification under both aerobic and anoxic conditions, also capable of growing satisfactorily at 10–30 °C, furthermore it was noted that it can grow at 5 °C. Acinetobacter johnsonii, as a psychrotolerant gram-negative bacterium, plays an important role in environmental engineering and contamination control on account of its peculiar metabolic and physiological features under cold shock conditions [13, 14, 27]. Cold shock proteins, composed of about 70 amino acids [19], are, by far, the most strongly induced proteins at low temperatures [4]. Data have shown that the expression of the cold shock proteins (Csps) reaches a maximum level during the phase of the cold shock adaptation, the so-called acclimation [26]. However, the researches on Csps of A. johnsonii have not been reported yet, so in this paper for better understanding the mechanisms of cold adaption for the psychrotolerant denitrifying bacterium A. johnsonii DBP-3, the Csp genes were searched using bioinformatics methods and cloned. The expression characteristics at transcriptional level of these Csps under cold shock conditions were investigated using qRT-PCR. Furthermore, SDS-PAGE, mass spectrometry, and Western blotting were performed to detect the Csps induced under cold shock conditions at translational level. This work may contribute to further studies focusing on the application of A. johnsonii at low temperature.
Materials and Methods
Bacterial Strain, Growth Characteristics, and Cold Shock Treatment
Acinetobacter johnsonii strain DBP-3 which was deposited in the China General Microbiological Culture Collection Center (CGMCC 4753) was cultured at 25 °C in the denitrification medium under shaking [14]. The growth kinetics of DBP-3 was investigated by measuring the OD600 of the 1 % inoculated cultures at different temperatures (5, 7.5, 10, 12.5, 15, 25, 30, 37, and 40 °C). An Arrhenius plot of the growth of strain DBP-3 was established for setting the cold shock temperature purpose as described previously [16]. For cold shock experiment, A. johnsonii cells were grown to the mid-exponential phase (OD600 0.6–0.8) at 25 °C in denitrification medium, after which 25 ml of the culture was incubated in 100 ml of pre-cooled (7.5 °C) fresh medium for 1 h and 3 h. Additionally, another 25 ml of the culture was incubated in 100 ml of non-cooled (25 °C) medium to use as control for qRT-PCR, SDS-PAGE, and Western blotting.
Screening and Identification of Acinetobacter johnsonii Cold Shock Proteins
According to the highly conserved RNA-binding motifs, i.e. RNP1 and RNP2 possessed by almost all cold shock proteins of microorganisms [8], seven Csps were found in A. johnsonii, then the genes were located in the genome of A. johnsonii SH046 (NZ_ACPL00000000) [18], and further checked in GenBank by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The seven gene fragments were amplified by PCR using primers in Table S 1 in the supplementary material and sequenced by Comate Bioscience Co., Ltd (Changchun, China).
SDS-PAGE and Protein Identification by Shotgun-LTQ Mass Spectrometry
A. johnsonii DBP-3 samples (the bacteria in the cold shock treated group and the control) were harvested at 1 h and 3 h post cold shock, and washed for four times with PBS, then the pellets were re-suspended in Triton X-100 lysis buffer. Cells were lysed using sonication (45 W, sonics VCX750, Vedeng, 4 times for 15 s with intermediate ice cooling) and centrifuged at 21,000×g for 30 min at 4 °C, and the supernatant was separated by SDS-PAGE using 15 % acrylamide gel. After Coomassie staining, the revealed differential band (1 h after cold shock) was excised and identified by shotgun-LTQ mass spectrometry at Shanghai Applied Protein Technology Co., Ltd (Shanghai, China).
Expression Analysis of the Csp Genes by qRT-PCR
Total RNA of the treatment and control samples were extracted using Trizol reagent (Takara, Dalian, China) according to the manufacturer’s instruction. The Primescript™ RT reagent kit (TaKaRa, Dalian, China) was used to perform the reverse transcription of RNA into cDNA. cDNA samples were then used as templates for qRT-PCR using SYBR® Premix Ex Taq™ (TIi RNaseH Plus) (Takara, Dalian, China) according to the manufacturer’s recommendations. Primers for qRT-PCR are listed in Table S 1 in the supplementary material. qRT-PCR was performed on the Bio-Rad iQ5 Multicolor Real-Time PCR detection system for 40 cycles (95 °C for 10 s; 50 °C for 15 s; and 72 °C for 30 s). All tests were conducted in three independent biological replicates. Quantification of mRNA was based on the threshold cycle (Ct) values. The Ct values of the seven Csp genes were normalized by the Ct value of the 16 s rRNA gene. Data analysis was performed using the comparative Ct (2−ΔΔCt) method [15]. Data obtained from qRT-PCR analysis were subjected to variance analysis to evaluate differences in the mean values among the treatments. Differences were considered significant at P < 0.05 and highly significant at P < 0.01. Statistical analysis was performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA).
Preparation of Polyclonal Antiserum
In order to produce the antibodies against Csp3 and Csp4 in A. johnsonii, Csp3 and Csp4 genes were amplified and cloned into the pET32a expression vector, separately. The recombinant pET32aCsp3/Csp4 were transformed into the E. coli BL21 (DE3) to produce the N-terminal trx-his-tag recombinant Csp3 and Csp4 proteins which were overproduced by inducing with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside). Proteins were then purified using HisPur™ Ni–NTA Resin (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The purified proteins were mixed with Freund’s complete adjuvant (Sigma, USA) and injected subcutaneously to rabbits in multiple places as described by Vaitukaitis [24]. Rabbits were boosted four times at 3-week intervals, and bleeding was done 10 days after. All experimental works with rabbits were approved by the Animal Care and Ethics Committee of Jilin University, China.
Western Blotting Analysis
For identifying the expressed proteins in strain DBP-3 under cold shock treatment, Western blotting was performed using rabbit antiserum (1:500) with antibodies against Csp3 and Csp4 as the primary antibodies and Horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies as the secondary antibodies (1:5000, Boster, China), respectively. Western blotting was visualized using the SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Amino Acid Sequences Analysis
The complete deduced protein sequences of the Csp3 and Csp4, together with the Csps of the strains possessing similar function with strain DBP-3 and representative bacterium selected from GenBank, were used for the multiple sequence alignment and phylogenetic tree construction. Multiple sequence alignments were performed using the ClustalX program [3]. The phylogenetic tree was constructed using the program Mega 6.0 [23]. Phylogenetic analysis was carried out by applying the neighbor-joining and maximum-likelihood algorithms to ensure coherency of the clusters formed. The bootstrapping supports for the trees were calculated from a sample of 1000 replicates.
Results
Growth Characteristics of A. johnsonii Strain DBP-3 at Different Temperatures
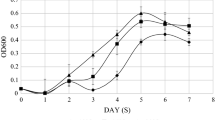
Growth kinetics was performed at nine temperatures for studying the growth characteristics of A. johnsonii strain DBP-3. As can be seen in Fig. 1a and b, the optimal and the minimal temperature for the growth of A. johnsonii were 25 °C and below 5 °C, respectively. The growth rate significantly decreased at 40, 15, 10, 7.5, and 5 °C than at 25 °C.
The growth characteristics of A. johnsonii. The growth curves of A. johnsonii strain DBP-3 at various temperatures which were 5–12.5 °C (a) and 15–40 °C (b). The means and standard deviations of the data showed in the figure were from triplicate experiments. c An Arrhenius plot of the relationship between growth rate (k) and temperature (kelvins) for A. johnsonii DBP-3
According to the Arrhenius plot (Fig. 1c), the temperatures over which A. johnsonii can grow were divided into the following ranges by the inflection points of 30, 15, and 10 °C: an optimum sub-range from 15 to 30 °C, within which the ln (k) value remain unchanged and suggested that the growth under the influence of the temperature from 15 to 30 °C is about the same; a cold shock sub-range below the optimum which exhibited two distinct slopes separated by the critical temperature 10 °C, within each slope the activation energy of growth is constant; and a heat shock sub-range (above 30 °C). A temperature of 7.5 °C in the higher slope of cold shock sub-range, which has a significant impact on the growth, was chosen for the cold shock experiments.
The Identification of Csps Genes in the Acinetobacter johnsonii DBP-3
Based on the genomic data of A. johnsonii SH046 (NCBI accession No. NZ ACPL00000000), seven Csps (temporarily named Csp1 to Csp7) which encode the proteins including the conserved nucleic acid-binding motifs RNP1 and RNP2 were found. These seven genes were amplified by PCR as shown in Fig. 2 and sequenced.
Identification of the seven Csp genes. Lane M, DL 2000 DNA marker (from top to bottom 2000 bp, 1500 bp, 1000 bp, 750 bp, 500 bp, 250 bp, and 100 bp); Lane 1–5, PCR products of the applied Csp1, Csp2, Csp3, Csp4, and Csp6 genes; Lane 6, PCR product of the applied Csp5 and Csp7 genes (the positions of Csp5 and Csp7 genes are close together in the genome of A. johnsonii, so the forward primer was located at the upstream of Csp5 gene and the reverse one was at the downstream of Csp7 gene)
Csp3 and Csp4 were Overproduced by the Temperature Downshift in the Acinetobacter johnsonii DBP-3
In order to investigate the expression characteristics of Csps responding to cold shock treatment, the protein expression patterns of whole-cell lysate from A. johnsonii with and without cold shock treatment were compared using SDS-PAGE. As indicated in Fig. 3a, a protein band of approximately 7 kDa was present in the lysate of the cold shock treated cells and appeared not to be present in the lysate of the untreated cells. For the ~7 kDa protein band, eight groups of peptide fragments of different proteins were detected by shotgun-LTQ mass spectrometry (Table 1), one group of peptide fragments (GPNAVNIVAI/L) was belonged to cold shock protein whose molecular size was about 7 kDa, which matched with the band size. Homologous analysis of sequences showed that they were Csp3 and Csp4 who shared high homology with CspE. Interestingly, qRT-PCR results showed that the expressions of Csp3 and Csp4 were significantly enhanced in the cold shock treatment group, which had an almost 400 fold change (P < 0.05), and no significant difference was observed on the other Csps (Fig. 3b, c). Additionally, Western blotting results showed that Csp3 and Csp4 were both overexpressed at low temperature (7.5 °C) while no significant band was observed in the normal-temperature cells (Fig. 4), which further confirmed that Csp3 and Csp4 were overproduced at 7.5 °C than at 25 °C in A. johnsonii DBP-3.
~7 kDa proteins of A. johnsonii overexpressed at cold shock treatment cells. a The comparison of proteins produced from the non-cold-shocked culture (lane 1) and from the cold-shocked culture for 1 h (lane 2) and 3 h (lane 3) of A. johnsonii DBP-3 in SDS-PAGE, the protein standards were shown in lane M. b qRT-PCR analysis of Csp1, Csp2, Csp5, Csp6, Csp7 mRNA expressions in response to the temperature downshift of the A. johnsonii DBP-3. c qRT-PCR analysis of Csp3 and Csp4 mRNA expressions in response to the temperature downshift of the A. johnsonii DBP-3. Standard error bars are shown. ***P < 0.001 compared with the control group
Analysis of the effect of cold shock stress on the expression of Csp3 and Csp4 proteins by Western blotting. Each lane contains equal amounts of total protein. Samples were obtained from the control (−) and cold shock treated (+) A. johnsonii DBP-3 cells, and reacted with the antibodies against Csp3 (left) and Csp4 (right)
Furthermore, deduced amino acid sequences of Csp3 and Csp4 of DBP-3, together with the Csps of the strains possessing similar function with strain DBP-3 and representative bacterium were selected to perform sequence alignment and constructed a phylogenetic tree. Multiple sequence alignment revealed that the amino acid sequences of Csp3 and Csp4 were homologous with the amino acid sequences of other known Csps. Besides, Csp3 and Csp4 also possessed RNP1 and RNP2 motifs that are known to be involved in binding to single-stranded nucleic acids (Fig. 5) [12]. Phylogenetic analysis showed that Csp3 and Csp4 had a close relationship and they were relatively close to Pseudomonas sp. Csps but were relatively distant form Listeria.sp and Bacillus sp. Csps, moreover they had a relatively closer relationship to CspE of different strains, which revealed that Csp3 and Csp4 belong to CspE (Fig. 6).
Amino acid sequence alignment of Csps of different strains. Sequence alignment of Csp3 and Csp4 of DBP-3 and the Csps of the strains possessing similar function with strain DBP-3 and representative bacterium was performed. Conserved nucleic acid-binding sequence motifs, the RNA-binding motifs RNP1 and RNP2, are labeled
Phylogenetic tree of Csps of different strains. Phylogenetic tree of Csp3 and Csp4 of DBP-3 and the Csps of the strains possessing similar function with strain DBP-3 and representative bacterium constructed using the program Mega 6.0. Numbers at each branch indicate the percentage of times a node was supported in 1000 bootstrap pseudo replications by neighbor joining. Accession numbers are shown in parentheses
Discussions
Bacteria respond to the decrease of temperature in a specific manner. The synthesis of most cellular proteins is inhibited after a decrease in temperature [10]. Psychrotolerant microorganisms are usually the organisms most frequently found in cold environments, perhaps because they may have better nutritional adaptability [25] or due to horizontal gene transfer from mesophiles [2]. In this article, the psychrotolerant denitrifying bacterium A. johnsonii DBP-3 grew well at 15–30 °C, and 7.5 °C was chosen as the cold shock temperature according to the Arrhenius plot.
Cold shock proteins, composed of about 70 amino acids [19], are the most strongly induced proteins at low temperatures [4]. The first cold shock protein was indentified in 1987 [5, 10], expression of the cold shock proteins reaches a maximum level during the phase of the cold shock adaptation, the so-called acclimation [26]. Most of the Csps share the conserved RNP1 and RNP2 motifs which are known to be involved in the binding with the single-stranded nucleic acids [9, 12, 22]. Furthermore, a multiple-deletion analysis showed that Csps play important roles not only during cold shock adaptation but also during the stationary phase or under nutrient stress [6].
In this article, Csp3 and Csp4 were detected to be overproduced by temperature downshift in strain DBP-3 by LTQ mass spectrometry, qRT-PCR, and Western blotting. The overall DNA sequence similarity percent between Csp3 and Csp4 was about 76.85 %, while the similarity percent of peptides between Csp3 and Csp4 was 84.51 %. Thus, in Western blotting, the antibodies of the two proteins reacting with each other was a possible situation, and could be explained by the phylogenetic analysis, which revealed that Csp3 and Csp4 had a close relationship with each other, and belonged to CspE as well. These results indicated that CspE may play an important role in the psychrotolerant mechanism of A. johnsonii DBP-3, which were in accordance with the findings of Kandasamy [11] who indicated that Enterobacter ludwigii PAS1 adapts quickly to low temperatures by a constitutive expression of the potentially cryoprotective CspE. Nevertheless, the understanding of the cold shock response in strain DBP-3 is still in infancy, further studies on the mechanisms employed by A. johnsonii DBP-3 to acclimate low temperature such as the interactions of different Csps, quantitative differences in Csps expression at the translational level under different conditions, and regulatory elements of Csps are needed to clarify.
In conclusion, the optimal growth temperature of A. johnsonii was 25 °C and the cold shock temperature was below 7.5 °C. Csp3 and Csp4 who belonged to CspE were overexpressed significantly under the cold shock treatment.
References
Bower CK, Daeschel MA (1999) Resistance responses of microorganisms in food environments. Int J Food Microbiol 50(1/2):33–44
Foght JM, Waterhouse EJ, Aislabie JM, Balks MR (2004) Hydrocarbon spills on Antarctic Soils: effects and Management. Environ Sci Technol 38(5):1265–1274
Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Graumann P, Marahiel MA (1996) Some like it cold: response of microorganisms to cold shock. Arch Microbiol 166(5):293–300
Graumann P, Marahiel MA (1998) A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci 23(8):286–290
Graumann P, Wendrich TM, Weber MH, Schröder K, Marahie MA (1997) A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol 25(4):741–756
Hall HK, Karem KL, Foster JW (1995) Molecular responses of microbes to environmental pH stress. Adv Microbiol Phys 37:229–264
Jeroen AW, Frank MR, Oscar PK, De Vos Willem M, Abee T (2000) The Role of Cold-Shock Proteins in Low-Temperature Adaptation of Food-Related Bacteria. System Appl Microbiol 23(2):165–173
Jones PG, Inouye M (1996) RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol 21(6):1207–1218
Jones PG, VanBogelen RA, Neidhardt FC (1987) Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol 169:2092–2095
Kandasamy P, Chaturvedi N, Sisodia BS, Shasany AK, Gahio S, Marla SS, Goel R (2013) Expression of CspE by a psychrotrophic bacterium Enterobacter ludwigii PAS1, isolated from Indian Himalayan soil and in silico protein modelling, prediction of conserved residues and active sites. Curr Microbiol 66:507–514
Landsman D (1992) RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res 20(11):2861–2864
Lee M, Woo SG, Ten LN (2012) Characterization of novel diesel-degrading strains Acinetobacter haemolysicus MJ01 and Acinetobacter johnsonii MJ4 isolated from oil-contaminated soil. World J Microbiol Biotechnol 28:2057–2067
Li MT, Liu JH, Zhao SJ, Wang ZX, Hao LL (2013) The characteristics of nitrate removal by the psychrotolerant denitrifying bacterium Acinetobacter johnsonii DBP-3, isolated from a low-temperature eutrophic body of water. J Environ Sci Heal B 48:885–892
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Mohr PW, Krawiec S (1980) Temperature characteristics and Arrhenius plots for nominal psychrophiles, mesophiles and thermophiles. J Gen Microbiol 121(2):311–317
Morita RY (1975) Psychrophilic Bacteria. Bacteriol Rev 39(2):144–167
Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L (2012) The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 7(10):e46984
Perl D, Welker C, Schindler T, Schröder K, Marahiel MA, Jaenicke R, Schmid FX (1998) Conservation of rapid two-state folding in mesophilic, thermophilic and hyperthermophilic cold shock proteins. Nat Struct Biol 5(3):229–235
Ray B (1986) Impact of bacterial injury and repair in food microbiology: its past, present and future. J Food Prot 49:651–655
Russell NJ, Evans RI, ter Steeg PF, Hellemons J, Verheul A, Abee T (1995) Membranes as a target for stress adaption. Int J Food Microbiol 28:255–261
Song WZ, Lin XZ, Huang XH (2012) Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G Polar Biol 35(10):1515–1524
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Vaitukaitis JL (1981) Production of antisera with small doses of immunogen: multiple intradermal injections. Methods Enzymol 73:46–52
Wynn-Williams DD (1990) Ecological aspects of Antarctic microbiology. Adv Microbial Ecol 11:71–146
Yamanaka K, Fang L, Inouye M (1998) The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol 27:247–255
Yao S, Ni J, Ma T, Li C (2013) Heterotrophic nitrification and aerobic denitrification at low temperature by a newly isolated bacterium, Acinetobacter sp. HA2. Bioresour Technol 139:80–86
Acknowledgments
This work was supported by the National Natural Science Foundation of China (51109089) and the Basic Scientific Research Fund of Jilin University (201003061).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Dan Su and Linlin Hao contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, D., Hao, L., Chen, F. et al. CspE is Overproduced by Temperature Downshift in the Acinetobacter johnsonii DBP-3. Curr Microbiol 72, 563–569 (2016). https://doi.org/10.1007/s00284-015-0979-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0979-3