Abstract
The present research aimed at evaluating the effects of urease enzyme and increasing pH on calcite nanocrystal formation. Unlike some researches, the results showed that CaCO3 precipitation is not a general phenomenon among the bacteria and if a bacterium has not this ability, it will not be able to produce calcite even with an increase in pH. All urease-positive bacteria had this ability, while only some urease-negative bacteria were able to produce calcite. Production and characterization of nanocrystals on precipitating medium were shown primarily by light microscopy and then confirmed by X-ray diffraction (XRD) analysis. Crystallite particle size was determined using Scherrer formula that was sub-100-nm in all samples. Based on qualitative and quantitative studies, strain C8 was selected as the best calcite-producing strain. Phylogenetic analysis indicated that this isolate has 99 % similarity with Enterobacter ludwigii. 16S rRNA sequence of isolate was deposited in GenBank with accession number JX666242. The morphology and exact composition of nanocrystalline particles were determined using scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX). According to data obtained by SEM, we suggest that nanocrystals of CaCO3 adhere to bacteria and each other to form small aggregates and then complex crystalline networks to trap bacteria. Many holes are present in these crystalline networks that seem to be due to the aggregation of nanocrystals.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microorganisms are active in a wide range of mineralization processes and have been involved in the deposition of minerals throughout the history of the earth. Bacteria from soils, freshwater, and saline habitats have frequently been reported to be able to precipitate calcite (calcium carbonate) both in natural and in laboratory conditions. This capability has been related to the formation of marine calcareous skeletons, carbonate sediments, and soil carbonate deposits [9]. Chemoorganotrophic microorganisms are major agents for distribution of carbon dioxide in nature by decomposition of organic matter during respiration. These microorganisms are also capable of mediating bioprecipitation of carbonate minerals that results from contact between carbon dioxide and calcium ion [20].
Proposed innovative applications of calcite mineralization by bacteria include biomimetic processes and examples of bioremediation in several fields ranging from applied environmental microbiology (leaching, solid-phase capture of inorganic contaminants), civil and environmental engineering (sediment dikes, bioplugging, biogrouting, and self-healing of concrete and limestone structures), and conservation of monumental calcareous stones [5, 13, 25]. In addition, high purity and quality calcite is required for better performance and it is a very laborious and expensive process to obtain calcite with a high degree of purity from natural sources. Thus, bacterially induced carbonate precipitation has received considerable attention as an environmentally friendly method for protecting and consolidating decayed ornamental carbonate stone, that has significant benefits and advantages compared to traditional restoration methods [18, 20].
Although there is no unified mechanism for the calcite formation by microorganisms, following conditions can cause calcite deposition: (1) extracellular polymeric substances (EPS), that carry a net negative charge and have the high binding ability to Ca2+ ions, can result in CaCO3 precipitation [1, 3, 7, 8, 22, 25, 29], (2) in the environment with higher content of dissolved inorganic carbon (DIC) [22], (3) autotrophic pathways that reduce local carbon dioxide in the bacterial environment [2], and (4) heterotrophic pathways that can lead to active or passive precipitation. Some bacteria through their metabolic processes (photosynthesis, ammonification, denitrification, sulfate reduction, and anaerobic sulfide oxidation and urea decomposition) can passively induce extracellular precipitation of calcite by producing ammonia as a metabolic end product which induces a pH increase. In active precipitation, the production of CO3 2− is due to ionic exchange through the cell membrane by calcium and/or magnesium ionic pump [2, 10, 12, 15, 22, 23].
Calcite precipitation in solution occurs via the overall equilibrium reaction of Ca2+ + CO3 2− ↔ CaCO3. Because the production of carbonate ions (CO3 2−) from bicarbonate (HCO3 1−) in water is strongly pH dependent, an increase in CO3 2− concentration occurs under alkaline conditions. Therefore, calcium carbonate precipitation readily occurs in alkaline environments and high concentrations of the calcium and carbonate ions [25].
The present research is aimed at: isolation of calcite forming bacteria from natural environments; comparison of ability of urease-negative and urease-positive bacteria in calcite precipitation; analyzing the type, shape, and size of calcium carbonate formed; and selection of the best strain for biotechnological applications.
Materials and Methods
Medium, Samples, and Their Maintenance
Soil, freshwater, chalk, cement, and activated sludge samples were collected in sterile tubes and kept at 4 °C until analyzed. Precipitation medium consists of 20 g/l urea, 2.12 g/l NaHCO3, 10 g/l NH4Cl, 3 g/l nutrient broth (NB), 25 g/l CaCl2·2H2O (Merck, Germany), and 15 g/l agar (AppliChem, Germany) [11]. The pH of medium was adjusted to 8 with addition 1 N NaOH.
Isolation of Calcite-Precipitating Bacteria
Samples (1 g) powered by mortar were suspended in 9 ml sterile saline solution and vortexed for 1 min [6]. Fifty microliter of the diluted samples was spread on precipitating agar plates and inoculated at 28 °C for 2 weeks. All experiments were carried out in triplicate. Colonies were purified based on observation of calcite precipitation by light microscope.
Studies of Crystal Formation
Each isolate was examined periodically with a light microscope every day for up to 2 weeks after inoculation, for comparing qualitative quantity of crystals produced by different bacteria.
Analysis of Nanocrystals
Once crystals were detected, the colonies were harvested and completely dried at 37 °C for X-ray diffraction (XRD) analysis. XRD measurements were done by a one-circle μ = μ diffractometer (XRD, D8 ADVANCE, Germany). The work condition of XRD was CuKα radiation via a rotating anode at 40 kV and 40 mA. The data were collected in step of 0.05° 2−1 s and range of scattering angles (2θ) from 5° to 80°. The crystalline phases were identified using the ICDD database (PDF2). Crystallite particle size was determined by XRD using Scherrer formula [32]. Dried samples were ground into fine powder using agate mortar and then coated with carbon for studying morphology and size characteristics of nanocrystals by scanning electron microscope (SEM) (LEO 1450 VP, England). The elementary composition of the nanocrystals was determined qualitatively with an IXRF500 EDS (energy-dispersive X-ray spectrometry) detector.
Identification of Calcifying Bacterial Strain
Genomic DNA was extracted in order to identify best precipitating bacterial strain.
DNA Extraction
DNA from culture was prepared by boiling. The samples were centrifuged at 9,600g for 10 min. The supernatant was eliminated, and the pellet was suspended in biology-grade water and centrifuged at 9,600g for 10 min. Cells were washed triple by this water. The pellet was twice freeze–thaw and resuspended in 1 ml of molecular biology-grade water and boiling at 100 °C in a water bath for 15 min, centrifuged at 6,700g for 5 min, and supernatant was stored at −20 °C [31].
PCR
PCR was initially done on DNA extracts using the universal primers RW01, 5′-AAC TGG AGG AAG GTG GGG AT-3′, and DG74, 5′-AGG AGG TGA TCC AAC CGC A-3′, which are highly conserved among all bacteria [21, 30]. The amplified PCR product is approximately 370 bp in length. Amplification reactions were done in a 25 μl reaction volume. Reaction tubes contained primers, 1 μl of each primer-pair (0.3 μg/μl), 0.5 μl of dNTPs (0.2 mM), X10 PCR buffer 2.5 μl, 25 mM MgCl2 0.75 μl, PCR H2O 17 μl, template DNA extracted from bacterium 2 μl, and Taq polymerase (5 U/μl) 0.25 μl. The PCR was performed in an Eppendorf Thermal Cycler using appropriate programs optimized for this primer. After denaturation of DNA through heating for 2 min at 94 °C, the PCR program involved 30 cycles; each cycle consisted of: denaturation at 94 °C for 2 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min. This was followed by a final elongation step for 2 min at 72 °C. The PCR products were separated on a 1.7 % agarose gel containing ethidium bromide in 1× TBE buffer, run at 100 V for 1.5 h, and the gel was visualized on a UV transilluminator. The purified PCR product was sequenced in both directions using an automated sequencer by Macrogen (Seoul, Korea). The sequences were edited using Finch TV V.1.4.0., and the BLASTN program was used for homology searches with the standard program default [27].
Urease Activity
All the isolates were tested for urease activity. Urea agar slants were inoculated with purified cultures. When organisms utilize urea, ammonia is produced that cause phenol red turns pink [11].
Nessler Assay Method
Effect of urease on calcite precipitation was studied by measurement of urea degradation rate in urease-positive isolates. The production of ammonia by different isolates was measured as a function of growth in liquid culture medium. Samples were taken from 3 biological replicates every 8 h for optical density (OD) and total ammonium nitrogen (TAN) measurements. In order to assess the growth, the optical density was measured with a spectrophotometer (Spectronic 21D, Milton Roy Company, USA) at 600 nm. TAN concentrations were determined colorimetrically by Nessler method. The specific urea degradation rate (SUD) was defined as the ratio of ammonium production (AP) per unit of bacterial growth in a given time (t) and is given by the following formula [14]:
Results and Discussion
Isolation of Calcite-Precipitating Bacteria
A total of 33 bacterial strains, C1–C31, were isolated from various environmental sources on precipitating medium. Strains C1–C5, C6–C7, C8–C24, C25–C27, and C28–C31 were isolated from freshwater, activated sludge, soil, chalk, and cement, respectively. Among thirty-one strains isolated, twenty-two strains had the ability to produce calcite nanocrystals.
Urea Degrading Capacity
Among 22 strains with the ability to produce calcite, 12 strains were urease-positive. Some strains have high urea affinities whereas some have lower affinities, by using the Nessler assay method, it was estimated that strains C8 and C18 have maximum urea degrading capacity. The specific urea degradation rate of the some strains is illustrated in Fig. 1a and b. Maximum urea degrading capacity is observed 18 h after inoculation for strains C4 (135 mg NH4 + l1− h1−), C7 (125 mg NH4 + l1− h1−), C8 (2,883 mg NH4 + l1− h1−), C21 (245 mg NH4 + l1− h1−), and C29 (127 mg NH4 + l1− h1−), after 24 h for strain C30 (226 mg NH4 + l1− h1−), and after 40 h for strain C18 (658 mg NH4 + l1− h1−). Urea degrading capacity in 5 other strains was significantly lower (not shown here).
Crystal Precipitation
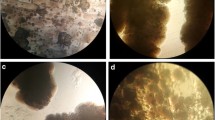
Microscopic observation showed that under laboratory conditions in precipitating medium, 71 % of the isolates (22 strains from 31 strains isolated during this study) were capable of forming crystalline CaCO3. As seen in Fig. 2, different types of calcite crystals can be produced by different bacterial strains, and bacteria with higher urease activity produce smaller crystalline aggregates as strain C8 with highest urease activity has the smallest aggregates in the size class and then, respectively, C18 and C7. Table 1 shows culture results of 14 strains (7 urease-positive strains and 7 urease-negative strains) on 4 different media of precipitating medium with urea (pH 7.0) and without urea (pH 7.0, 8.0, and 9.0).
Unlike Rivadeneyra [24], Hammes [17], and Banks’ [4] researches, we suggest that CaCO3 precipitation (calcification) is not a general phenomenon among the bacteria but can be produced by some bacteria with different mechanisms. If a bacterium has not this ability, it will not be able to produce calcite even with increasing pH. The strains C5, C12, C17, and 7 other strains (not shown in this paper) among urease-negative bacteria have the ability to produce calcite. Most calcite production occurs at alkaline pH and in the presence of urea in urease-negative and some urease-positive bacteria, respectively. Amount of calcite production in presence of urea is the same with alkaline pH in strains with low urease activity.
Although Fujita [16] and Chahal [11] believe that the rate of calcite precipitation directly associated with the rate of urea hydrolysis and therefore calcite precipitation is reinforced in presence of urea hydrolyzing bacteria by alkalinizing sedimentary environment, in this study it was demonstrated that the effect of urea hydrolysis on calcite production is more associated with carbonate ions. On the other hand, the ability of a urease-positive strain for nanocrystal production is directly related to urease activity as the strain C8, isolated from Iranian Northern Soil, with highest urease activity has maximum production and strains C4, C7, and C29 with low urease activity have minimum production among the isolated urease-positive strains in this study. There is a direct relation between the ability of a strain for hydrolysis of urea and the increase in pH (due to the production of ammonia) and carbonate ion concentration, while based on obtained results, the role of pH is much less than carbonate ions because significant change is not seen in amount of crystal production between pH 8.0 and pH 9.0. Therefore, increasing pH is not only consequence of urea hydrolysis in bacterial strains with high urease activity.
Some urease-negative strains are able to produce calcite only in the colony center (Fig. 2b) but some of them (Fig. 2a, c) and all of urease-positive strains (Fig. 2d–f) produce calcite in medium too. Therefore, some urease-negative strains likely have ability to secrete different kinds of enzymes that not introduced by previous researchers. More research is needed to explore these enzymes.
Comparison of XRD Patterns Among Different Treatments
In the XRD patterns, the characteristic diffraction peaks occurred at 2θ = 23.3°, 29.6°, 36.2°, 39.6°, 43.4°, 47.7°, and 48.7°, and the strongest reflection occurred at 2θ = 29.6. In all samples except C7, nanocrystals formed by bacteria were calcite, as indicated by X-ray diffraction. Vaterite was precipitated only by strain C7 (Fig. 3), isolated from the activated sludge, at 28 °C. Vaterite is only transiently found in nature. The crystallite size of nanoparticles was sub-100-nm as shown in Table 2.
Characterization and Identification of Strain C8
16S rRNA gene sequencing and biochemical characteristics were used to identify strain C8. The results of biochemical tests (e.g., positive results for myo-inositol and 3-O-methyl-d-glucopyranose) and BLAST analysis showed that strain C8 is more than 99 % identical of Enterobacter ludwigii; therefore, this bacterium is a strain of E. ludwigii with positive urease activity. The accession number of G11 which submitted to the database of GenBank is JX666242.
SEM and EDX Analysis
Scanning electron microscopy (SEM) showed that calcite nanocrystals produced by strain E. ludwigii are heterogeneous in shape and size. Nanocrystals were seen in two shapes of rhombohedra and rod-shaped. According to the information contained from XRD analysis, particle size was 35–79 nm (Table 2) but since bacterial cells act as nucleation sites in the precipitation of the calcite nanocrystals, over time these particles are placed on bacterial surfaces and so we see complex of bacteria covered with nanocrystals by SEM (Fig. 4).
Jimenez-Lopez [19] showed that bacterial-oriented calcite crystals typically appear as oriented compact aggregates of rhombohedra, and the bacterial surfaces were covered and limited by the newly formed crystals that help identifying bacterial carbonate cements. Moreover, Tong [26] suggested an assumption of limited-fusion or stepwise aggregation mechanism for calcite precipitation from the view of the small biomolecules polarity that can control over the growth of the crystals and the formation of the microcrystals. Similar to those, we believe that nanocrystals of CaCO3 adhere to bacteria and each other to form small aggregates, and then these small aggregates attract each other and form complex crystalline networks to trap bacteria. Many holes are present in these crystalline networks (Fig. 4f) that seems to be due to the aggregation of nanocrystals. On the other hand, since this strain can produce two types of crystals, at least two different kinds of pathways may be used and therefore urease is not the only enzyme to induce CaCO3 synthesis.
Amorphous phases cannot be detected directly by XRD analysis, because they do not produce additional visible reflexes in the XRD diagram, but only increase the background [28], and since there may be in the sample, we used EDX in conjunction with SEM to ensure that chemical composition of materials will be CaCO3. EDX spectrum of precipitates taken from the region shown in Fig. 4f was typical for CaCO3.
Conclusions
Based on qualitative and quantitative studies, E. ludwigii was selected as the best calcite-producing strain and used for biosynthesis of calcite nanoparticles as a biological and safe synthetic method. We concluded that CaCO3 precipitation (calcification) is not a general phenomenon among the bacteria and if a bacterium has not this ability, it will not be able to produce calcite even with increasing pH. On the other hand, the ability of a urease-positive strain for nanocrystal production is directly related to its urease activity. Actually, higher production ability corresponds to higher urease activity. In this way, the importance of pH is much less than carbonate ions. The composition of crystals was determined by XRD and EDX. Based on data obtained from SEM, nanocrystals of CaCO3 associate with each other for formation of complex crystalline networks to trap bacteria and are heterogeneous in shape, therefore, urease is not the only enzyme to induce CaCO3 synthesis in this strain of E. ludwigii and enzymes or other factors will be involved in this phenomenon.
References
Aloisi G, Gloter A, Kruger M, Wallmann K, Guyot F, Zuddas P (2006) Nucleation of calcium carbonate on bacterial nanoglobules. Geology 34:1017–1102
Al-Thawadi SM (2011) Ureolytic bacteria and calcium carbonate formation as a mechanism of strength enhancement of sand. J Adv Sci Eng Res 1:98–114
Arp G, Hofmann J, Reitner J (1998) Microbial fabric formation in spring mounds (‘microbialites’) of alkaline salt lakes in the Badain Jaran Sand Sea, PR China. Palaios 13:581–592
Banks ED, Taylor NM, Gulley J, Lubbers BR, Giarrizzo JG, Bullen HA, Hoehler TM, Barton HA (2010) Bacterial calcium carbonate precipitation in cave environments: a function of calcium homeostasis. Geomicrobiol J 27:444–454
Barabesi C, Galizzi A, Mastromei G, Rossi M, Tamburini E, Perito B (2007) Bacillus subtilis gene cluster involved in calcium carbonate biomineralization. J Bacteriol 189:228–235
Barton HA, Spear JR, Pace NR (2001) Microbial life in the underworld: biogenicity in secondary mineral formations. Geomicrobiol J 18:359–368
Bissett A, de Beer D, Schoon R, Shiraishi F, Reimer A, Arp G (2008) Microbial mediation of stromatolite formation in karst-water creeks. Limnol Oceanogr 53:1159–1168
Braissant O, Decho AW, Przekop KM, Gallagher KL, Glunk C, Dupraz C, Visscher PT (2009) Characteristics and turnover of exopolymeric substances in a hypersaline microbial mat. FEMS Microbiol Ecol 67:293–307
Cacchio P, Ercole C, Cappuccio G, Lepidi A (2003) Calcium carbonate precipitation by bacterial strains isolated from a limestone cave and from a loamy soil. Geomicrobiol J 20:85–98
Castanier S, Le Metayer-Levrel G, Perthuisot JP (1999) Ca-carbonates precipitation and limestone genesis: the microbiologist point of view. Sediment Geol 126:9–23
Chahal N, Rajor A, Siddique R (2011) Calcium carbonate precipitation by different bacterial strains. Afr J Biotechnol 10:8359–8372
Chen L, Shen YH, Xie AJ, Huang B, Jia R, Guo RY, Tang WZ (2009) Bacteria-mediated synthesis of metal carbonate minerals with unusual morphologies and structures. Cryst Growth Des 9:743–754
De Muynck W, De Belie N, Verstraete W (2010) Microbial carbonate precipitation in construction materials: a review. Ecol Eng 36:118–136
Dick J, De Windt W, De Graef B, Saveyn H, Van der Meeren P, De Belie N, Verstraete W (2006) Bio-deposition of a calcium carbonate layer on degraded limestone by Bacillus species. Biodegradation 17:357–367
Ercole C, Cacchio P, Cappuccio G, Lepidi A (2001) Deposition of calcium carbonate in karst caves: role of bacteria in Stiffe’s Cave. Int J Speleol 30:69–79
Fujita Y, Ferris FG, Lawson RD, Colwell FS, Smith RW (2000) Calcium carbonate precipitation by ureolytic subsurface bacteria. Geomicrobiol J 17:305–318
Hammes F, Verstraete W (2002) Key roles of pH and calcium metabolism in microbial carbonate precipitation. Rev Environ Sci Biotechnol 1:3–7
Jimenez-Lopez C, Rodriguez-Navarro C, Pinar G, Carrillo-Rosua FJ, Rodriguez-Gallego M, Gonzalez-Munoz MT (2007) Consolidation of degraded ornamental porous limestone stone by calcium carbonate precipitation induced by the microbiota inhabiting the stone. Chemosphere 68:1929–1936
Jimenez-Lopez C, Jroundi F, Pascolini C, Rodriguez-Navarro C, Pinar-Larrubia G, Rodriguez-Gallego M, Gonzalez-Munoz MT (2008) Consolidation of quarry calcarenite by calcium carbonate precipitation induced by bacteria activated among the microbiota inhabiting the stone. Int Biodeterior Biodegrad 62:352–363
Lee YN (2003) Calcite production by Bacillus amyloliquefaciens CMB01. J Microbiol 41:345–348
Leong DU, Greisen KS (1993) PCR detection of bacteria found in cerebrospinal fluid. In: Persing DH, Smith TF, Tenover FC, White TJ (eds) Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC, pp 300–309
Li W, Liu LP, Zhou PP, Cao L, Yu LJ, Jiang SY (2011) Calcite precipitation induced by bacteria and bacterially produced carbonic anhydrase. Curr Sci 100:502–508
McConnaughey TA, Whelan JF (1997) Calcification generates protons for nutrient and bicarbonate uptake. Earth Sci Rev 42:95–117
Rivadeneyra MA, Perez-Garcia I, Salmeron V, Ramos-Cormenzana A (1985) Bacterial precipitation of calcium carbonate in presence of phosphate. Soil Biol Biochem 17:171–172
Rodriguez-Navarro C, Rodriguez-Gallego M, Ben Chekroun K, Gonzalez-Munoz MT (2003) Conservation of ornamental stone by Myxococcus xanthus-induced carbonate biomineralization. Appl Environ Microbiol 69:2182–2193
Tong H, Ma W, Wang L, Wan P, Hu J, Cao L (2004) Control over the crystal phase, shape, size and aggregation of calcium carbonate via a l-aspartic acid inducing process. Biomaterials 25:3923–3929
Uh Y, Lee CH, Lee MK, Park IG, Park DW, Yoon KJ (1998) Diagnosis of bacteremia by universal primer of eubacteria. Korean J Clin Pathol 18:195–200
Walenta G, Fullmann T (2004) Advances in quantitative XRD analysis for clinker, cements, and cementitious additions. ICDD Adv X-ray Anal 47:287–296
Wen ZF, Zhong JH, Li Y, Guo ZQ, Gao JB, Xu XL (2004) Current study on genesis and formation conditions of stromatolites. Geol J China Univ 10:418–428
Yaaghoobi M, Emtiazi G, Roghanian R (2012) A novel approach for aerobic construction of iron oxide nanoparticles by acinetobacter radioresistens and their effects on red blood cells. Curr Nanosci 8:286–291
Yang JL, Wang MS, Cheng AC, Pan KC, Li CF, Deng SX (2008) A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J Gastroenterol 14:2872–2876
Yena FS, Chang PL, Yu PC, Yang RJ (2007) Characterization on microstructure homogeneity of θ-Al2O3 powder systems during phase transformation. Key Eng Mater 351:81–87
Acknowledgments
The current study was supported by Grant from the University of Isfahan to Sara Ghashghaei for obtaining M.Sc. degree. In addition, we would like to thank Gh. R. Ghezelbash from University of Isfahan for helping.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghashghaei, S., Emtiazi, G. Production of Calcite Nanocrystal by a Urease-Positive Strain of Enterobacter ludwigii and Study of Its Structure by SEM. Curr Microbiol 67, 406–413 (2013). https://doi.org/10.1007/s00284-013-0379-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0379-5