Abstract
TIGIT is a lymphocyte surface receptor, which is mainly expressed on the surface of CD8+T cells. The role of TIGIT in colorectal cancer and its expression pattern in colorectal cancer infiltrating lymphocytes are still controversial. This study aimed at identifying the function of TIGIT in colorectal cancer. Patients with colorectal cancer showed significantly higher TIGIT+CD8+T cell infiltration in tumor tissues, metastases compared with paired PBMC and normal tissues through flow cytometry. TIGIT+CD8+T cells showed an exhausted phenotype and expressed low levels of killer cytokines IFN-γ, IL-2, TNF-α. In addition, more inhibitory receptors such as PD-1, LAG-3, and TIM-3 were expressed on the surface of TIGIT+CD8+T cells. TGF-β1 could promote the expression of TIGIT and inhibit CD8+T cell function in vitro. Moreover, the accumulation of TIGIT+T cells in tumors was associated with advanced disease, predicted early recurrence, and reduced survival rates in colorectal cancer patients. Our results indicate that TIGIT can be a biological marker for the prognosis of colorectal cancer, and TIGIT can be used as a potential target for treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is currently the third most common cancer in the world and the second most common cause of cancer-related deaths [1]. Immunotherapy has developed into an important treatment for advanced tumors [2]. Utilizing the expression of immune checkpoints is revolutionizing cancer immunotherapy by inducing meaningful clinical responses in a variety of cancer types [3, 4]. Immunotherapies targeting PD-1 and PD-L1 have been widely used in the treatment of advanced tumors [5, 6]. However, only a few patients who contain microsatellite instability in colorectal cancer can benefit from anti-PD-1 and anti-PD-L1 immunotherapy [7, 8]. The combined application of immune checkpoint blocking therapy may improve the effect of colorectal cancer immunotherapy, and potential immune checkpoint in colorectal cancer should be discovered. Research has found that many other suppressive immune checkpoints exist in colorectal cancer tissues, including TIGIT [9].
T cells play a major role in cellular immunity. It contains a variety of biological functions, such as directly killing target cells, assisting, or inhibiting B cells from producing antibodies, responding to specific antigens and mitogens, and producing cytokines [10]. Many studies have confirmed the existence of a large number of T cells in tumor tissues, and cellular immunity plays important roles in tumor progression[11,12,13]. However, these T cells have shown different phenotypes and functions. CD8+T cell is an important group of cells that play the role of immune surveillance and antitumor immune response [14, 15]. The function of CD8+T cells in tumor-infiltrating lymphocytes (TILs) is an important part that affects the balance of the tumor immune microenvironment and deserves attention [16].
TIGIT (T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domain) is a co-inhibitory receptor expressed on the surface of killer T cells, Tregs, and NK cells, which competes with the co-stimulatory receptor CD226, binding to the receptor CD155 [17,18,19,20]. The expression of TIGIT on CD8+T and NK cells leads to a cellular immunosuppressive phenotype and is related to the production of cytokines. TIGIT plays an important regulatory role in the occurrence and development of autoimmune diseases such as SLE and viral infections [21, 22]. A recent study has confirmed that TIGIT is upregulated in a variety of tumor immune microenvironments, such as AML, CML, lung cancer, and melanoma, and promotes tumor development[23]. However, the role of TIGIT in colorectal cancer and its expression pattern in tumor-infiltrating lymphocytes are still controversial.
In this study, we found that the expressions of TIGIT in CRC tissues and metastases were significantly upregulated compared to adjacent normal tissues and peripheral blood mononuclear cells (PBMCs). Our result showed that TIGIT was a co-inhibitory receptor on the surface of CD8+T lymphocytes and could inhibit the ability of CD8+T cells to secrete cytokines. Furthermore, we confirmed that colorectal cancer cells could regulate the expression of TIGIT through the TGF-β1 pathway and further affect the function of CD8+T lymphocytes. The expression of TIGIT was positively correlated with the poor prognosis of patients with colorectal cancer. These findings indicate that TIGIT can be a biological marker for the prognosis of colorectal cancer and can be used as a potential target for treatment.
Methods and materials
Patients and tissue samples
Fresh paired normal and tumor tissues samples used to analyze the TIGIT expression between normal tissues and tumors were obtained from 22 colorectal cancer patients who underwent surgical treatment in the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) from the year 2019 to 2020 during surgery (Supplementary Table S1 cohort1). And PBMCs, tumor tissues, and lymph node metastases from 10 patients were used to analyze the TIGIT expression in PBMCs, tumor tissues, and lymph node metastases (Supplementary Table S1 cohort2). The tissues were obtained within half an hour after resection. Peripheral blood samples were also collected from healthy donors. And 139 paraffin specimens of tumors from colorectal cancer patients who underwent surgery during 2009–2013 were applied to immunohistochemistry (Supplementary Table S1 cohort3). In addition, we collected clinical data of these patients and maintained following-up until 2018. These patients were subjected to diagnosis using conventional histology. Prior to surgery, no patients received other antitumor treatments, and clinical stages were classified according to the guidelines of the International Union Against Cancer. According to the ethics committee of the Third Affiliated Hospital of Sun Yat-sen University, all patients signed the informed consent. A tissue microarray (BIOTECH WELL, Cat#ZL-CocMet56) contained primary colorectal cancer, and liver metastases were used to evaluate the expression of TIGIT in liver metastases.
Isolation of peripheral blood mononuclear cells and tumor-infiltrating lymphocytes
Peripheral blood samples from healthy people were collected from healthy donors, and peripheral blood from patients with colorectal cancer was collected before surgery in the Third Affiliated Hospital of Sun Yat-sen University. For peripheral blood mononuclear cells isolation, a standard Ficoll procedure was utilized as described previously [24]. To isolate tumor-infiltrating lymphocytes, the fresh tissue samples were collected within half an hour after surgery, washed with saline, and then transported to the laboratory within 2 h. The tissues were cut into 0.5 mm specimens followed by digestion with collagenase D(Roche Cat# 11,088,858,001)and DNaseI(Roche Cat# 10,104,159,001)for 1 h in 37 °C. Dissociated cells were filtered through 70 um mesh and separated by percoll centrifugation. The mononuclear cells were washed by PBS and resuspended in 1640 medium for the next experiment.
Flow cytometry
The PBMC and TILs were then stained with fluorochrome-conjugated anti-CD3, CD4, CD8, CD56, TIGIT, PD-1, TIM-3, LAG-3, CD226 antibodies for 30 min in 4 °C. After washing three times, samples were collected and analyzed using flow cytometry. For intracellular staining, the mononuclear cells were fixed and permeabilized for 30 min by fixed and permeabilized kit (BD Biosciences Cat#554,714) after extracellular staining, and then incubated with the corresponding antibodies (fluorochrome-conjugated anti-IFN-γ, IL-2, TNF-α antibodies) for 30 min at 4 °C for intracellular staining. The staining cells were analyzed using a FACS Calibur flow cytometer (Becton Dickinson, the USA), and the data were analyzed with FlowJo software. The antibodies used in this study are shown in Supplementary Table S2.
ELISA assay
Mononuclear cells were isolated, and then, TIGIT+CD8+T and TIGIT−CD8+T cells were collected by fluorescence-activated cell sorting (FACS). To measure the concentration of IFN-γ, IL-2, and TNF-α, TIGIT+CD8+T and TIGIT−CD8+T cells were cultured in 24 well plates at a density of 106/ml and stimulated with a-CD3 and a-CD28. After cultured for 24, 48, and 72 h, the culture mediums were collected for ELISA. The concentration of IFN-γ、IL-2, and TNF-α was detected by ELISA kits (KeyGEN BioTECH China) according to the manufacturer’s instructions.
Preparation of culture supernatant from primary colorectal cancer and co-culture
Culture supernatants were acquired by the culture of completely digested colorectal cancer tissues. The digested tumor cells were washed with PBS and then resuspended in RPMI 1640 (Gibco, the USA) medium containing 10% fetal bovine serum (FBS, Gibco) at 37 °C under 5% CO2. After 72 h, the supernatant was collected for further experiment. The PBMCs were collected after isolation and seeded in a 24-well plate coated with α-CD3 (Biolegend Cat# 317,326) and α-CD28(Cat# 302,933) for stimulating culture, TGF-β1 (PeproTech Cat# AF-100-21C-2), TGF-β1 inhibitor (SB431542), (Meilunbio Cat# MB5459), and tumor supernatant were added separately to the medium. After 72 h, PBMCs were collected and stained with anti-CD8, anti-TIGIT, and anti-IFN-γ antibodies followed by flow cytometry analysis.
Immunohistochemistry staining
Formalin-fixed paraffin-embedded tissue was cut into 4 μm sections. The sections and tissue microarray were deparaffinized and rehydrated after heated in 65 °C for 1 h. Antigen retrieval was performed by EDTA (Solarbio Cat# C1034) and non-specific antigen blocking by 10%BSA (Gibco, the USA). The sections were then incubated with primary antibody against TIGIT (Abcam Cat# ab243903) in 4 °C overnight. After incubated with HPR-second antibody (Affinity Biosciences Cat# S0001), the staining was detected by the DAB system (Beyotime Cat# P0202). The integrated optical density (IOD) was quantified using ImagePro Plus software (Media Cybernetics, the USA) in a blinded manner.
GEPIA2 analysis
GEPIA2 is an interactive web portal that allows in-depth analysis of TCGA gene expression data, which contains RNA sequencing data and clinical data from 31 cancer[25]. It can analyze the relative expression of query genes between tumors and normal samples in various tumors. GEPIA2 is publicly available at http://gepia2.cancer-pku.cn/#general.
Data analysis
The GraphPad Prism 7 and SPSS22 statistical analysis software programs were used for statistical analysis of the experimental data, and results were shown as means ± SEM. Significant differences between the two unpaired groups were determined by log-rank test or Student's t test. Significant differences between two paired groups were determined by paired t-tests. Cut off value of TIGIT IOD was determined by the receiver operating characteristic curve. Kaplan–Meier survival curves were plotted using patient survival data and tested by log-rank test. All data were analyzed using a two-sided test, and P < 0.05 was considered as a statistically significant criterion. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Results
TIGIT is upregulated in colorectal cancer and metastases.
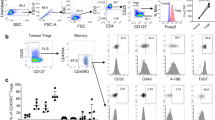
To clarify the expression pattern of TIGIT in lymphocytes, we evaluated the expression of TIGIT in 22 colorectal cancer tissues (including tumor tissues and corresponding normal tissue) by flow cytometry, and the gating pattern is shown in Supplementary Fig. S1. In colorectal cancer patients, the proportion of TIGIT+CD3+T in tumor tissues is significantly higher than in normal tissues (Fig. 1a). The above result was further confirmed by flow cytometry fluorescence analysis (Fig. 1b). Based on the above results, we also studied the difference in the expression of TIGIT in CD4+T and CD8+T cells. Among CD4+T cells, the proportion of TIGIT+CD4+T cells in tumor tissues was significantly higher than normal tissues (Fig. 1c). And the mean fluorescence intensity of TIGIT in CD4 showed the same trend (Fig. 1d). Similar results also appeared in the CD8+T cell subset (Fig. 1e). The mean fluorescence intensity in tumor tissues was also significantly higher than normal tissues (Fig. 1f). We further compared the expressions of TIGIT on the surface of NK cells in tumor tissues and normal tissues. TIGIT expressions on NK cells were also significantly upregulated in tumor tissues (Supplementary Fig. S2a). And we applied the TCGA datasets to assess the RNA pattern of TIGIT in colorectal cancer; the result showed a trend consistent with our results though it did not reach significance (Supplementary Fig. S2b). Compared to PBMC, TIGIT+ cells occupied a larger proportion in tumor-infiltrating CD3, CD4, and CD8+T cells (Fig. 1g), and the representative FACS plots are shown in Supplementary Fig. S3a-c. Furthermore, the expressions of TIGIT in lymph node metastases were higher than paired tumor tissues (Fig. 1h). Then, we compared the expressions of TIGIT in tumor tissues and liver metastases by immunohistochemistry staining and found TIGIT was higher in liver metastases (Supplementary Fig. S3d). In order to more intuitively reflect the expressions of TIGIT in tissues, we performed immunohistochemical labeling of TIGIT on 139 tumor tissues and 68 normal tissues (Fig. 2a). We verified a significant increase in TIGIT protein in colorectal cancer tissues (Fig. 2b). Next, we analyzed the expressions of TIGIT in 68 pairs of tumor tissues and normal tissues, and the results still suggest that the expressions of TIGIT in tumor tissues were significantly higher than the matched normal tissues (Fig. 2c).
TIGIT is upregulated in colorectal cancer TILs and metastases, especially in CD8+T cells a. Different expressions of TIGIT expression in CD3+T cells between normal tissue and tumor tissues. n = 12. b. The mean fluorescence intensity of TIGIT in CD3+T cells. n = 12. c. Different expressions of TIGIT expression in CD4+T cells between normal tissue and tumor tissues. n = 22. d. The mean fluorescence intensity of TIGIT in CD4+T cells. n = 22. e. Different expressions of TIGIT expression in CD8+ cells between normal tissues and tumor tissues. n = 22. f. The mean fluorescence intensity of TIGIT in CD8+T cells. n = 22. g. Different expressions of TIGIT expression in CD3, CD4, and CD8+T cells between PBMC and tumors. n = 10. h. Different expressions of TIGIT expression in CD3, CD4, and CD8+T cells between tumors and lymph node metastases. n = 6. (PBMC, peripheral blood mononuclear cell; LN-M, lymph node metastases). Error bars represent mean ± SEM. Two-tailed Student’s t test and paired t-test were performed for statistical analysis; ns: not significant; *:p < 0.05; **:p < 0.01; ***:p < 0.001; ****: P < 0.0001
The protein expression pattern of TIGIT is different in human colorectal cancer a. Representative IHC staining of TIGIT in human colorectal cancer tissues and adjacent noncancerous tissues. Scale bars: 400 × 20 μm. b. TIGIT protein expression of 139 colorectal cancer tissues and 68 adjacent noncancerous tissues by IHC in tissue paraffin sections. c. Paired comparison of TIGIT protein expression of 68 paired cancer tissues and noncancerous tissues. Error bars represent mean ± SEM. Two-tailed Student’s t test and paired t-test were performed for statistical analysis; ns: not significant; *:p < 0.05; **:p < 0.01; ***:p < 0.001; ****: P < 0.0001
TIGIT+CD8+T cells exhibit a unique suppressive function
The expressions of TIGIT in colorectal cancer tissues were elevated, and it can be preliminarily judged that the expression of TIGIT is related to lymphocyte exhaustion. Lymphocytes play a killing function mainly by secreting cytokines, and CD8+T is a vital subset that plays a killing role in lymphocytes. Therefore, we investigated the effect of TIGIT expression on cytokine secretion by infiltrating CD8+T lymphocytes. We detected the proportion of TIGIT expression in CD8+T cells by flow cytometry and then detected the expressions of related cytokines IFN-γ, IL-2, and TNF-α by intracellular staining. Next, we divided the CD8+T cells into two groups as TIGIT+CD8+T and TIGIT−CD8+T cells and tested the ability secreting cytokines of these two subsets. The results showed that the ability of TIGIT+CD8+T cells to secrete IFN-γ, IL-2, and TNF-α was significantly lower than TIGIT−CD8+T cells (Fig. 3a, b, c). To further clarify the effect of TIGIT on the secretion of functional cytokines by CD8+T cells, the concentrations of IFN-γ, IL-2, and TNF-α in the culture mediums of TIGIT+CD8+T and TIGIT−CD8+T cells were measured by ELISA kits. The results showed the concentrations of IFN-γ, IL-2, and TNF-α of TIGIT+CD8+T cells were at lower levels (Fig. 3d). The expression of TIGIT led to the depletion of CD8+T cell functions.
TIGIT+CD8+T cells exhibit a unique suppressive function a. IFN-γ expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 17. b. IL-2 expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 14. c. TNF-α expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 15. d. The concentration of IFN-γ, IL-2, and TNF-α in the culture medium of TIGIT−CD8+T cells and TIGIT+CD8+T cells after culturing for 24, 48, 72 h. n = 3 biologically independent experiments. Error bars represent mean ± SEM. Two-tailed Student’s t test and paired t-test were performed for statistical analysis; ns: not significant; *P< 0.05; **p < 0.01; ***p < 0.001; ****P < 0.0001
TIGIT+CD8+T cells exhibit a unique suppressive phenotype
The expression of TIGIT can restrict T lymphocytes' ability to secrete cytokines and cause CD8 + T cells functional exhaustion. The functional status of lymphocytes is mostly affected by surface receptors. To further clarify that TIGIT expression promotes CD8 + T cell depletion, we tested different T cell receptors' expression status by flow cytometry. PD-1 is a major receptor for T cell exhaustion. We found that PD-1 was significantly increased on the surface of TIGIT+CD8+T cells (Fig. 4a). TIM-3 showed a significant increase in TIGIT+CD8+T cells compared to TIGIT−CD8+T cells (Fig. 4B). LAG-3 also appeared to be elevated in TIGIT+CD8+T cells (Fig. 4c). Interestingly, the expression of co-stimulatory receptors CD226 showed no significant difference between TIGIT+CD8+T cells and TIGIT−CD8+T cells (Supplementary Fig. S4). Besides, we compared the expression patterns of PD-1, TIM-3, LAG-3, and CD226 on TIGIT+CD8+T cells between normal tissues and tumors. The expressions of PD-1, TIM-3, and LAG-3 showed a rising trend in tumors, while CD226 showed an adverse trend (Fig. 4d). This is consistent with the suppressive state of the tumor microenvironment. The expressions of co-inhibitory receptors were upregulated. The expression of co-stimulatory receptors was reduced, indicating that TIGIT can affect the expression of receptors on CD8+T cells and cause CD8+T cells exhaustion.
TIGIT+ CD8+T cells exhibit a unique suppressive phenotype a. PD-1 expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 13. b. TIM-3 expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 16. c. LAG-3 expression pattern between TIGIT−CD8+T cells and TIGIT+CD8+T cells. n = 17. d. PD-1, TIM-3, LAG-3, and CD226 expression on TIGIT+CD8+T cells between normal tissues and tumors. Error bars represent mean ± SEM. Two-tailed Student’s t test and paired t-test were performed for statistical analysis; ns: not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****P < 0.0001
Colorectal cancer promotes TIGIT+CD8+T cell expansion via TGF-β1
In previous studies, we found that TIGIT expression was upregulated in colorectal cancer tumor tissues. A previous study has confirmed that TGF-β1 can promote the expression of PD-1[26]. And TGF-β1 is a crucial component of the colorectal cancer tumor microenvironment, so we chose to explore its role in TIGIT expression. The co-culture of T cells with TGF-β1 enhanced TIGIT expression on CD8+T cells in a concentration-dependent manner (Fig. 5a). Next, we investigated whether blockade TGF-β1 could reverse the TGF-β1 dependent TIGIT enhancement. TGF-β1 inhibitor, SB431542, could inhibit TGF-β1 dependent TIGIT enhancement (Fig. 5b). We found that the supernatant of tumor cells can promote the expression of TIGIT in vitro culture (Fig. 5 c, d). Then, after adding the TGF-β1 inhibitor to the culture system, TIGIT expression decreased (Fig. 5c, d). In addition, we studied the effect of TGF-β1 on CD8+T cell function. After adding TGF-β1 or tumor supernatant to the culture medium, the level of IFN-γ in TIGIT+CD8+T cells showed a severe decline, which could be reversed by TGF-β1 inhibitor (Fig. 5e, f). These results indicated that TGF-β1 secreted by tumor cells could cause TIGIT+CD8+ T cells expansion and CD8+T cells exhaustion.
Colorectal cancer promotes TIGIT+CD8+T cell expansion via TGF-β1 a. TIGIT expression on CD8+T in different TGF-β1 concentrations. n = 3 biologically independent experiments. b. TIGIT expression on CD8+T in different TGF-β1 inhibitor (SB431542) concentrations with TGF-β1(1 ng/ml). n = 3 biologically independent experiments. c. Representative contour plots for the percentage of TIGIT+CD8+T cells in TGF-β1(1 ng/ml), TGF-β1(1 ng/ml) + inhibitor(10uM/ml), tumor supernatant, and tumor supernatant + inhibitor r(10uM/ml) conditional medium systems. d. Quantification of FACS analysis for the percentage of TIGIT+CD8+T cells. n = 3 biologically independent experiments. e. Representative contour plots for the percentage of IFN-γ+TIGIT+CD8+T cell in TGF-β1(1 ng/ml), TGF-β1(1 ng/ml) + inhibitor(10uM/ml), tumor supernatant, and tumor supernatant + inhibitor r(10uM/ml) conditional medium systems. f. Quantification of FACS analysis for the percentage of IFN-γ+TIGIT+CD8+T cells. n = 3 biologically independent experiments. Error bars represent mean ± SEM. Two-tailed Student’s t test was performed for statistical analysis; ns: not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****P < 0.0001
Upregulated TIGIT expression is correlated with poor prognosis of colorectal cancer
Depletion of lymphocytes and reduced tumor-killing capacity will inevitably promote tumor progression. In order to clarify the impact of TIGIT expression on the prognosis of colorectal cancer, we evaluate TIGIT expression by immunohistochemistry. The expressions of TIGIT in tumor tissues showed different patterns (Fig. 6a). The expressions of TIGIT in patients with stage I–II were significantly lower than that in patients with stage III–IV (Fig. 6b). The expressions of TIGIT in the tissues in patients with recurrence were significantly higher than patients without recurrence (Fig. 6c). In addition, the expressions of TIGIT in dead and survival patients were further analyzed. The expression of TIGIT showed an increased pattern in tumor tissues of patients who died due to tumor (Fig. 6d). Compared the expression of TIGIT in patients with different levels of tumor differentiation, TIGIT expression was lower in highly differentiated tumors (Supplementary Table S3). Then, colorectal cancer patients were divided into two groups with high TGIT expression and low TIGIT expression according to ROC (receiver operating characteristic curve) (Fig. 6e), and we compared the difference in tumor recurrence time and survival time between the two groups. TIGIT high expression group contained a higher proportion of recurrence and shorter time to recurrence (Fig. 6f). The overall survival of patients with high TIGIT expression was significantly reduced (Fig. 6g), suggesting that TIGIT was associated with a poor prognosis for colorectal cancer. It suggested that the expression of TIGIT is an independent influencing factor for the prognosis of colorectal patients and can be used as a predictor of the outcome for colorectal cancer patients.
Upregulated TIGIT expression is correlated with poor prognosis of colorectal cancer a. Low expression pattern of TIGIT in human colorectal cancer tissues. Scale bars: 100 × 100 μm, 400 × 20 μm. b. High expression pattern of TIGIT in human colorectal cancer tissues. Scale bars: 100 × 100 μm, 400 × 20 μm. c. TIGIT protein expression in stage I–II and stage III–IV colorectal cancer. stage I–II n = 54, stage III–IV n = 85. d. TIGIT protein expression in non-relapse and relapsed colorectal cancer. non-relapse n = 94, relapse n = 45. e. TIGIT protein expression in survival and dead colorectal cancer patients. survival n = 103, dead n = 36. f. ROC of TIGIT expression. Analyzed by the log-rank test. g. Kaplan–Meier analysis of relapse-free survival according to low and high TIGIT protein expression in 139 colorectal cancer patients. h. Kaplan–Meier analysis of overall survival according to low and high TIGIT protein expression in 139 colorectal cancer patients. Error bars represent mean ± SEM. Two-tailed Student’s t test was performed for statistical analysis; ns: not significant; *:p < 0.05; **:p < 0.01; ***:p < 0.001; ****: P < 0.0001
Discussion
TIGIT has been discovered in recent years as a new immune checkpoint, which is mainly expressed on the surface of lymphocytes, including killer T cells, helper T cells, and NK cells. The existing study has confirmed that TIGIT is a co-inhibitory receptor, and its corresponding ligand is CD155[27]. The expression of TIGIT can cause depletion of effector T cells and NK cells. In the field of oncology, the function of TIGIT in many tumors has been confirmed, such as liver cancer[28], head and neck squamous cell carcinoma[29], and lung cancer[30]. However, in colorectal cancer, the function of TIGIT is still controversial. Some studies have found that TIGIT expression is elevated in colorectal cancer[31], while another study has found that TIGIT expression is downregulated in PBMC of advanced colorectal cancer but showed an opposite trend in tumor tissues [32]. And Kitsou et al. showed that TIGIT expression seemed to be associated with better overall survival of colorectal cancer but did not reach significance[33]. Therefore, more researches are needed to confirm the role of TIGIT in colorectal cancer.
Our research suggested that TIGIT was upregulated in colorectal cancer infiltrating lymphocytes, including CD3, CD4, CD8, and NK cells. Compared with the corresponding PBMC and normal tissues, TIGIT in tumor tissues was significantly increased, especially in CD8+T cells. And the expressions of TIGIT in metastases were higher than primary tumors. According to the TCGA database data and the immunohistochemical analysis of tissue samples, the mRNA and protein levels of TIGIT in colorectal cancer tumor tissues were higher than normal tissues, consistent with the previous report[31]. We also found that the expression of TIGIT in tumor tissues was related to the recurrence and survival of patients with colorectal cancer. Furthermore, we discovered that the expression of TIGIT in advanced colorectal cancer was significantly higher than that in early stage colorectal cancer. These results suggested that the expression of TIGIT can affect the prognosis of colorectal cancer.
CD8+T cells are a group of immune cells that play immune surveillance and killing roles in several tumors [14]. The existing study has confirmed that the expression of co-inhibitory receptors affects the function of CD8+T cells and promotes the exhaustion of CD8+T cells[34]. TIGIT is a co-inhibitory receptor that limits antitumor and other CD8+T cell-dependent immune responses, and TIGIT blockade can restore CD8+ T cell function and IFN-γ release[35]. Our results suggested that TIGIT-expressing CD8 + T cells' ability to secrete IFN-γ, IL-2, and TNF-α was significantly reduced. Whether in normal tissues or tumor tissues, the expression of TIGIT can cause a decrease in the ability of CD8+T cells to secrete cytokines, which is manifested as functional exhaustion. CD8+T cell exhaustion is also manifested in its surface co-inhibitory receptor expression. We explored the effect of TIGIT expression on CD8+T cells expressing other co-inhibitory or stimulation receptors. Surprisingly, CD8+T cells expressing TIGIT also highly express other co-inhibitory receptors, such as PD-1, TIM -3, and LAG-3, while the expression of co-stimulatory receptor CD226 is not affected by it. T cell exhaustion is mainly caused by several aspects, such as poor effector function, continuous expression of inhibitory receptors, and transcriptional status different from functional effector T cells. And immune checkpoint expression can also impact cytokine secretion and gene transcription[34, 36]. Our study showed that TIGIT+CD8+T cells exhibit T cell exhaustion features, including up-regulation of several inhibitory regulators and decreased production of cytokines. We suspect that TIGIT may promote the exhaustion of CD8+T cells by up-regulating the co-inhibitory receptors on the surface of CD8+T cells and attenuating their function of secreting cytokines.
The immune microenvironment of colorectal cancer is composed of multiple components. On the one hand, immune cells play a major role in immune surveillance and kill tumor cells; on the other hands, tumor cells regulate immune cell functions. Tumor cells can bind to immune cell surface receptors through ligands to regulate immune cells. Moreover, tumor cells can indirectly regulate immune cells' function and phenotype by secreting cytokines and other substances. TGF-β is one of many cytokines secreted by tumor cells[37]. Studies have confirmed that TGF-β plays an important role in immune suppression within the tumor microenvironment and regulating the phenotype of immune cells[38, 39]. A previous study has verified that TGF-β can regulate the expression of lymphocyte surface receptors[26]. Our study confirmed that TGF-β1 could upregulate the expression of TIGIT, and TGF-β1 inhibitors could inhibit this up-regulation. Besides, our result showed that tumor cells could upregulate the expression of TIGIT on the surface of CD8+T cells by secreting TGF-β1. Furthermore, TGF-β1 inhibited the IFN-γ secreted by TIGIT+CD8+T cells, which inhibited the function of CD8+T cells. This result suggested that tumor cells can upregulate the expression of TIGIT through TGF-β1 and promote CD8+T cell function exhaustion, thereby evading immune surveillance and promoting tumor growth. However, the specific mechanism that affects TIGIT expression in colorectal cancer may be complicated, and there might be many factors that regulate its expression. Our study suggested that TGF-β1 secreted by tumor cells could upregulate the expression of TIGIT in colorectal cancer. It indicated that TGF-β1 might be one of the factors regulating TIGIT expression. But the molecular mechanism that regulates the expression of TIGIT needs to be further explored.
In summary, TIGIT is upregulated in colorectal cancer tissues and metastases, especially in CD8+ T cells. The expression of TIGIT can not only directly affect the secretion of killer cytokines IFN-γ, IL-2, and TNF-α from CD8+T cells, but also upregulate the expression of PD-1, TIM-3, and LAG-3 on the surface of CD8+T cells. The exhaustion of CD8+T cell function causes tumor cell immune escape and promotes tumor progression, positively correlated with the poor prognosis of tumor patients. Tumor cells can upregulate the expression of TIGIT by secreting TGF-β1 to promote the exhaustion of CD8+T cells and promote tumor immune escape. Therefore, TIGIT is a potential target for the treatment of colorectal cancer and can be regulated through the TGF-β1 pathway. Our research provides a theoretical basis for applying TIGIT as a potential target of immunotherapy for colorectal cancer.
Data availability
The data used in this study are available from the corresponding author on reasonable request.
Abbreviations
- CRC:
-
Colorectal cancer
- IFN-γ:
-
Interferon γ
- IL-2:
-
Interleukin-2
- LAG-3:
-
Lymphocyte-activation gene 3
- NK cells:
-
Natural killer cells
- PBMC:
-
Peripheral blood mononuclear cell
- PD-1:
-
Programmed cell death protein 1
- PD-L1:
-
Programmed cell death ligand 1
- TGF-β:
-
Transforming growth factor-β
- TIGIT:
-
T cell immunoreceptor with Ig and ITIM domains
- TILs:
-
Tumor-infiltrating lymphocytes
- TIM-3:
-
T cell immunoglobulin and mucin domain-3
- TNF-α:
-
Tumor necrosis factor-α
- Tregs:
-
Regulatory T cells
References
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70(1):7–30. https://doi.org/10.3322/caac.21590
Yang Y (2015) Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest 125(9):3335–3337. https://doi.org/10.1172/jci83871
Chen DS, Mellman I (2017) Elements of cancer immunity and the cancer-immune set point. Nature 541(7637):321–330. https://doi.org/10.1038/nature21349
Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH (2016) Coinhibitory pathways in immunotherapy for cancer. Annu Rev Immunol 34:539–573. https://doi.org/10.1146/annurev-immunol-032414-112049
Gentzler R, Hall R, Kunk PR, Gaughan E, Dillon P, Slingluff CL Jr, Rahma OE (2016) Beyond melanoma: inhibiting the PD-1/PD-l1 pathway in solid tumors. Immunotherapy 8(5):583–600. https://doi.org/10.2217/imt-2015-0029
Kluger HM, Chiang V, Mahajan A, Zito CR, Sznol M, Tran T, Weiss SA, Cohen JV, Yu J, Hegde U, Perrotti E, Anderson G, Ralabate A, Kluger Y, Wei W, Goldberg SB, Jilaveanu LB (2019) Long-term survival of patients with melanoma with active brain metastases treated with pembrolizumab on a phase II trial. J Clin Oncol 37(1):52–60. https://doi.org/10.1200/jco.18.00204
Dt Le, Jn Uram, Wang H, Bartlett Br et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency. New Engl J Med 372(26):2509–20. https://doi.org/10.1056/NEJMoa1500596
Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, Mcdermott R, Hill A, Sawyer MB, Hendlisz A, Neyns B, Svrcek M, Moss RA, Ledeine JM, Cao ZA, Kamble S, Kopetz S, André T (2018) Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol 36(8):773–779. https://doi.org/10.1200/jco.2017.76.9901
Harjunpää H, Guillerey C (2020) TIGIT as an emerging immune checkpoint. Clin Exp Immunol 200(2):108–119. https://doi.org/10.1111/cei.13407
Kumar BV, Connors TJ, Farber DL (2018) Human T Cell development, localization, and function throughout Life. Immunity 48(2):202–213. https://doi.org/10.1016/j.immuni.2018.01.007
Solinas C, Pusole G, Demurtas L, Puzzoni M, Mascia R, Morgan G, Giampieri R, Scartozzi M (2017) Tumor infiltrating lymphocytes in gastrointestinal tumors: controversies and future clinical implications. Crit Rev Oncol Hematol 110:106–116. https://doi.org/10.1016/j.critrevonc.2016.11.016
Stanton SE, Disis ML (2016) Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer 4:59. https://doi.org/10.1186/s40425-016-0165-6
Bremnes RM, Busund LT, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, Hald S, Khanehkenari MR, Cooper WA, Kao SC, Dønnem T (2016) The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol 11(6):789–800. https://doi.org/10.1016/j.jtho.2016.01.015
Effros RB (2004) Replicative senescence of CD8 T cells: potential effects on cancer immune surveillance and immunotherapy. Cancer Immunol Immunother 53(10):925–933. https://doi.org/10.1007/s00262-004-0508-x
Farhood B, Najafi M, Mortezaee K (2019) CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol 234(6):8509–8521. https://doi.org/10.1002/jcp.27782
Gajewski TF, Schreiber H, Fu YX (2013) Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14(10):1014–1022. https://doi.org/10.1038/ni.2703
Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, Eaton D, Grogan JL (2009) The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 10(1):48–57. https://doi.org/10.1038/ni.1674
Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, Wang Z, Wu Q, Peng H, Wei H, Sun R, Tian Z (2018) Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. https://doi.org/10.1038/s41590-018-0132-0
Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, Sharpe AH, Quintana FJ, Mathis D, Benoist C, Hafler DA, Kuchroo VK (2014) Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 40(4):569–581. https://doi.org/10.1016/j.immuni.2014.02.012
Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA (2012) The TIGIT/CD226 axis regulates human T cell function. J Immunol 188(8):3869–3875. https://doi.org/10.4049/jimmunol.1103627
Schorer M, Rakebrandt N, Lambert K, Hunziker A, Pallmer K, Oxenius A, Kipar A, Stertz S, Joller N (2020) TIGIT limits immune pathology during viral infections. Nat Commun 11(1):1288. https://doi.org/10.1038/s41467-020-15025-1
Mao L, Hou H, Wu S, Zhou Y, Wang J, Yu J, Wu X, Lu Y, Mao L, Bosco MJ, Wang F, Sun Z (2017) TIGIT signalling pathway negatively regulates CD4(+) T-cell responses in systemic lupus erythematosus. Immunology 151(3):280–290. https://doi.org/10.1111/imm.12715
Blessin NC, Simon R, Kluth M, Fischer K, Hube-Magg C, Li W, Makrypidi-Fraune G, Wellge B, Mandelkow T, Debatin NF, Höflmayer D, Lennartz M, Sauter G, Izbicki JR, Minner S, Büscheck F, Uhlig R, Dum D, Krech T, Luebke AM, Wittmer C, Jacobsen F, Burandt EC, Steurer S, Wilczak W, Hinsch A (2019) Patterns of TIGIT expression in lymphatic tissue, inflammation, and cancer. Dis Markers 2019:5160565. https://doi.org/10.1155/2019/5160565
Kizhakeyil, Atish, Ong, Seow Theng et al (2019) Isolation of Human Peripheral Blood In: T-Cell Motility. Methods in Molecular Biology. pp 11-17. doi:https://doi.org/10.1007/978-1-4939-9036-8_2
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102. https://doi.org/10.1093/nar/gkx247
Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, Winter ME, Huynh TV, Sebald SM, Lee SJ, Pan F, Pardoll DM, Cox AL (2016) TGFbeta1-Mediated SMAD3 enhances PD-1 expression on antigen-specific T Cells in cancer. Cancer Discov 6(12):1366–1381. https://doi.org/10.1158/2159-8290.cd-15-1347
Kucan Brlic P, Lenac Rovis T, Cinamon G, Tsukerman P, Mandelboim O, Jonjic S (2019) Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell Mol Immunol 16(1):40–52. https://doi.org/10.1038/s41423-018-0168-y
Liu X, Li M, Wang X, Dang Z, Jiang Y, Wang X, Kong Y, Yang Z (2019) PD-1(+) TIGIT(+) CD8(+) T cells are associated with pathogenesis and progression of patients with hepatitis B virus-related hepatocellular carcinoma. Cancer Immunol Immunother 68(12):2041–2054. https://doi.org/10.1007/s00262-019-02426-5
Wu L, Mao L, Liu JF, Chen L, Yu GT, Yang LL, Wu H, Bu LL, Kulkarni AB, Zhang WF, Sun ZJ (2019) Blockade of TIGIT/CD155 signaling reverses t-cell exhaustion and enhances antitumor capability in head and neck squamous cell carcinoma. Cancer Immunol Res. https://doi.org/10.1158/2326-6066.Cir-18-0725
Sun Y, Luo J, Chen Y, Cui J, Lei Y, Cui Y, Jiang N, Jiang W, Chen L, Chen Y, Kuang Y, Tang K, Ke Z (2020) Combined evaluation of the expression status of CD155 and TIGIT plays an important role in the prognosis of LUAD (lung adenocarcinoma). Int Immunopharmacol 80:106198. https://doi.org/10.1016/j.intimp.2020.106198
Zhou, X., Ding, X., Li, H., Yang, C., Ma, Z., Xu, G., Yang, S., Zhang, D., Xie, X., Xin, L., and Luo, X., Upregulation of TIGIT and PD-1 in Colorectal Cancer with Mismatch-repair Deficiency. Immunol Invest, 2020: p. 1-18.DOI: https://doi.org/10.1080/08820139.2020.1758130
Saleh R, Taha RZ, Toor SM, Sasidharan Nair V, Murshed K, Khawar M, Al-Dhaheri M, Petkar MA, Abu Nada M, Elkord E (2020) Expression of immune checkpoints and T cell exhaustion markers in early and advanced stages of colorectal cancer. Cancer Immunol Immunother. https://doi.org/10.1007/s00262-020-02593-w
Kitsou M, Ayiomamitis GD, Zaravinos A (2020) High expression of immune checkpoints is associated with the TIL load, mutation rate and patient survival in colorectal cancer. Int J Oncol 57(1):237–248. https://doi.org/10.3892/ijo.2020.5062
Kurachi M (2019) CD8(+) T cell exhaustion. Semin Immunopathol 41(3):327–337. https://doi.org/10.1007/s00281-019-00744-5
Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL (2014) The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 26(6):923–937. https://doi.org/10.1016/j.ccell.2014.10.018
Ej W (2011) T cell exhaustion. Nat Immunol 12(6):492–499. https://doi.org/10.1038/ni.2035
Terra M, Oberkampf M, Fayolle C, Rosenbaum P, Guillerey C, Dadaglio G, Leclerc C (2018) Tumor-derived TGFβ alters the ability of plasmacytoid dendritic cells to respond to innate immune signaling. Cancer Res 78(11):3014–3026. https://doi.org/10.1158/0008-5472.Can-17-2719
Batlle E, Massagué J (2019) Transforming growth factor-β signaling in immunity and cancer. Immunity 50(4):924–940. https://doi.org/10.1016/j.immuni.2019.03.024
Yang L, Pang Y, Moses HL (2010) TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 31(6):220–227. https://doi.org/10.1016/j.it.2010.04.002
Acknowledgements
Not applicable.
Funding
This work was supported in part by grants from the Science and Technology Planning Project of Guangdong Province (2017B020227009 to Bo Wei).
Author information
Authors and Affiliations
Contributions
R.L. and B.W. contributed to conceived and designed the experiments; R.L., X.Z., and B.W. were involved in analyzed the data; R.L. and X.Z. contributed to performed the experiments and writing — original draft; B.W. was involved in writing —review and editing and funding acquisition; T.L., D.D., X.Y., and J.S contributed to assisted during the experiment; H.W. was involved in given guidance on research ideas during the research process; and Z.Z., T.C., Y.H., and J.L. contributed to provided clinical samples and information.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethics approval and consent to participate
This study was executed under ethical approvals from The Third Affiliated Hospital of Sun Yat-Sen University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, R., Zhu, X., Lan, T. et al. TIGIT promotes CD8+T cells exhaustion and predicts poor prognosis of colorectal cancer. Cancer Immunol Immunother 70, 2781–2793 (2021). https://doi.org/10.1007/s00262-021-02886-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-021-02886-8