Abstract
Purpose
The PET scanners with long axial field of view (AFOV) having ~ 20 times higher sensitivity than conventional scanners provide new opportunities for enhanced parametric imaging but suffer from the dramatically increased volume and complexity of dynamic data. This study reconstructed a high-quality direct Patlak Ki image from five-frame sinograms without input function by a deep learning framework based on DeepPET to explore the potential of artificial intelligence reducing the acquisition time and the dependence of input function in parametric imaging.
Methods
This study was implemented on a large AFOV PET/CT scanner (Biograph Vision Quadra) and twenty patients were recruited with 18F-fluorodeoxyglucose (18F-FDG) dynamic scans. During training and testing of the proposed deep learning framework, the last five-frame (25 min, 40–65 min post-injection) sinograms were set as input and the reconstructed Patlak Ki images by a nested EM algorithm on the vendor were set as ground truth. To evaluate the image quality of predicted Ki images, mean square error (MSE), peak signal-to-noise ratio (PSNR), and structural similarity index measure (SSIM) were calculated. Meanwhile, a linear regression process was applied between predicted and true Ki means on avid malignant lesions and tumor volume of interests (VOIs).
Results
In the testing phase, the proposed method achieved excellent MSE of less than 0.03%, high SSIM, and PSNR of ~ 0.98 and ~ 38 dB, respectively. Moreover, there was a high correlation (DeepPET: \({R}^{2}\)= 0.73, self-attention DeepPET: \({R}^{2}\)=0.82) between predicted Ki and traditionally reconstructed Patlak Ki means over eleven lesions.
Conclusions
The results show that the deep learning–based method produced high-quality parametric images from small frames of projection data without input function. It has much potential to address the dilemma of the long scan time and dependency on input function that still hamper the clinical translation of dynamic PET.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) plays an important role in molecular imaging, which quantitatively reveals the tissue metabolism and neurochemistry in vivo and has been widely used in humans and animals [1, 2]. In clinical routine, a semi-quantitative index, namely standardized uptake value (SUV), is deemed as the routine interpretation of PET images [3]. However, there are a number of factors, such as the amount of tracer injected and uptake time after injection, that affect the accuracy of image evaluation and diagnosis [4]. In order to enable the absolute quantitative analysis, dynamic PET scan following kinetic modeling has been applied to provide useful physiological parameters of interest such as blood flow and metabolism, providing complementary information for clinical diagnosis and therapy [5, 6]. Conventionally, the approaches to produce parametric images rely on independently reconstructing a series of dynamic images from sinogram data first and then fitting the time activity curves (TACs) through kinetic models, in which the linear graphical analyses, e.g., Patlak/Logan plot and non-linear compartment models, were acknowledged [6]. However, the noise distribution in iteratively reconstructed dynamic images is usually space variant, objective dependent, and difficult to characterize, resulting in inaccurate estimation of parametric images in this indirect approach [7, 8]. The parametric image reconstruction tackles this problem by directly generating parametric images from measured raw sinograms where the noise distribution is a well-defined Poisson distribution [9]. It has the advantages to reduce the noise propagation and influence [10] and therefore improves the quality of the parametric images [11] as well as the physiological quantification [12].
In spite of its promising image results and potential clinical applications, dynamic PET imaging still has been hampered by some limitations: (i) long acquisition time, (ii) accurate measurement of arterial input function (AIF) is needed, and (iii) large data sizes due to number of frames [3,4,5, 13]. In current standard axial field-of-view PET scanners, dynamic whole-body imaging can be achieved by using a protocol of multi-bed multi-pass, due to the small axial field of view (AFOV) and low sensitivity of the PET scanner itself [11, 14, 15]. Usually, a routine dynamic scan starts after tracer injection and lasts for more than 1 h to guarantee adequate photon counts and avoid noisy image results. Such long acquisitions result in inevitable physiological motion [2] and low-throughout PET scan for hospitals [16], as well as discomforting conditions for patients. Moreover, parametric image reconstruction methods require an accurate estimation of AIF, for which an invasive blood sampling through a catheter in the arterial or arterialized venous [17] was performed in early research, but it is invasive and costly for patient and clinical staff. Therefore, several alternative non-invasive methods have been proposed, including the population-based [18], factor analysis [19], image-driven input function (IDIF) [20,21,22], simultaneous estimation [23], and recent machine learning methods [24]. IDIF is the most common non-invasive method and needs to measure the activity distribution of like the ascending or descending aorta, and left ventricle (LV). The characterization of dynamic PET scan implied many data frames required so that large dataset became a tough issue to be overcome [25].
Recent advancements in long axial field-of-view (LAFOV) PET scanners such as the uEXPLORER (United Imaging Shanghai, China), PennPET Explorer, and Biograph Vision Quadra (Siemens Healthineers, Hoffman Estates, IL, USA) provide new possibilities and challenges for parametric imaging [26,27,28,29], making a single-bed single-pass whole-body dynamic scan possible [30, 31]. The large coverage and high sensitivity make it convenient for blood input function measurement, more accurate tracer kinetic modeling, and high-quality parametric imaging [32]. It also enabled the potential use of abbreviated dynamic imaging protocols [33]. Nevertheless, the estimation of AIF is still necessary in current dynamic protocol of either conventional or novel total-body PET scanners; many short time frame data were acquired leading to heavy storage and computation burden for PET system. Therefore, the methodology avoiding AIF measurement and saving storage is urgently needed.
In recent years, deep learning has been applied to many kinds of tasks in medical imaging, such as noise reduction [34,35,36,37], image segmentation [38, 39], and image reconstruction [40,41,42,43,44]. Using convolutional neural networks (CNNs) or generative adversarial networks (GAN) could get comparable and superior results compared to traditional algorithms, along with a fast computation speed.
In particular, researches about using CNNs as regularization term in reconstruction model [40] or directly transforming the PET projection data into image through CNNs [43, 44] draw much attention in deep learning–based PET image reconstruction. The work in [44] proposed a convolutional encoder–decoder (CED) model, i.e., DeepPET, to reconstruct the PET sinogram into a high-quality image successfully without time-consuming back-projection steps. Therefore, motivated by the powerful representation ability and the end-to-end training pattern of DeepPET, we intended to realize fast parametric imaging with not only high image quality but also no need to apply an IDIF. Specifically, we modified the original DeepPET architecture and introduced self-attention modules to reconstruct the dynamic multi-frame sinograms into the direct Patlak plot images. The experiment was implemented on a total-body PET scanner, the Biograph Vision Quadra. Twenty patients were recruited for an 18F-FDG dynamic scan. During training, the acquired sinograms in partial scan time were set as input and the conventionally reconstructed direct Patlak Ki images were as ground truth. As a preliminary study, this work mainly attempted to demonstrate the feasibility of fast parametric reconstruction without input function using deep learning technology.
Materials and methods
Data preparation
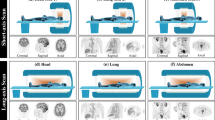
Biograph Vision Quadra is a LAFOV PET scanner with a high sensitivity (176 cps/KBq) [29] which has the potential to accelerate data acquisition [31], and the long axial length (106 cm) covers the critically important organ of interest, enabling parametric imaging of major organs of interest in a single-bed position. Twenty patients were recruited for an 18F-FDG dynamic scan. The local Institutional Review Board approved the study (KEK 2019–02,193), and all patients provided informed consent. As the Patlak graphical method is commonly used to extract the late-time linear phase of a graphical plot, we chose the last 5-frame (25 min, 40–65 min post-injection) sinograms as the training input dataset, in which the sinograms were crystal-based and only random correction was applied by subtracting the delayed sinograms. Meanwhile, they were reconstructed into parametric image by a direct parametric image reconstruction method, the nested EM algorithm (8 iterations, 5 subsets, and 30 nested loops) with an IDIF measured from the descending aorta. A Gaussian filter with 2-mm FWHM was applied to the final reconstructed parametric images [13, 32].
Parametric image reconstruction model
In a dynamic PET scan, measured data \(y\) is following a Poisson distribution as below:
where \({p}_{lj}\) specified the PET system matrix, \(l,j\) is the index of sinogram bins and image pixel, \(m\) means the index of the frame, \(r\) and \(s\) are the measured random noise and scatter events during data acquisition, and \(x\) is the activity map. For conventional parametric imaging reconstruction in this work, linear Patlak modeling was used, which is the most widely used graphical analysis technology for irreversible tracers, like 18F-FDG. In this model, the activity map \(x\) at the time \(t\) can be modeled below [45]:
where \({t}^{*}\) is the equilibrium time, \({K}_{i}\) means the uptake rate of tracer into the irreversibly bound compartment, and the intercept \(DV\) means the initial volume of distribution. \({C}_{p}\) represents the plasma input function obtained by the aforementioned invasive blood sampling or non-invasive approaches.
To estimate the \({K}_{i}\) and \(DV\) directly from projection data, a nested EM algorithm [46] was employed, in which the activity image update and parameter estimation are decoupled into the following steps ((4)–(6)) iteratively[13]:
where the sub-loop or namely nested loop in (5) is embedded in the main loop from (4) to (6). In this work, we targeted the Patlak Ki image.
CNN framework
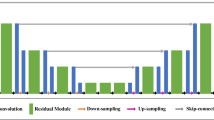
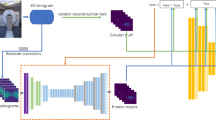
In this study, we constructed a deep CNN network motivated by DeepPET [44]; it employed a CED architecture to reconstruct projection data into an image. Compared to the traditional iterative methods, e.g., maximum-likelihood expectation maximization (MLEM), DeepPET reconstruction was implemented by learning a mapping or an operator from projection into image by plenty of training datasets. Adequately diverse and extensive training data is the key consideration mapping an unseen data input to an unknown ground truth [47]. Therefore, we attempted to construct a DeepPET-like structure for the task of parametric imaging. Figure 1 illustrates the schematic view of the CNN framework used in this study, which consists of encoding, transformation, and decoding parts, as well as a domain transformation module that reconstructs the input sinograms into dynamic images by the ordered subset expectation maximization (OSEM) algorithm, and then introduces the dynamic image information into the decoding part. The final output is the predicted parametric image. Introducing dynamic image information can promote the network to learn richer features such that to improve the generalization ability itself. The multi-frame sinograms were fed into the encoding phase in a way of multi-slice input and the direct reconstructed Patlak Ki images were set as the training label. While, due to the characteristics of parametric reconstruction, we introduced a self-attention module to capture the spatial and temporal features in spatial and channel dimensions. Traditional convolution operations process a local receptive field by customized-size kernels (e.g., 3 × 3, 5 × 5) and lack the ability to capture global information or long-range dependency [48, 49]. Therefore, we replaced the transformation layer between encoder and decoder in origin DeepPET with spatial attention and temporal/channel attention modules to improve the feature representation, as can be seen on the right of Fig. 1.
As shown in Fig. 1, the multi-frame sinograms went through the encoding phase, and then into a latent space representation, and were rebuilt stepwise into a dataset of image domain in the decoding phase. In detail, each layer of the network consists of a convolutional layer (Conv), batch normalization layer (BN), and activation layer (ReLU). At first, sinograms were convoluted with two layers having a kernel size of 7 × 7, and then processed by two down-sampling blocks with five 5 × 5 convolution layers and the other layers having a kernel size of 3 × 3. As mentioned above, we adopted two structures to be the transformation phase; one was the module used in DeepPET, and the other was the self-attention module. In DeepPET, all features in the transformation layer were same size of 16 × 16 and the structure consists of consecutive three, five, and three convolution layers, respectively. As for the details of self-attention module, shown in Fig. 2, it depicts that there are two parallel attention modules connecting the encoder and decoder. After the encoder phase, the feature maps were first fed into a convolution module to get high-level features. Then, the parallel spatial and channel attention modules were employed to obtain the attention matrix representing the spatial dependency within each slice and the interdependency between channel maps, respectively. The following steps were a matrix multiplication between the attention matrix and the high-level features and an element-wise sum between two multiplied matrixes. Prior to the decoder phase, the summed result was fed into a convolution module again. The difference between spatial and channel attention and the calculation details were referenced from a scene segmentation task, namely DANet [50]. Finally, in the decoding phase, the feature maps were decreased by a series of up-sampling and Conv-BN-ReLU blocks, and the last 3 × 3 convolution layer delivered one feature map.
Optimization
In the optimization step, the mean absolute error (MAE) was adopted as a loss function, described below:
where the \({y}_{i}\) means Patlak Ki, the label data, \({x}_{i}\) means sinogram, and \(f\) represents the neural network. To encourage the network to generate the realistic textures and details to label, we introduced a perceptual loss [51], and the expression is as follows:
For the mapping function \(\phi\), we chose a pre-trained VGG16 network [52]. We extracted the second and fifth pooling layer outputs and calculated their MAE loss for consideration of both low-level and high-level features, and details can be seen in Fig. 3. Overall, the total loss function is as follows:
where \(\alpha\) and \(\beta\) are the weighting parameters and control the MAE loss and perceptual loss, respectively. We evaluated the performance of the proposed network trained with different combinations of \(\alpha\) and \(\beta\) to determine the final loss function. The value of \(\beta\) was first set to 0 and \(\alpha\) was chosen from {0.01, 0.1, 1, 10, 50}. After fixing the optimal value for \(\alpha\), \(\beta\) was chosen from {0.01, 0.1, 0.5, 1}. The effect of \(\alpha\) and \(\beta\) values on predicted results is shown in Fig. 4. The mean square error (MSE) between predicted Ki and label Ki was set as the criterion. Finally, the minimum of MSE was found when \(\alpha\) and \(\beta\) were set to 10 and 0.01, respectively.
Training details
During network training and testing, the sinograms and Patlak Ki images were set as input and label data, respectively. The dimension of the original sinogram was 520 × 50 × 5 and the Patlak Ki was 440 × 440. We resized the sinogram and Ki images into 256 × 256 × 5 and 256 × 256 by an interpolation algorithm, respectively. Sixteen patient data were used in training and four in testing. Data pairs of sinograms and direct Patlak Ki images were involved in network training and optimization; the whole workflow can be seen in Fig. 5. The network was implemented using Python3.8 and Pytorch1.8. The training and testing processes were implemented on Ubuntu 20.04. For the optimization of our network, we chose an Adam optimizer with a learning rate of 0.0001; the batch size was set to 48. The epoch number of 300 was chosen, where the model converged. In order to inspect the performance of the CNN-based method on lesion volume, a qualified nuclear medicine physician assisted to identify the 18F-FDG avid malignant lesions and tumor volume of interests (VOIs) using a professional tool (PMOD v.4.1) setting a threshold with 50% of max in SUV images.
Evaluation metrics
To perform a quantitative evaluation of the CNN-based methods, MSE, structural similarity index measure (SSIM), and peak signal-to-noise ratio (PSNR) were calculated.
where \({u}_{x}\) and \({u}_{y}\) are the mean value of network output and label, \({\sigma }_{xy}\) means covariance and \(\sigma\) is variance, and \({c}_{1}\) and \({c}_{2}\) are two constants.
Results
General results
To assess the performance of CNN-based reconstruction, six normal 2D slices representing multiple body parts from four test patient data were shown to prove how capable the CNN output is compared to the conventional reconstructed direct Ki. The comparisons between the DeepPET and proposed self-attention DeepPET were also carried out, as shown in Fig. 6; these two networks were dubbed DeepPET and proposed in all figures and tables, respectively. From top to bottom, Fig. 6 shows the results of DeepPET, self-attention DeepPET, and label Ki images. In order to observe more details, we zoomed in the local region where the red-frame rectangle was in the label Ki image for each result. Overall, as seen in Fig. 6, the CNN-based results can produce similar image structures to the nested EM results. The results of self-attention DeepPET outperformed those of the DeepPET in detail for which the fully 2D convolution operations with a limited receptive field are insufficient to capture global information. Especially in the high activity regions, self-attention DeepPET showed a closer structure profile and value distribution to label Ki images than the original DeepPET framework. Taking the cardiac area, for example, the predicted results of DeepPET seemed to overestimate Ki showing a broader distribution in high Ki region for slice 3, while underestimate Ki for slice 4, compared with the self-attention DeepPET. In slice 2, the DeepPET results even had small structures missing. To quantitatively compare their differences, MSE, PSNR, and SSIM values between predicted results and label images were listed on the lower left of each image slice. The low MSE (< 0.1%), high PSNR (> 30 dB), and SSIM (> 0.9) could be observed in both CNN-based methods. It demonstrated that the CNN-based framework can achieve excellent image quality as traditional direct parametric image reconstruction and the performance of self-attention DeepPET is better than that of the DeepPET framework.
In quantitative analysis, the average MSE, SSIM, and PSNR values were calculated over all the test datasets to evaluate the performance of CNN-based parametric reconstruction, as listed in Table 1; also, a more clear demonstration can be seen in Fig. 7. From Table 1, it is apparent that both CNN-based methods got a small MSE of about 0.03% and a high SSIM of about 0.98, as well as a considerable PSNR. Additionally, between DeepPET and self-attention DeepPET, the MSE value was 0.032% for the former and 0.028% for the latter, and PSNR for the latter is ~ 0.7 dB higher than the former, whereas both predicted images had a quite similar statistical result on SSIM value. Besides that, as one of the concerns in our work, the reconstruction time between the CNN-based methods is shown in Table 2. Here, we regarded the sum of the model loading time (nearly 3.0 s) and image generation time of an individual volume (619 slices per patient) as the reconstruction time. The CNN-based methods took less than 20 s to reconstruct an individual volume. Since self-attention DeepPET replaced the very deep convolution layer in the transformation part of DeepPET with self-attention modules that only involved few convolution and matrix operations, it took less time than DeepPET.
Lesion analysis
According to the lesion segmentation results, we got 11 VOIs from the test dataset and selected six slices to show, as seen in Fig. 8, which shows the results of DeepPET, self-attention DeepPET, and label Ki from top to bottom. The values of related evaluation metrics including MSE, PSNR, and SSIM were listed, and the local regions were zoomed in. Qualitatively and quantitatively, compared to the Patlak Ki images reconstructed by the nested EM algorithm, the predicted Ki could recover the most details of lesion. Moreover, the proposed method using self-attention module produced better result than the DeepPET framework in terms of MSE, PSNR, and SSIM. Similarly, the results of DeepPET had a larger error than that of self-attention DeepPET in case of the same learning rate and epoch number implemented. Like the normal slices, self-attention DeepPET delineated a more accurate profile and value distribution for each lesion than DeepPET. For example, in lesion 1, the DeepPET result overestimated the Ki values on the edge of the lesion, in which the higher Ki means a higher tracer influx rate, while the proposed self-attention framework showed lesion morphological structures closer to the label Ki.
To quantify the performance of the CNN-based method on lesion detection, we calculated the Ki means with standard deviations over a total of 11 lesion VOIs and listed the statistical result in Table 3; the unit of Ki is mL/g/min. Additionally, the histogram and linear regression results are shown in Fig. 9. In the regression plot, the value in the horizontal axis is true Ki and in the vertical axis is predicted Ki from CNN-based methods. No significant difference between CNN-based and traditional reconstructed results was found, which suggested that the CNN-based method is implementable in parametric reconstruction and could produce the same high-quality images as direct reconstructed images. The high correlation between CNN-based and nested EM methods verified this conclusion, and the \({R}^{2}\) was 0.73 for DeepPET and 0.82 for proposed self-attention DeepPET.
The histogram result with standard deviation (a) and the linear regression between predicted Ki and label Ki values (b) based on Table 4
In Fig. 10, we selected four larger lesions to evaluate the correlation between predicted Ki and true Ki. Based on the lesion segmentation masks, we calculated the Ki mean in each slice within each lesion volume. It means that the number of calculated Ki mean is equal to the number of slices a lesion volume covers. A linear regression process was applied between predicted Ki and true Ki. In each subplot, the left presented the sagittal (top), coronal (middle), and transverse (bottom) planes, and the lesions were labeled in red and the right presented the regression result. As seen in Fig. 10, there was a significant correlation between predicted Ki and true Ki found on most lesions. Additionally, the proposed self-attention DeepPET showed better result than the DeepPET.
Moreover, to further investigate the ability of CNN-based parametric imaging in small lesion, three small lesions with diameter less than 10 mm were chosen from the twenty patients’ data. The new training and testing were performed, and the training details were the same as above. As shown in Fig. 9, they are the nodule located in the posterior lower segment of the right liver lobe, the nodule in apical segment of the left lung, and the lymph node in the right axilla, respectively. The diameters of 8.9 mm, 8.0 mm, and 6.0 mm were measured on static PET transverse view, respectively, as seen in Fig. 11a. As can be seen from Fig. 11b, the predicted Ki results indicated that the CNN-based methods could detect the small lesion successfully. With the lesion segmentation mask, we calculated the Ki means within these three lesions for both CNN-based results and label data, as shown in Table 4. From the results, the predicted Ki images preserved the lesion details and had comparable statistic values, which is meaningful for clinical oncology research. Meanwhile, with the self-attention mechanism introduced, the predicted results behaved better than DeepPET.
Discussion
In this work, we estimated the parametric images using a CNN-based method for the total-body PET scanner. Based on previous work such as DeepPET and DPIR-Net [43, 44] that successfully produced static PET images directly from raw projection data, we proposed a deep convolutional encoder–decoder network for dynamic parametric reconstruction.
Apart from the raw projection data, we involved the low-resolution dynamic images in the decoding phase to facilitate the network to converge to optimal results under the circumstance of a limited dataset. In previous research about DeepPET [43, 44], a large number of datasets including simulation phantoms were used. In this study, present results have proven that utilizing sinogram and dynamic images simultaneously could deliver high-quality parametric images for the DeepPET-like network. In addition, we explored the feasibility of CNN-based parametric image generation from static or dynamic PET images only [53, 54]. A 2D U-Net CNN [55] was adopted to map static or dynamic PET images into parametric images. The static PET image (256 × 256, 60–65 min post-injection) and dynamic PET images (256 × 256 × 5, 40–65 min post-injection) were sent into U-Net CNN and trained separately. Compared with the proposed DeepPET-based structures, the parameters except for learning rate remained during the training of U-Net. A learning rate of 0.0002 was chosen for U-Net to achieve the optimal results. There are three examples seen in Fig. 12. The first column shows static PET images, the middle four columns show predicted Ki images from different CNN structures, and the last column shows the Patlak Ki images. The predicted Ki results obtained from U-Net trained with static/dynamic images looked inferior, especially in low Ki regions, compared to DeepPET-based networks trained with sinogram and dynamic images, and in magnified regions, the latter results presented a closer structure and value distribution to label Ki than the former. Figure 13 shows the quantitative results of the test dataset among four different CNN-based methods in terms of MSE, PSNR, and SSIM. The two DeepPET-based methods achieved lower MSE, higher PSNR, and SSIM than U-Net. Meanwhile, training U-Net with dynamic PET images achieved better results than that with static images. This may be because the multi-frame input can be regarded as feature augmentation and introduces time-varying tracer distribution information.
Three slices of static PET, predicted Ki, and Patlak Ki images from left to right. The predicted Ki images are the results obtained from U-Net trained by static images, U-Net trained by dynamic images, and DeepPET and self-attention DeepPET trained by sinograms along with dynamic images, respectively
Around the deep learning–based parametric imaging researches, a CNN module was embedded into reconstruction model, like CT-guided Logan plot [56], in which an iterative reconstruction framework with a deep neural network as a constraint was implemented. This kind of method no longer need the large number of training pairs, but the corresponding anatomical image from CT or MRI. Another approach is mapping indirect Patlak images to direct ones by CNN, whereas prior to CNN was a procedure of indirect Patlak reconstruction [57]. Anyway, for this deep learning–based parametric reconstruction, it is necessary to acquire blood input function non-invasively or invasively. While, the proposed CNN-based method worked well without other anatomical images and blood input function, delivering high-quality Patlak Ki estimations comparable to the standard nested EM algorithm.
Recently, there has been an attractive interest in the total-body PET scanner. The LAFOV offers large anatomical coverage with excellent sensitivity. In previous scanners, the poor sensitivity of less than 1% has long been a challenge that results in poor signal-to-noise ratio (SNR) in images. LAFOV PET approach addressed this dilemma. Up to now, several studies have demonstrated that total-body PET leads to an approximately 40-fold increment in effective sensitivity and enables shorter times [58]. The PET scanner with higher sensitivity than conventional scanner has significant potential to promote the development of fast dynamic scans and lower radiation scans. However, with it comes dramatically increased volume and complexity of dynamic data. With respect to this motivation, studies about parametric imaging of early kinetics of 18F-FDG have demonstrated the feasibility of estimating parametric images using only the first 90 s of post-projection scan data on the total-body PET scanner [25]. In this study, we used the last five frames as data to be reconstructed, which not only saves the data volume but also conforms to the conclusion that Patlak graphical method is commonly used to extract the late-time linear phase of a graphical plot.
All the results demonstrated that the CNN-based method could achieve an equivalent image quality to direct parametric reconstruction results using the nested EM algorithm. It is evidenced suggesting that deep learning methods potentially can generate total-body PET parametric images using data from Biograph Vision Quadra and LAFOV PET scanner. For the dynamic protocols on Biograph Vision Quadra, a total of 62 frames were reconstructed leading to a large data size in excess of one gigabyte, and it takes considerable time to perform both indirect and direct reconstruction. Therefore, a deep learning–based approach may be appropriate and could significantly save the reconstruction time and complexity.
Compared with static PET scans, dynamic PET kinetic analysis reveals the tracer kinetics and has a temporal dimension. In CNN, multi-frame sinograms were fed into a network and the temporal information was convoluted in channel dimension. To account for the characteristics of parametric reconstruction, we replaced the deep convolution layer in the transformation part of DeepPET with two parallel self-attention modules: spatial and channel attention. The results reveal that only using 2D convolution operations would miss the global information of features and lead to insufficient performance on detail structure in the final predicted Ki images. Moreover, in this work, we only targeted the Patlak graphical plot, which is mainly used in an irreversible or nearly irreversible radiotracer, e.g., 18F-FDG. As for the other tracers like gallium-68 (68 Ga)-labeled prostate-specific membrane antigen (68 Ga-PSMA) or the non-linear compartment model, there is also an important issue for further research. Meanwhile, because of the limited dataset at present, we introduced a domain transformation module to constrain the network training process. Despite its simplicity, noise propagates from emission images to final estimated Ki images. With this consideration, a more diverse and extensive simulation or real datasets are required that would make CNN sufficiently represent the possible features of the input domain. Additionally, due to the limitation of current academic computational resources, the proposed networks only tackle the 2-D parametric reconstruction ignoring the spatial information and leading to inconsecutive predicted results across slices [59]. Nevertheless, with the further increasing of AI computational power, the 3-D network combining with the major parts of this work, such as loss function and attention mechanism, may be feasible in the future for the task of 3-D parametric imaging.
Conclusion
The purpose of this study is to demonstrate the feasibility of CNN-based parametric imaging on a total-body PET scanner, Biograph Vision Quadra. We proposed an encoder–decoder framework with spatial and channel self-attention modules to generate high-quality Patlak Ki images from dynamic data. We only used few frames of data but with adequate quality, which owes to the high sensitivity of scanner. The results show that the CNN-based method can produce high-quality parametric images from few projection data. In all test datasets, the proposed method achieves excellent MSE of less than 0.03%, high SSIM, and PSNR of ~ 0.98 and ~ 38 dB, respectively. Meanwhile, no input function used in the CNN-based method and the dramatic reduction of reconstruction time have much potential to make dynamic PET scan more acceptable clinically.
References
Muehllehner G, Karp JS. Positron emission tomography. Phys Med Biol. 2006;51(13);R117–R137. https://doi.org/10.1088/0031-9155/51/13/R08.
Beyer T, Bidaut L, Dickson J, Kachelriess M, Kiessling F, Leitgeb R, et al. What scans we will read: imaging instrumentation trends in clinical oncology. Cancer Imaging. 2020;20:1–38.
Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, Rahmim A. Dynamic whole-body PET parametric imaging: I Concept, acquisition protocol optimization and clinical application. Phys Med Biol. 2013;58:7391–418.
Huang SC. Anatomy of SUV. Nucl Med Biol. 2000;27:643–6.
Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging. 2021;48:21–39.
Carson RE. Tracer kinetic modeling in PET. Positron Emiss Tomogr. 2006;127–59.
Tsoumpas C, Turkheimer FE, Thielemans K. A survey of approaches for direct parametric image reconstruction in emission tomography. Med Phys. 2008;35:3963–71.
Tsoumpas C, Turkheimer FE, Thielemans K. Study of direct and indirect parametric estimation methods of linear models in dynamic positron emission tomography. Med Phys. 2008;35:1299–309.
Karakatsanis NA, Casey ME, Lodge MA, Rahmim A, Zaidi H. Whole-body direct 4D parametric PET imaging employing nested generalized Patlak expectation-maximization reconstruction. Phys Med Biol. 2016;61:5456–85. https://doi.org/10.1088/0031-9155/61/15/5456.
Rahmim A, Tang J, Zaidi H. Four-dimensional (4D) image reconstruction strategies in dynamic PET: beyond conventional independent frame reconstruction. Med Phys. 2009;36:3654–70.
Reader AJ, Verhaeghe J. 4D image reconstruction for emission tomography. Phys Med Biol. 2014;59:R371-418.
Cheng X, Bayer C, Maftei CA, Astner ST, Vaupel P, Ziegler SI, et al. Preclinical evaluation of parametric image reconstruction of [18F]FMISO PET: correlation with ex vivo immunohistochemistry. Phys Med Biol. 2014;59:347–62.
Hu J, Panin V, Smith AM, Spottiswoode B, Shah V, CA von Gall C, et al. Design and implementation of automated clinical whole body parametric PET with continuous bed motion. IEEE Trans Radiat Plasma Med Sci. 2020;4:696–707
Dias AH, Pedersen MF, Danielsen H, Munk OL, Gormsen LC. Clinical feasibility and impact of fully automated multiparametric PET imaging using direct Patlak reconstruction: evaluation of 103 dynamic whole-body 18F-FDG PET/CT scans. Eur J Nucl Med Mol Imaging. 2021;48:837–50.
Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. J Nucl Med. 2018;59:3–12.
Zhang X, Zhou J, Cherry SR, Badawi RD, Qi J. Quantitative image reconstruction for total-body PET imaging using the 2-meter long EXPLORER scanner. Phys Med Biol. 2017;62:2465–85.
Van der Weerdt AP, Klein LJ, Visser CA, Visser FC, Lammertsma AA. Use of arterialised venous instead of arterial blood for measurement of myocardial glucose metabolism during euglycaemic-hyperinsulinaemic clamping. Eur J Nucl Med. 2002;29:663–9.
Chen K, Bandy D, Reiman E, Huang SC, Lawson M, Feng D, et al. Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. J Cereb Blood Flow Metab. 1998;18:716–23.
Wu HM, Hoh CK, Choi Y, Schelbert HR, Hawkins RA, Phelps ME, et al. Factor analysis for extraction of blood time-activity curves in dynamic FDG-PET studies. J Nucl Med. 1995;36:1714–22.
Croteau E, Lavallée É, Labbe SM, Hubert L, Pifferi F, Rousseau JA, et al. Image-derived input function in dynamic human PET/CT: methodology and validation with 11C-acetate and 18F-fluorothioheptadecanoic acid in muscle and 18F-fluorodeoxyglucose in brain. Eur J Nucl Med Mol Imaging. 2010;37:1539–50.
Sari H, Erlandsson K, Law I, Larsson HBW, Ourselin S, Arridge S, et al. Estimation of an image derived input function with MR-defined carotid arteries in FDG-PET human studies using a novel partial volume correction method. J Cereb Blood Flow Metab. 2017;37:1398–409.
Sundar LKS, Muzik O, Rischka L, Hahn A, Rausch I, Lanzenberger R, et al. Towards quantitative [18F]FDG-PET/MRI of the brain: automated MR-driven calculation of an image-derived input function for the non-invasive determination of cerebral glucose metabolic rates. J Cereb Blood Flow Metab. 2019;39:1516–30.
Roccia E, Mikhno A, Ogden RT, Mann JJ, Laine AF, Angelini ED, et al. Quantifying brain [18F]FDG uptake noninvasively by combining medical health records and dynamic PET imaging data. IEEE J Biomed Heal Informatics. 2019;23:2576–82.
Kuttner S, Wickstrm KK, Kalda G, Dorraji SE, Martin-Armas M, Oteiza A, et al. Machine learning derived input-function in a dynamic 18F-FDG PET study of mice. Biomed Phys Eng Express. 2020;6(1):015020. https://doi.org/10.1088/2057-1976/ab6496.
Feng T, Zhao Y, Shi H, Li H, Zhang X, Wang G, et al. Total-body quantitative parametric imaging of early kinetics of 18 F-FDG. J Nucl Med. 2021;62:738–44.
Badawi RD, Shi H, Hu P, Chen S, Xu T, Price PM, et al. First human imaging studies with the explorer total-body PET scanner. J Nucl Med. 2019;60:299–303.
Spencer BA, Berg E, Schmall JP, Omidvari N, Leung EK, Abdelhafez YG, et al. Performance evaluation of the uEXPLORER total-body PET/CT scanner based on NEMA NU 2–2018 with additional tests to characterize PET scanners with a long axial field of view. J Nucl Med. 2021;62:861–70.
Pantel AR, Viswanath V, Daube-Witherspoon ME, Dubroff JG, Muehllehner G, Parma MJ, et al. PennPET explorer: human imaging on a whole-body imager. J Nucl Med. 2020;61:144–51.
Prenosil GA, Sari H, Fürstner M, Afshar-Oromieh A, Shi K, Rominger A, et al. Performance characteristics of the Biograph Vision Quadra PET/CT system with long axial field of view using the NEMA NU 2–2018 Standard. J Nucl Med. 2021;121:261972.
Zhang X, Xie Z, Berg E, Judenhofer MS, Liu W, Xu T, et al. Total-body dynamic reconstruction and parametric imaging on the uEXPLORER. J Nucl Med. 2020;61:285–91.
Alberts I, Hünermund JN, Prenosil G, Mingels C, Bohn KP, Viscione M, et al. Clinical performance of long axial field of view PET/CT: a head-to-head intra-individual comparison of the Biograph Vision Quadra with the Biograph Vision PET/CT. Eur J Nucl Med Mol Imaging. 2021;48:2395–404.
Sari H, Mingels C, Alberts I, Hu J, Buesser D, Shah V, et al. First results on kinetic modelling and parametric imaging of dynamic 18F-FDG datasets from a long axial FOV PET scanner in oncological patients. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-021-05623-6.
Viswanath V, Sari H, Pantel AR, Conti M, Daube-Witherspoon ME, Mingels C, et al. Abbreviated scan protocols to capture 18F-FDG kinetics for long axial FOV PET scanners. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-022-05747-3.
Xu J, Gong E, Pauly J, Zaharchuk G. 200x Low-dose PET reconstruction using deep learning. 2017. http://arxiv.org/abs/1712.04119. Accessed 8 Dec 2017.
Cui J, Gong K, Guo N, Wu C, Kim K, Liu H, et al. Populational and individual information based PET image denoising using conditional unsupervised learning. Phys Med Biol. 2021;66(15). https://doi.org/10.1088/1361-6560/ac108e.
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64(16):165019.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46:2780–9.
Niyas S, Pawan SJ, Kumar MA, Rajan J. Medical image segmentation using 3D convolutional neural networks: a review. 2021. http://arxiv.org/abs/2108.08467. Accessed 9 Apr 2022.
Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, et al. CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans Med Imaging. 2019;38:2281–92.
Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2019;38:675–85.
Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol. 2018;63.
Sun Y, Xu W, Zhang J, Xiong J, Gui G. Super-resolution imaging using convolutional neural networks. Lect Notes Electr Eng. 2020;516:59–66.
Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, et al. DPIR-Net: direct PET image reconstruction based on the Wasserstein Generative Adversarial Network. IEEE Trans Radiat Plasma Med Sci. 2021;5:35–43.
Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62.
Wang G, Qi J. Direct estimation of kinetic parametric images for dynamic PET. Theranostics. 2013;3:802–15.
Scipioni M, Giorgetti A, Della Latta D, Fucci S, Positano V, Landini L, et al. Accelerated PET kinetic maps estimation by analytic fitting method. Comput Biol Med. 2018;99:221–35. https://doi.org/10.1016/j.compbiomed.2018.06.015.
Reader AJ, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci. 2020;5:1–25.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;5999–6009.
Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: Self-Attention Convolutional Neural Network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans Med Imaging. 2020;39:2289–301.
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, et al. Dual attention network for scene segmentation. Proc IEEE Conf Comput Vis Pattern Recognit. (CVPR). 2017;299–307.
Sajjadi MSM, Scholkopf B, Hirsch M. EnhanceNet: single image super-resolution through automated texture synthesis. Proc IEEE Int Conf Comput Vis. (ICCV). 2017;4491–500.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 3rd Int Conf Learn Represent ICLR 2015 - Conf Track Proc. 2015;1–14.
Zaker N, Haddad K, Faghihi R, Arabi H, Zaidi H. Direct inference of Patlak parametric images in whole-body PET/ CT imaging using convolutional neural networks. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-022-05867-w.
Huang Z, Wu Y, Fu F, Meng N, Gu F, Wu Q, et al. Parametric image generation with the uEXPLORER total-body PET/CT system through deep learning. Eur J Nucl Med Mol Imaging. 2022;2482–92.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. MICCAI. Lect Notes Comput Sci. 2015;351. https://doi.org/10.1007/978-3-319-24574-4_28.
Cui J, Gong K, Guo N, Kim K, Liu H, Li Q. CT-guided PET parametric image reconstruction using deep neural network without prior training data. Proc Med Imag: Phys Med Imag. 2019;34.
Xie N, Gong K, Guo N, Qin Z, Wu Z, Liu H, et al. Rapid high-quality PET Patlak parametric image generation based on direct reconstruction and temporal nonlocal neural network. Neuroimage. Elsevier. 2021;240:118380.
Zhang X, Cherry SR, Xie Z, Shi H, Badawi RD, Qi J. Subsecond total-body imaging using ultrasensitive positron emission tomography. Proc Natl Acad Sci U S A. 2020;117:2265–7.
Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62.
Funding
This work was supported in part by the National Key Research and Development Program of China (No: 2020AAA0109502), by the National Natural Science Foundation of China (No: U1809204, 61701436), Zhejiang Provincial Natural Science Foundation of China (No: LY22F010007), by the Talent Program of Zhejiang Province (2021R51004), and by the Key Research and Development Program of Zhejiang Province (No: 2021C03029). Swiss National Science Foundation (SNF No. 188914) and Germaine de Staël Programm of Swiss Academy of Engineering Science (SATW).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. H. Sari and S. Xue acquired and pre-processed the data, Y. Li trained network and analyzed data, J.Hu segmented the lesions, and R. Ma and S. Kandarpa assisted in network training. Y. Li drafted the manuscript and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The local Institutional Review Board approved the study (KEK 2019–02193), and written informed consent was obtained from all patients. The study was performed in accordance with the Declaration of Helsinki.
Conflict of interest
H. Sari is a full-time employee of Siemens Healthineers. The other authors have no conflicts of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Hu, J., Sari, H. et al. A deep neural network for parametric image reconstruction on a large axial field-of-view PET. Eur J Nucl Med Mol Imaging 50, 701–714 (2023). https://doi.org/10.1007/s00259-022-06003-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-06003-4