Abstract
Purpose
In selective internal radiation therapy (SIRT), an accurate total liver segmentation is required for activity prescription and absorbed dose calculation. Our goal was to investigate the feasibility of using automatic liver segmentation based on a convolutional neural network (CNN) for CT imaging in SIRT, and the ability of CNN to reduce inter-observer variability of the segmentation.
Methods
A multi-scale CNN was modified for liver segmentation for SIRT patients. The CNN model was trained with 139 datasets from three liver segmentation challenges and 12 SIRT patient datasets from our hospital. Validation was performed on 13 SIRT datasets and 12 challenge datasets. The model was tested on 40 SIRT datasets. One expert manually delineated the livers and adjusted the liver segmentations from CNN for 40 test SIRT datasets. Another expert performed the same tasks for 20 datasets randomly selected from the 40 SIRT datasets. The CNN segmentations were compared with the manual and adjusted segmentations from the experts. The difference between the manual segmentations was compared with the difference between the adjusted segmentations to investigate the inter-observer variability. Segmentation difference was evaluated through dice similarity coefficient (DSC), volume ratio (RV), mean surface distance (MSD), and Hausdorff distance (HD).
Results
The CNN segmentation achieved a median DSC of 0.94 with the manual segmentation and of 0.98 with the manually corrected CNN segmentation, respectively. The DSC between the adjusted segmentations is 0.98, which is 0.04 higher than the DSC between the manual segmentations.
Conclusion
The CNN model achieved good liver segmentations on CT images of good image quality, with relatively normal liver shapes and low tumor burden. 87.5% of the 40 CNN segmentations only needed slight adjustments for clinical use. However, the trained model failed on SIRT data with low dose or contrast, lesions with large density difference from their surroundings, and abnormal liver position and shape. The abovementioned scenarios were not adequately represented in the training data. Despite this limitation, the current CNN is already a useful clinical tool which improves inter-observer agreement and therefore contributes to the standardization of the dosimetry. A further improvement is expected when the CNN will be trained with more data from SIRT patients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Selective internal radionuclide therapy (SIRT) or radioembolization aims at treating surgically non-resectable primary or metastatic liver tumors. In SIRT, yttrium-90 (90Y) microspheres are injected into the hepatic artery [1], which is the predominant vessel for blood supply to liver tumors [2]. By selecting the appropriate branch of the hepatic artery, the radioactivity can be selectively administered to the targeted tumors, which results in high-dose tumoral irradiation, while keeping the dose to the healthy liver below the tolerance level [3,4,5]. In the pre-treatment study, macro-aggregated albumin particles labeled with technetium-99m (99mTc-MAA) are injected and whole-body planar imaging and SPECT/CT are performed within the hour after injection [6] to estimate the lung shunt fraction and predict the intra-hepatic distribution of 90Y microspheres inside the liver. Following the injection of 90Y microspheres, a post-treatment study is performed to obtain 90Y-PET images (PET/MR or PET/CT). The actual distribution of 90Y microspheres can be determined by these images [7, 8] to verify the treatment.
In both pre- and post-treatment studies, the absorbed dose to the tumor(s) and the normal liver parenchyma is estimated or measured to predict or verify the treatment’s result on the tumor and on the healthy liver [9]. Several approaches are being used for this purpose, including mono-compartment [10] and multi-compartment [11] methods and voxel-based approaches [10]. For all these methods, accurate contours of the liver and tumors are required for dosimetric analysis. Therefore, good liver and tumor delineation plays an important role in these dosimetric activity prescription methods [12].
Manual liver segmentation is tedious and time-consuming and suffers from inter-observer variability [13]. This segmentation variability may increase the variability of the absorbed dose computed by dosimetric methods. In recent years, CNNs have been increasingly used in the medical field for segmenting different organs, such as the liver or kidney [14, 15]. Many grand challenges for automatic liver segmentation, organized by the scientific and technical communities, made available CT or MRI datasets with reference liver delineations on the internet. The participants trained their algorithms on the training datasets and tested them on the test datasets. The test results were evaluated through several metrics and the final ranking of the algorithms was published. The Liver Tumor Segmentation Challenge (LiTS) held in 2017 involves the tasks of liver and tumor segmentation on CT data. The best algorithm, trained on 131 datasets and tested on 70, achieved a dice similarity coefficient (DSC) of 0.96 [16]. Most algorithms adopted U-net-derived architectures and used 2D or so-called 2.5D images due to the long training time and high resource requirements [16]. Chelbus et al. implemented three orthogonal 2D U-net-like CNN models trained with axial, coronal, and sagittal image patches from the MR data of SIRT patients [13]. They reported good liver segmentation results with a mean DSC of 0.95. According to their experimental results, manual correction of the CNN segmentation resulted in a much lower inter-observer variability than taht of manual routine segmentations [13].
The aim of our study is to develop an automatic method of liver segmentation on CTs for SIRT patients, including both contrast-enhanced CTs and non-contrast-enhanced CTs, the latter obtained during PET/CT. This method should be generic, robust, and applicable to CT images with various contrast and irregular liver shapes. For this purpose, we modified a 3D CNN structure named DeepMedic [17] for the task of liver segmentation. Our hypotheses were that the liver segmentations from the CNN can be good enough for clinical application with limited adjustments and will reduce the inter-observer variability of liver segmentation for CT. SIRT patients usually underwent a variety of preceding treatments (e.g., liver resection, chemotherapy, tumor ablation) and have abnormal liver shapes and high disease burden. Therefore, applying the CNN model to liver segmentation of CTs for these patients is more challenging than for those data from public challenges. Besides, automatic liver segmentation using the CNN has the potential of speeding up the segmentation process by minimizing manual interaction from the medical doctors and technologists and standardizing the clinical workflow. Its practical value remains to be investigated in a more clinical context.
Material and methods
Data
In our study, most training datasets were from several international challenges of liver and tumor segmentation for CT, including SLIVER07 challenge (20 datasets), Liver Tumor Segmentation (LiTS17) challenge (131 datasets), and Medical Segmentation Decathlon (MSD) challenge (131 datasets). In the LiTS17 challenge, 20 datasets were discarded due to errors in their image headers. The image in-plane resolution of the challenge datasets ranged from 0.56 to 1.0 mm and the slice thickness ranged from 0.7 to 5.0 mm. During the process of our research, 65 CT datasets from the SIRT patients with liver delineation were collected in our hospital. Their image in-plane resolution ranged from 0.65 to 1.37 mm and their slice thickness ranged from 1.0 to 5.0 mm.
When carrying out the experiments, the datasets from the MSD challenge were found to be identical to those from the LiTS17 challenge, which as far as we could see is not clearly indicated on the respective websites. As a result, 91 of these datasets from the two challenges had twice the weight of the other ones during training. Since the same network trained with uniform weights did not perform better (the difference was within the variation caused by the random CNN initialization), we continued using the original model.
The number of the training, validation, and test data is shown in Table 1. The characteristics of the 38 SIRT patients from whom the 40 test SIRT datasets come and the 25 SIRT patients for training and validation are presented in Table 2. The challenge datasets for training and validation are anonymous and publicly available. Their patient characteristics are not available.
Two clinical experts manually segmented the livers in CT images from the test set. In addition, they also performed manual corrections to the segmentations produced by the CNN. All SIRT datasets for this research were evaluated at the KU Leuven after approval by the Ethics Committee Research of UZ/KU Leuven.
CNN development
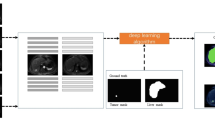
The CNN model used in the paper is a modified version of the dual pathway, 11-layer deep, three-dimensional structure (named DeepMedic) designed for the task of brain lesion segmentation [17]. The network adopts a hybrid scheme between the common patch-wise training (the CNN model only predicts the central voxel of the input image patch) and the so-called dense training on the full image (the network outputs the prediction for all voxels in the input image) [18]. If the input of CNN is the whole 3D image, the dense training setting is mainly constrained by the limited GPU memory. In the patch-wise setting, the same voxels in the overlapping patches are repeatedly involved in the convolutional computations for the prediction of different central voxels, which is inefficient in making full use of the computational power and memory of GPU. The DeepMedic structure overcomes these problems by using image segments with a size larger than the receptive field as the CNN input. This scheme enables the network to output the prediction for multiple voxels in the image segment in one forward pass.
Furthermore, the DeepMedic network introduces the multi-scale processing technique by using parallel convolutional pathways at different resolutions. The contextual information inside the CNN’s receptive field plays an important role in the CNN inference. The more spatial context is incorporated in the inference process, the more comprehensive understanding of the detected object the network can obtain. However, more incorporated contextual information means increasing computation and memory demands if the images with the normal resolution are used. The DeepMedic structure employs a clever way to incorporate both the local and global contexts by adding a low-resolution pathway operating on down-sampled images. In this way, the receptive field of the low-resolution pathway is enlarged greatly at the cost of resolution. But this cost can be compensated by combining the low- and high-resolution pathways, since the local information is preserved in the high-resolution pathway.
Architecture
In the modified CNN structure, a third pathway with lower resolution than the second is added (see Fig. 1). Considering that the liver is much larger than a brain lesion, this third pathway is introduced to help the CNN learn the context information from the whole abdominal region, which is essential for reducing errors. The down-sample rates of the three pathways are 1, 5, and 15, respectively. The kernel size used by the convolutional layers in the three pathways is 3 × 3 × 3. To give more weight on the context information from the second and the third pathways, the number of features is increased in the deeper layers. The outputs of the second and third pathways are up-sampled by 5 and 15, respectively. Then, the outputs of the three pathways are treated as three features which are combined by the next two layers with a 1 × 1 × 1 convolutional kernel. Through one classification layer, the CNN outputs the probability map, where each voxel represents its probability of belonging to the liver.
Overview of the modified DeepMedic structure. The model consists of three pathways (10 convolutional layers in each pathway) followed by a common pathway with two fully connected layers and one classification layer. The input image has the voxel size of 1.4 × 1.4 × 1.4 mm3. The input image segments of three pathways are randomly sampled from the images downsampled by 1, 5, and 15 during the training process. The output segment has the size of 15 × 15 × 15
Training
The CNN model was trained on 3D image segments randomly sampled from the 3D image with a batch size of 16. The model used binary cross entropy as the loss function with the stochastic gradient descent optimizer. The initial learning rate is 0.007 and decreased every 32 epochs. The model quality was evaluated every 8 epochs on the full segmentation of the validation set using the DSC. The training process took 26.75 h using a GPU of NVIDIA P100 with 16 Gb DRAM. The time for the CNN prediction of 40 test SIRT datasets ranged from 11 to 55 s using the GPU. When using a CPU of Intel Xeon E5-2699, the time for the CNN prediction of 40 test SIRT datasets was between 3 and 13 min.
Data preprocessing
The 3D CT images were cropped so that they included the whole abdominal region in each transaxial slice. In an earlier version of the network, we used images containing only the liver. However, we found that, when the images were enlarged such as to contain the entire abdomen in every transaxial slice, the liver segmentation performance of the CNN increased substantially. The cropped images were resampled to 1.4-mm isotropic voxel size so that the CNN could learn about the size of the liver and the surrounding organs. After that, the resampled images were median filtered and normalized by a linear mapping of the Hounsfield units (HU) of the CT images between − 200 and 200 to the range of − 0.2 to 0.2.
Data augmentation
The voxel intensities in the lower-contrast CT images from SIRT patients are often lower than those in the contrast-enhanced CT images from the challenges. To ensure the robustness of the CNN model to variations in the amount of contrast enhancement, a random intensity shift was applied to modify (and usually decrease) the intensity of the training images. This was done by adding a single random value, drawn from a Gaussian distribution with a mean of – 40 HU and a standard deviation of 40 HU, to all the voxel values of a particular training image. Additionally, a random flipping with probability of 0.5 along the x- and y-axis and random elastic deformations were applied.
Data postprocessing
The output of the CNN model was a probability map. It was transformed into a binary mask of liver with the threshold of 0.5. To verify our threshold choice, a simple experiment was done to find an optimal threshold which maximizes the DSCs of the training datasets. After that, the optimal threshold of 0.32 was applied to the validation datasets. The median DSCs of the challenge datasets for validation were around 0.97 for both thresholds and the median DSC of the SIRT datasets for validation using the threshold of 0.32 was 0.6% higher than that using the threshold of 0.5. Because the improvement using the optimal threshold is small and the network output is supposed to be a probability map, we prefer to use a threshold of 0.5, which is the natural choice because it selects the voxels which are more likely to belong to the liver than not. The binary mask was eroded to disconnect the regions with weak connection. Then, the largest connected region in the binary mask was selected while other small islands were not included in the liver volume of interest. The largest connected region was dilated back to its original size and then was taken as the final result of liver segmentation.
Experiments
Comparison between the CNN segmentation, manual segmentation, and adjusted segmentation
To evaluate the liver segmentation quality of our CNN model, an experienced radiographer (WC) was asked to delineate the liver segmentation for 40 test datasets from SIRT patients in our hospital, with his choice of appropriate software available to them in the clinic at the time. These segmentations were performed semi-automatically using Siemens Syngo MMWP Volume software (Siemens Healthcare, Erlangen, Germany). After that, the CNN segmentation and the manual segmentation were compared with each other through several metrics. To analyze the errors of the CNN segmentation and its possibility of being used in clinical application, the expert was also asked to adjust the liver segmentation from CNN for all 40 test SIRT datasets. The adjustment was done using MIM software (MIM Software Inc., Cleveland, OH). When the expert did the adjustment, he was asked to score the CNN segmentation from 1 to 5 with a minimum interval of 0.5. The criteria used by the first expert for scoring are listed in Table 3. By comparing the CNN segmentation and adjusted segmentation, the areas where the errors of CNN segmentation often occur can be found, which is helpful for the further improvement of the CNN model and the selection of training datasets.
Inter-observer variability
To evaluate the influence of the CNN segmentation on the inter-observer variability of liver segmentation, a nuclear medicine physician (CMD) with over 10-year experience in SIRT also provided manual liver segmentations and manual adjustments to the segmentations from the CNN. To shorten the processing time and reduce the expert’s workload, 20 datasets were randomly selected from 40 test SIRT datasets for de novo segmentation and adjustment. For both tasks, MIM software was used. Out of 40 test SIRT datasets, there were 2 SIRT datasets where the CNN model had a very poor liver segmentation (several large parts of the liver were missing). These two segmentations were excluded intentionally when picking out the 20 datasets. After that, the difference between the 20 manual segmentations from two experts was compared with the difference between their adjusted segmentations through several metrics. The criteria used by the second expert for scoring the CNN segmentation are similar to the criteria used by the first expert but more detailed for each single score (see Table 3).
Analysis of manual adjustment
The adjusted segmentations from the two experts were compared with the CNN segmentations for the 20 test SIRT datasets through visual inspection to look into the regions most frequently corrected by the experts.
Evaluation metrics
In our experiment, the difference between segmentations was measured through several metrics calculated in 3D, including dice similarity coefficient (DSC), volume ratio (RV), mean surface distance (MSD), and Hausdorff distance (HD).
Dice similarity coefficient
DSC is used to measure the volume-based similarity between two segmentations [19]. The more overlap the two segmentations have, the larger DSC is. The value of DSC is always between 0 and 1.
Volume ratio
RV computes the ratio of the liver volumes from two segmentations, defined as RV(seg1, seg2) = V1/V2, where V1 and V2 are the volumes of two segmentations.
Mean surface distance and Hausdorff distance
MSD and HD are designed to measure the surface-based difference between two segmentations [20]. MSD computes the average distance between the two segmentation surfaces, whereas HD computes the largest distance between them.
Results
Comparison between the CNN segmentation, manual segmentation, and adjusted segmentation
The median DSC, RV, MSD, and HD between the CNN segmentation and manual segmentation were 0.94, 0.93, 2.1 mm, and 29.2 mm (see Fig. 2). The median DSC, RV, MSD, and HD between the CNN segmentation and adjusted segmentation were 0.98, 0.98, 1.0 mm, and 30.1 mm (see Fig. 2). The median DSC, RV, MSD, and HD between the manual segmentation and adjusted segmentation were 0.95, 1.04, 1.7 mm, and 23.5 mm (see Fig. 2). From the results of DSC and RV, it is evident that the liver volume from the adjusted segmentation agrees more with the liver volume from the CNN than that from the manual segmentation. According to Fig. 2c, the liver surfaces from most adjusted segmentations are more similar to the liver surfaces from the CNN than those from the manual segmentations. The Hausdorff distance between the CNN segmentation and adjusted segmentation is slightly larger than that between the manual segmentation and adjusted segmentation in Fig. 2d. This is explainable because the CNN model has some errors in its liver segmentations, due to the inclusion of other tissues or to the exclusion of some parts of the liver.
The scores assigned to the 40 CNN segmentations are presented in Table 4. According to the scores given by the first expert, 40% (16/40) of liver segmentations from the CNN are very good and can be used for clinical application with slight or no adjustment from the expert. The CNN segmentations of 47.5% (19/40) SIRT datasets require limited adjustment and are then ready for clinical use. There are 12.5% (5/40) poor liver segmentations from the CNN which should not be applied in clinical use. The scores from the expert verify that 87.5% (35/40) of liver segmentations from CNN are good enough for clinical use with some additional adjustment. Some examples of the liver segmentations from CNN with different scores are presented in Fig. 3.
Examples of the CNN segmentation (magenta) compared with the manual segmentation (yellow) and adjusted segmentation (cyan) from the first expert. In the following, “CNN,” “manual,” and “adjust” represent the CNN segmentation, the manual segmentation, and the adjusted segmentation, respectively. a Case where the CNN segmentation was scored 4.5: the DSCs (CNN vs manual, CNN vs adjust, manual vs adjust) were 0.92, 0.99, and 0.92. b Case where the CNN segmentation was scored 3: the DSCs (CNN vs manual, CNN vs adjust, manual vs adjust) were 0.91, 0.95, and 0.93. c Case where the CNN segmentation was scored 1: the DSCs (CNN vs manual, CNN vs adjust, manual vs adjust) were 0.84, 0.89, and 0.92
When looking into the reasons why the CNN model produced poor segmentations on some datasets, we identified the following scenarios which were present in the SIRT datasets but very infrequent in the training datasets: low contrast or low dose, lesions with large density difference from their surroundings, extreme liver position and shape. Some examples of these cases are presented in Fig. 3. In Fig. 3b, one round lesion with low density is seen in the second image and part of the left lobe is located in the extreme left lateral position within the abdomen. The CT shown in Fig. 3c has very low dose and low contrast.
Inter-observer variability
The median DSC, RV, MSD, and HD between the 20 manual segmentations were 0.94, 1.08, 2.0 mm, and 25.0 mm (see Fig. 4). The median DSC, RV, MSD, and HD between the 20 adjusted segmentations were 0.98, 1.01, 0.6 mm, and 21.0 mm (see Fig. 4). According to the results of DSC and RV, the volume difference between the adjusted segmentations was much smaller than that between the manual segmentations. Similarly, the mean surface distance between the two adjusted liver contours was reduced to a large extent compared with the manual contours from the two experts. The relative decrease of HD was not as large as that of the other three metrics after adjustment. It is mainly because a large discrepancy of delineation between two experts exists in the regions of vessels or ligaments, where the delineation criteria are not clearly defined. This discrepancy cannot be eliminated using the CNN segmentation as a baseline.
Besides, the scores of the 20 test SIRT datasets from the two experts are presented in Fig. 5. The score difference remains within 0.5 for 16 patients. However, a large score difference of over 0.5 exists for the other 4 patients, although the two experts used similar scoring criteria. It is caused by the subjectivity existing in the criteria and in the judgment from the experts.
Analysis of manual adjustment
The frequency of every corrected region for each expert was recorded for the 20 test SIRT datasets (see Fig. 6). From the figure, it is evident that the inferior vena cava (IVC) is the region corrected by both experts most frequently. In the training datasets, a part of IVC adjacent to the liver was included in the liver delineation in some datasets while not in the other datasets. As a result, the CNN segmentation appears random and irregular in the IVC region. For the portal vein, expert 1 tended to include it in the liver delineation, while expert 2 agreed more with the CNN segmentation to exclude the portal vein from the liver segmentation. Besides, CNN segmentation errors in some regions required additional adjustment from the experts. For example, the left tip of the liver is the third most frequently adjusted region since shape abnormality often occurs in this region. The lesions with large density difference from their surroundings are the fourth most frequently corrected regions. The regions between the liver and the surrounding organs (e.g., heart, stomach, duodenum, colon) are frequently corrected due to CNN segmentation errors caused by their small density difference in low-contrast CTs.
Time used for manual segmentation and adjustment
The time spent on manual segmentation and adjustment of the CNN segmentation for expert 1 (40 test SIRT datasets) and expert 2 (20 test SIRT datasets) is presented in Fig. 7. For expert 1, the time for manual segmentation is always within 5 min, which is shorter than the time for adjustment. The time for adjustment ranges from 3.17 to 32.75 min with a median of 9.18 min. Expert 2 spent much less time on adjustment than on manual segmentation. The time for manual segmentation ranges from 22.32 to 64.82 min with a median of 28.53 min and the time for adjustment ranges from 2.15 to 20.45 min with a median of 6.72 min.
Discussion
Our modified CNN model mainly trained on public datasets of liver cancer achieved good results on the SIRT CT images with good image quality, relatively normal liver shapes, and low disease burden. The CNN segmentation achieved a median DSC of 0.94 with the manual segmentation and of 0.98 with the adjusted segmentation, respectively. Only 2 out of the 40 test SIRT datasets had a RV outside the range from 0.9 to 1.1 between the CNN segmentation and adjusted segmentation. It indicates that the difference of injected activity caused by CNN segmentation errors is within 10% for 95% of the 40 test SIRT datasets when using the mono-compartment method. 87.5% (35/40) of automatic liver segmentations from CNN are eligible for clinical use with limited adjustment from the expert in the judgment of 2 experienced liver delineators. This implies a promising future for applying deep learning to the traditional liver segmentation task in the clinical routine of SIRT.
However, the current CNN model may fail in the following cases: poor image quality (low-dose or low-contrast CT), lesions with large density difference from their surroundings, and extreme liver position and shape. Each of the above cases has many different variations. A small density difference can occur among most organs in the abdomen or between the liver and a neighboring organ. The lesions in the liver may appear homogeneous and round with very low density, large and diffuse, or with high vascularity. The liver can be extremely large or compressed in the sagittal plane and the left lobe may occur in the very left position of the abdomen. These variations and their combinations make them difficult to be defined and quantified. Through visual inspection, it was found that the above three cases and their variations occurred in the training datasets (mainly the challenge datasets) with low frequency. Besides, the DSCs of the challenge datasets for validation and the SIRT datasets for validation are 0.97 and 0.94 respectively when comparing the CNN segmentation with the manual segmentation from the radiographer. This further proves that some discrepancy exists between the SIRT datasets and the challenge datasets.
By using the CNN segmentation as a baseline for adjustment, the inter-observer variability was reduced to a large extent compared with starting the manual liver segmentation from scratch. It can help reduce the random and subjective errors in absorbed dose calculation introduced by inconsistent liver volumes and contours from different observers. The ratio of RVs outside the range from 0.9 to 1.1 is 0% (0/20) between the adjusted segmentations from the two experts and 20% (4/20) between their manual segmentations. This implies that the adjusted segmentations keep the difference of injected activity caused by the inter-observer variability of the liver segmentation within 10% if the mono-compartment model is used.
Currently, the corrections from the experts mainly happen in the regions including the vessels (IVC, portal vein, sushepatic vein) and in the regions where the CNN model has a poor delineation. Since there are no criteria defining the way of including or excluding these vessels for the liver delineation, the experts make the decision based on their own experience and background (e.g., radiographer vs nuclear medicine physician). On CT without intravenous contrast enhancement, the IVC is difficult to discern from normal liver tissue, contrary to contrast-enhanced CT. This further increases the difficulty of liver delineation near the vessel regions. Although the contour difference caused by these vessels does not have an evident influence on dosimetry, it decreases the consistency of liver delineation. This can be solved by proposing a criterion for vessels’ exclusion agreed upon by the physicians.
It is remarkable that expert 1 needed more time for adjusting a segmentation than for drawing one from scratch, whereas the opposite was the case for expert 2. For this experiment, we allowed the experts to use the segmentation software of their choice. Expert 1 is a radiographer who is doing clinical segmentations since many years, and he did the manual segmentations with the software which he uses also clinically: the Siemens Syngo MMWP Volume software. However, he found that this software is less suited for correcting existing segmentations and therefore used the MIM software for that, which he had not used before. Expert 2 is a nuclear medicine physician, who is not used to providing manual organ segmentations. He chose the MIM software for both tasks. Consequently, we attribute this discrepancy to the many years of experience of expert 1 with the Siemens software. We cannot claim that correcting a segmentation is always faster than providing one from scratch, as these times depend heavily on the software used for that task and the talents of the operator for using that software efficiently. However, we would argue that when the software is optimized for the task, a skilled operator should be faster at correcting a fairly good segmentation than at creating a new one, since the former task is simpler in principle.
We will introduce our CNN-based correction tool into the SIRT workflow and possibly other clinical workflows involving liver segmentation. Once the experts get used to this tool, shorter time may be spent on liver delineation with better accuracy. As a result, it will become easier for the experts to provide a large amount of liver contours eligible for training the CNN model, further improving the CNN performance. As assistance from the current CNN already improved the inter-observer agreement, we believe this CNN-assisted liver segmentation will contribute to improving and standardizing the liver contours used in SIRT planning and help nuclear medicine physicians to obtain more precise dose predictions and better treatment verification.
In summary, we believe that the performance of our current CNN makes it a useful tool for clinical SIRT image analysis. In addition, further improvements are anticipated by including more representative SIRT work-up datasets for training, which will reduce the discrepancy between the characteristics of the training images and those of the typical SIRT images. Besides, the potential of the CNN model to reduce the segmentation time remains to be fully studied in the future. A CNN model for MRI liver segmentation is planned to be developed in the future. The reduction of inter-observer variability for MR is also anticipated.
Conclusion
The CNN-based automatic liver segmentation achieved good results for CT images from SIRT patients, who usually have abnormal liver shapes and high tumor burden. 87.5% of the 40 CNN liver segmentations were considered eligible for clinical use with limited adjustment from the expert. The inter-observer variability of liver segmentation was reduced considerably when the CNN segmentation was used as a baseline for manual adjustments. As a result, the CNN-based automatic liver segmentation is anticipated to become a valuable tool for clinical routine in the near future.
References
Dezarn WA, Cessna JT, DeWerd LA, et al. Recommendations of the American Association of Physicists in Medicine on dosimetry, imaging, and quality assurance procedures for 90Y microsphere brachytherapy in the treatment of hepatic malignancies. Med Phys. 2011;38:4824–45.
Breedis C, Young G. The blood supply of neoplasms in the liver. Am J Pathol. 1954;30:969–77.
Gray BN, Burton MA, Kelleher D, Klemp P, Matz L. Tolerance of the liver to the effects of yttrium-90 radiation. Int J Radiat Oncol Biol Phys. 1990;18:619–23.
Cremonesi M, Chiesa C, Strigari L, Ferrari M, Botta F, Guerriero F, et al. Radioembolization of hepatic lesions from a radiobiology and dosimetric perspective. Front Oncol. 2014;4:210. https://doi.org/10.3389/fonc.2014.00210.
Kennedy AS, Nutting C, Coldwell D, Gaiser J, Drachenberg C. Pathologic response and microdosimetry of (90)Y microspheres in man: review of four explanted whole livers. Int J Radiat Oncol Biol Phys. 2004;60:1552–63.
De Gersem R, Maleux G, Vanbilloen H, et al. Influence of time delay on the estimated lung shunt fraction on 99mTc-labeled MAA scintigraphy for 90Y microsphere treatment planning. Clin Nucl Med. 2013;38:940–2.
Maughan NM, Eldib M, Faul D, et al. Multi institutional quantitative phantom study of Yttrium-90 PET in PET/MRI: the MR-QUEST study. EJNMMI Phys. 2018;5:7.
Wright CL, Binzel K, Zhang J, Wuthrick EJ, Knopp MV. Clinical feasibility of 90Y digital PET/CT for imaging microsphere biodistribution following radioembolization. Eur J Nucl Med Mol Imaging. 2017;44:1194–7.
Garin E, Lenoir L, Rolland Y, et al. Dosimetry based on 99mTc-macroaggregated albumin SPECT/CT accurately predicts tumor response and survival in hepatocellular carcinoma patients treated with 90Y-loaded glass microspheres: preliminary results. J Nucl Med. 2012;53:255–63.
Bastiaannet R, Kappadath SC, Kunnen B, Braat AJAT, Lam MGEH, de Jong HWAM. The physics of radioembolization. EJNMMI Phys. 2018;5:22.
Ho S, Lau WY, Leung TW, et al. Partition model for estimating radiation doses from yttrium-90 microspheres in treating hepatic tumours. Eur J Nucl Med. 1996;23:947–52.
Jafargholi Rangraz E, Coudyzer W, Maleux G, Baete K, Deroose CM, Nuyts J. Multi-modal image analysis for semi-automatic segmentation of the total liver and liver arterial perfusion territories for radioembolization. EJNMMI Res. 2019;9:19. https://doi.org/10.1186/s13550-019-0485-x.
Chlebus G, Meine H, Thoduka S, et al. Reducing inter-observer variability and interaction time of MR liver volumetry by combining automatic CNN-based liver segmentation and manual corrections. PLoS One. 2019;14:e0217228.
Wang K, Mamidipalli A, Retson T, et al. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol: Artif Intell. 2019;1:180022.
Sharma K, Rupprecht C, Caroli A, et al. Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Sci Rep. 2017;7:1–10.
Bilic P, Christ P, Vorontsov E, Chlebus G, Chen H, Dou Q, et al. The liver tumor segmentation benchmark (LiTS). 2019. arXiv:1901.04056.
Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78.
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, 2015.
Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index1. Acad Radiol. 2004;11:178–89.
Heimann T, van Ginneken B, Styner M, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging. 2009;28:1251–65.
Funding
This project is funded by the H2020-ITN (MSCA 764458) project Hybrid and by the Research Foundation Flanders (FWO) project G082418N.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Georg Schramm is supported by NIH Grant 1P41EB017183-01A1 CAI2R TRDP #3. David Robben is employed by icometrix, Leuven, Belgium. Christophe M. Deroose is a Senior Clinical Investigator at the Research Foundation Flanders (FWO). Mark Gooding is employed by Mirada Medical Ltd, Oxford, UK, a medical software company. The department of nuclear medicine at KU Leuven receives support from GE for image reconstruction research. No other potential conflicts of interest relevant to this article exist.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee Research of UZ/KU Leuven and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Rights and permissions
About this article
Cite this article
Tang, X., Jafargholi Rangraz, E., Coudyzer, W. et al. Whole liver segmentation based on deep learning and manual adjustment for clinical use in SIRT. Eur J Nucl Med Mol Imaging 47, 2742–2752 (2020). https://doi.org/10.1007/s00259-020-04800-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04800-3