Abstract
Purpose
To assess the predictive power of pre-therapy 18F-FDG PET/CT-based radiomic features for epidermal growth factor receptor (EGFR) mutation status in non-small cell lung cancer.
Methods
Two hundred and forty-eight lung cancer patients underwent pre-therapy diagnostic 18F-FDG PET/CT scans and were tested for genetic mutations. The LIFEx package was used to extract 47 PET and 45 CT radiomic features reflecting tumor heterogeneity and phenotype. The least absolute shrinkage and selection operator (LASSO) algorithm was used to select radiomic features and develop a radiomics signature. We compared the predictive performance of models established by radiomics signature, clinical variables, and their combinations using receiver operating curves (ROCs). In addition, a nomogram based on the radiomics signature score (rad-score) and clinical variables was developed.
Results
The patients were divided into a training set (n = 175) and a validation set (n = 73). Ten radiomic features were selected to build the radiomics signature model. The model showed a significant ability to discriminate between EGFR mutation and EGFR wild type, with area under the ROC curve (AUC) equal to 0.79 in the training set, and 0.85 in the validation set, compared with 0.75 and 0.69 for the clinical model. When clinical variables and radiomics signature were combined, the AUC increased to 0.86 (95% CI [0.80–0.91]) in the training set and 0.87 (95% CI [0.79–0.95]) in the validation set, thus showing better performance in the prediction of EGFR mutations.
Conclusion
The PET/CT-based radiomic features showed good performance in predicting EGFR mutation in non-small cell lung cancer, providing a useful method for the choice of targeted therapy in a clinical setting.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is a malignant tumor with the highest morbidity and mortality in the world. Non-small cell lung cancer (NSCLC) accounts for more than 85% of lung cancer patients, adenocarcinoma being the most common pathological type [1]. In recent years, with the development of molecular biology, targeted therapy has gained increasing attention, in addition to more traditional treatment methods. In particular, epidermal growth factor receptor tyrosine kinase inhibitors (TKIs) have been widely used in the treatment of NSCLC and were shown to effectively prolong the survival of lung adenocarcinoma patients with epidermal growth factor receptor (EGFR) gene mutations [2]. Accordingly, it is important to determine whether a patient has EGFR mutations prior to targeted therapy. Currently, tissue or cytological specimens are commonly used for genetic testing, which are mostly invasive, making them unavailable to many NSCLC patients due to technical difficulties, cost, patient health status, tumor heterogeneity, and other reasons [3]. Therefore, it is necessary to develop noninvasive, rapid, and simple methods for gene mutation detection.
Radiomics is an emerging field in which a large number of objective and quantitative imaging features are explored to select those most significantly associated with clinical, pathological, molecular, and genetic characteristics, so as to improve the diagnostic and prognostic accuracy and the evaluation of therapeutic efficacy [4]. At present, radiomics research for predicting lung cancer gene mutations is mostly based on CT images [5,6,7]. However, there are few studies on 18F-FDG PET/CT in this field, often based on visual analysis or conventional quantitative indicators such as SUV values, leading to unsatisfactory predictive ability [8,9,10]. In this paper, 18F-FDG PET/CT radiomics, which extracts the texture features of PET and CT images from the same volume of interest (VOI), and combines the texture features of metabolism and structure, is applied to develop a radiomics signature for the prediction of EGFR mutation status. Therefore, the aim of this study was to build and validate a PET/CT radiomics-based model to predict EGFR mutation status in patients with lung adenocarcinoma.
Materials and methods
Patient selection
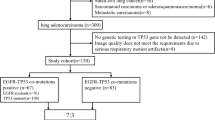
This was a retrospective single-center study. We included consecutive patients with histologically proven lung adenocarcinoma, who had undergone pre-therapy 18F-FDG PET/CT scan between January 2015 and January 2019 in our department, and satisfying the following inclusion criteria: (1) lung adenocarcinoma confirmed by pathology, (2) 18F-FDG PET/CT examination performed within 1 month before surgery or biopsy, (3) no anti-tumor treatment received before PET/CT examination, (4) no history of other malignant tumors, (5) single lesion with a maximum diameter of > 1 cm to avoid partial volume effects. The exclusion criteria were (1) no genetic testing for EGFR or other mutations and (2) pure ground-glass nodule (pGGN) without FDG metabolism.
According to the above criteria, 248 lung cancer patients were included, including 135 males and 113 females, with an average age of 62.43 ± 9.19 years (range 29–84). The sample included 133 EGFR mutant and 115 EGFR wild-type cases. The patients were randomly divided into two sets, in the ratio of 7:3, with 175 cases assigned to the training set and 73 to the validation set. The study was approved for retrospective analysis by the institutional ethics committee and the requirement of informed consent was waived. The patient selection process is shown in Fig. 1.
EGFR mutation detection
EGFR mutation detection was performed on histological specimens obtained by surgical resection or puncture. EGFR mutation detection was conducted by real-time fluorescence PCR method to detect mutations of exons 18–21 of the EGFR gene. The Roche Cobas DNA sample preparation and EGFR mutation detection kit were used, with the specific steps carried out in accordance with the kit instructions. PCR analysis was performed using Roche Cobas Z480. If any exon mutation in EGFR exons 18–21 was detected, the tumor was considered EGFR mutant. Otherwise, the tumor was classified as EGFR wild type.
Image acquisition
The imaging acquisition protocol was established following the Image Biomarker Standardisation Initiative (IBSI) reporting guidelines [11]. All the details of the procedures are provided in the electronic supplementary material. 18F-FDG PET/CT (GEMINI GXL16, PHILIPS, The Netherlands) examination was performed within 1 month before treatment. Briefly, 18F-FDG PET/CT images were acquired 60 ± 5 min after 18F-FDG injection in accordance with the European Association of Nuclear Medicine (EANM) guidelines, version 1.0 (version 2.0 from February 2015) [12]. A 3D LOR reconstruction algorithm was used to reconstruct the PET images. The attenuation correction of PET images was carried out with CT data, and the corrected PET images were fused with CT images.
Radiomic feature extraction
The LIFEx package (version 4.00, http://www.lifexsoft.org) was used to extract the texture features of PET/CT images of lesions in the same VOI [13]. The PET/CT image of the patient in the DICOM format was imported into the software. Two experienced PET/CT diagnostic physicians semi-automatically delineated the region of interest of the target lesion using a threshold of 40% of the maximum standardized uptake value (SUVmax) [14]. Then, the software program automatically calculates and extracts 47 PET radiomic features and 45 CT radiomic features, which are provided in the Supplementary Material.
Radiomic feature selection and model establishment
In our study, the number of radiomic features was large, while the number of cases was relatively small. In order to avoid overfitting of the model, we first selected the features with significant differences between EGFR mutant and wild type in the training set using the Mann–Whitney U test, obtaining a total of 53 features with p value < 0.05. Then, the least absolute shrinkage and selection operator (LASSO) algorithm was used to select the optimal predictive features among the 53 selected in the training set [15]. The LASSO algorithm adds a L1 regularization term to a least square algorithm to avoid overfitting. A prediction model was established by logistic regression, and the radiomics signature score (rad-score) for each patient was calculated based on the selected discriminating radiomic features. The model performance was tested in the validation set. The receiver operating characteristic (ROC) curve and the area under the curve (AUC) were used to evaluate the model performance in the training and validation sets.
Rad-score and clinical information variables were combined to establish a multivariate logistic regression model (the complex model) and to develop a nomogram, which can intuitively display the prediction result for each patient. Moreover, calibration curves were plotted to improve the nomogram’s prediction accuracy, which was verified by the Hosmer–Lemeshow test.
Statistical analysis
Statistical analyses were performed with the R software (version 3.4.3, http://www.R-project.org), the EmpowerStats software (www.empowerstats.com, X&Y solutions, Inc. Boston MA), and SPSS Statistics for Windows (version 21.0, IBM, Armonk, NY, USA). The R package “glmnet” was used to perform LASSO binary logistic regression analysis; the “rms” package, to create the nomogram; and the “pROC” package, to analyze ROC curves. Independent sample t tests or Mann–Whitney U tests were used to compare continuous variables, while the chi-squared test was used for the comparison of categorical variables. A p value < 0.05 was considered statistically significant.
Results
Clinical characteristics of patients
The baseline characteristics of the patients in the training and validation sets are summarized in Table 1. There were no statistical differences in age (p = 0.3849), gender (p = 0.4397), smoking history (p = 0.5652), pathological stage (p = 0.8117), or carcinoembryonic antigen (CEA) level (p = 0.2721) between the training and the validation sets. Gender and smoking history were significantly different between the EGFR mutation and EGFR wild-type groups in both the training (both p < 0.0001) and the validation set (p < 0.0001 and p = 0.0003, respectively). EGFR mutations were more common in women and never smokers, while the EGFR wild type status was more common in men and smokers in both sets. Age, CEA level, and pathological stage were not significantly different between the EGFR mutation and EGFR wild-type groups in either the training or the validation set. Univariate analyses revealed that gender (OR = 3.9, 95% CI [2.1–7.3], p < 0.001) and smoking history (OR = 0.1, 95% CI [0.1–0.3], p < 0.001) were significant predictors of EGFR mutation.
Feature extraction and selection
The LASSO algorithm and 10-fold cross-validation were used to extract the optimal subset of radiomic features. Eventually, 10 radiomic features were extracted to build the radiomics signature based on the 175 patients included in the training set (Fig. 2). The 10 selected radiomic features were SUVpeaksphere, SHAPE_Compacity, Grey-Level Co-occurrence Matrix_Energy (GLCM_Energy), Grey-Level Zone Length Matrix_Short-Zone Emphasis (GLZLM_SZE), Grey-Level Zone Length Matrix_Zone Percentage (GLZLM_ZP), Maximum, SHAPE_Sphericity, Grey-Level Run Length Matrix_High Grey-level Run Emphasis (GLRLM_HGRE), Neighborhood Grey-Level Different Matrix_Busyness (NGLDM_Busyness), and Grey-Level Zone Length Matrix_Zone Length Non-Uniformity (GLZLM_ZLNU). Among the above features, the first 5 are PET features and the last 5 are CT features. A radiomics signature score (rad-score) for each patient was calculated using the following formula:
The LASSO algorithm and 10-fold cross-validation were used to extract the optimal subset of radiomic features. a Optimal feature selection according to AUC value. When the value ln (λ) increased to − 4.071, the AUC reached the peak corresponding to the optimal number of radiomic features. b LASSO coefficient profiles of the 53 radiomic features. The vertical line was drawn at the value selected by 10-fold cross-validation, where the optimal λ resulted in 10 nonzero coefficients
Rad-score = – 8.60837 + 0.08830 × SUVpeaksphere + 1.95669 × SHAPE_Compacity + 10.20396 × GLCM_Energy – 2.81476 × GLZLM_SZE – 1.35737 × GLZLM_ZP – 0.00724 × Maximum + 7.25372 × SHAPE_Sphericity + 0.00016 × GLRLM_HGRE – 24.53804 × NGLDM_Busyness – 0.00687 × GLZLM_ZLNU
The median and the interquartile range for the 10 selected radiomic features and the calculated rad-score are shown in Table 2. The rad-score and the 10 selected features were significantly different between the EGFR mutant and the EGFR wild-type groups in both the training and the validation sets (p < 0.05). Specifically, lung adenocarcinomas with EGFR mutations had higher rad-score than those with wild-type EGFR in both the training (rad-score = 0.908 vs − 0.154) and validation sets (rad-score = 0.710 vs − 0.257). The rad-score for each patient in the two sets is displayed in the form of a bar graph in Fig. 3a and b.
Performance of the radiomics signature and clinical features
To evaluate the performance of the radiomic features in predicting EGFR mutation status, we compared models based on the radiomics signature, clinical variables (gender, smoking history), and their combinations (Fig. 4). The radiomics model had good predictive performance, and its AUCs in differentiating EGFR mutation status were 0.79 (95% CI [0.73–0.86]) in the training set and 0.85 (95% CI [0.76–0.94]) in the validation set. The clinical model had AUCs of 0.75 (95% CI [0.68–0.82]) and 0.69 (95% CI [0.58–0.81]) in the training and validation set, respectively. The AUC values of these two models were not significantly different in the training set (p = 0.3212) and marginally significant in the validation set (p = 0.0529). The complex model, based on the radiomics signature score (rad-score) and clinical variables, had higher AUCs, namely 0.86 (95% CI [0.80–0.91]) and 0.87 (95% CI [0.79–0.95]) in the training and validation set, respectively. There were statistically significant differences in AUC between the complex and the clinical model in both the training and the validation sets (both p < 0.0001). In the training set, the AUCs of the complex and the radiomics models were also significantly different (p = 0.0194). However, in the validation set, there was no statistically significant difference in AUC between the radiomics and the complex model (p = 0.6974). The predictive abilities of the three models, including sensitivity, specificity, positive predictive value, negative predictive value, and accuracy, are shown in Table 3.
Individualized nomogram construction and validation
Considering that the complex model based on both rad-score and clinical variables (gender, smoking history) had better ability to predict gene mutation, we created a nomogram representing the individualized predictions based on the training set, which can visualize the prediction result and the proportion of each factor (Fig. 5a). The calibration curve of the nomogram is presented in Fig. 5b and c and shows good agreement between the predicted and observed values in the training and validation sets. The Hosmer–Lemeshow test was not significant in either the training (χ2 = 3.568, p = 0.894) or the validation set (χ2 = 11.196, p = 0.191), demonstrating a good fit.
Development and performance of a nomogram. a Nomogram based on rad-score and clinical factors (gender and smoking history). Calibration curves of the nomogram in the training (b) and validation sets (c). The horizontal axis is the predicted incidence of the EGFR mutation. The vertical axis is the observed incidence of the EGFR mutation. The red line on the diagonal is the reference line, indicating that the predicted value is equal to the actual value. The black line is the calibration curve, and the yellow area on both sides represents the 95% CI
Discussion
Given the well-established role of tyrosine kinase inhibitors in the treatment of lung adenocarcinoma with EGFR mutations, EGFR genotyping is crucial for targeted treatment. The traditional identification of the EGFR genotype is invasive and likely limited by several factors, such as patient refusal to undergo an invasive biopsy, the location or size of the tumor, difficulties in biopsy sampling, lack of available tissue, poor health status of the patient, tumor heterogeneity in space and time [16,17,18]. In this study, we established a model based on 10 radiomic features derived from pre-therapy 18F-FDG PET/CT images to predict EGFR mutations in lung adenocarcinoma patients, which showed good predictive performance.
Among the 10 selected features in our study, SUVpeak is a basic PET feature, while Maximum is a basic CT feature. In both the training and validation sets, SUVpeak and Maximum were lower in the EGFR mutation group than in the EGFR wild group. SUVpeak reflects the mean SUV in a sphere with a volume of 1 mL, located so that the average value in the VOI is at maximum and is less affected by noise than SUVmax. Kim et al. showed that the values of all the metabolic and volumetric 18F-FDG PET/CT parameters were significantly lower in EGFR mutant than in EGFR wild type lung adenocarcinomas and that SUVpeak was an independent prognostic factor. EGFR mutations were associated with low metabolic activity of localized lung adenocarcinoma on 18F-FDG PET/CT [19]. Some scholars believe that EGFR tyrosine kinase can promote the glucose uptake of tumor cells: tumor cells with high active glucose metabolism have sufficient glucose uptake, so they have less demand for EGFR tyrosine kinase than tumor cells with low metabolism. Therefore, the EGFR mutation rate of non-small cell lung cancer patients with high SUVmax is lower [20]. Our findings are consistent with such results.
In our study, the Maximum feature was significantly lower in the EGFR mutation group than in the wild-type group, which may be related to the fact that the EGFR mutation group had more ground-glass opacity (GGO) components and less solid and pulmonary fibrosis components [21,22,23]. Sacconi et al. found that the mean, standard deviation, and skewness of CT quantitative parameters were associated with EGFR mutations in advanced lung adenocarcinoma and that the mean CT values of the EGFR mutation group were lower than those of the wild-type group [24], in agreement with our results. However, in our study, the mean CT value showed marginal significance between the EGFR mutation group and the EGFR wild-type group. Finally, Maximum, which showed a significant difference, was selected as the modelling index by LASSO regression, which might be related to differences in the number of enrolled patients between the two studies.
SHAPE_Sphericity describes how spherical a volume of interest is and is equal to 1 for a perfect sphere. SHAPE_Compacity reflects how compact the volume of interest is. According to Desseroit et al., the 3D shape features of lesions, such as irregularity and sphericity, extracted from PET and low-dose CT images, are highly repeatable, so that they can provide morphological indexes of lesions with good accuracy [25]. In our study, lung adenocarcinomas of the EGFR mutant type had higher sphericity and were more compact than those with wild-type EGFR.
The remaining 6 texture features, including three PET metabolic texture features (GLCM_Energy, GLZLM_SZE, GLZLM_ZP) and three CT texture features (GLRLM_HGRE, NGLDM_Busyness, GLZLM_ZLNU), are all associated with image uniformity and heterogeneity. In this study, lung adenocarcinomas with EGFR mutations were more heterogeneous than those with wild-type EGFR. Many studies have appeared on heterogeneity in radiomics. Aerts et al. found that radiomic features could reflect the inherent heterogeneity of lung tumors and were related to the tumor types based on gene expression [26]. Yip et al. analyzed the 18F-FDG PET/CT images of 348 NSCLC patients and assessed 21 imaging features, finding that the PET radiomic features could characterize the differences in tumor metabolic phenotypes caused by EGFR mutations, and might potentially be used to develop noninvasive imaging biomarkers for somatic mutations [27]. Rios Velazquez et al. analyzed the radiomic characteristics of 763 patients with lung adenocarcinoma from 4 medical centers and their relationship with EGFR and KRAS mutations. Univariate analysis showed that 16 features were associated with EGFR mutations, all of which suggested that tumors with EGFR mutations were more heterogeneous [28]. Our results are consistent with those of the studies mentioned above: We found that lung adenocarcinomas with EGFR mutation had lower metabolism and density, and PET and CT images were more heterogeneous than those of EGFR wild-type lung cancer.
Large-scale studies indicate that EGFR mutations are more likely to occur in Asians, adenocarcinomas, females, and non-smokers [29, 30]. Gender and smoking history are also predictors of EGFR mutation status in our study. The radiomics model we developed showed a significant ability to discriminate between EGFR mutation and EGFR wild type, and when combined with clinical features, its prediction accuracy was significantly improved, supporting the complementarity of radiomics and clinical signatures. Furthermore, we created a nomogram including the radiomics signature score and clinical risk factors (gender, smoking history), which can visualize the prediction results and provide an easy-to-use method for individualized prediction of EGFR mutations.
The studies by Liu et al. [31] and Zhang et al. [32] also demonstrated that combining radiomics features with clinical risk factors can provide added predictive value for EGFR mutant status. However, their radiomics studies on the prediction of lung cancer gene mutations are all based on CT images only. In our study, the radiomics method is applied to 18F-FDG PET/CT images to extract the texture features of PET and CT images, and to integrate the texture features of metabolism and structure. Simply by visual observation, it can be noted how the model generated by PET/CT radiomic features alone is superior to the models generated by CT radiomic features alone in predicting EGFR mutations. Notwithstanding these differences, the results of our study together with those mentioned above indicate the fact that the combination of radiomics features and clinical variables has a complementary and synergistic effect in predicting gene mutations.
The potential clinical significance of our study is two-fold: (1) It provides a relatively accurate, convenient, and noninvasive method for predicting EGFR mutation status, applicable to patients with contraindications for biopsy. (2) The EGFR mutation status can be dynamically observed, as can be the effectiveness of targeted drug therapy, through the changes of PET and CT texture features. Therefore, the radiomics method based on PET/CT is expected to solve the problem of conducting multiple lesion biopsies in clinical work.
Our study has some limitations: (1) Its single-center design and the relatively small sample size may compromise the model’s generalization ability and affect its sensitivity and specificity. Therefore, it is necessary to formulate a unified standard for multi-center studies and to establish and test multi-center data through radiomics methods to ensure better robustness of the model. (2) EGFR mutation status of only one tissue type (lung adenocarcinoma) has been studied, and the predictive performance of this model in other lung cancer types should be further validated. (3) This study proves that the radiomics model has good prediction performance: In the future, deep learning models based on PET/CT images should be established. Models established by different methods should be compared and combined to develop a model with optimal prediction performance.
In conclusion, the model developed by pre-therapy 18F-FDG PET/CT-based radiomic features has good predictive performance. 18F-FDG PET/CT radiomics provides a relatively accurate, convenient, and noninvasive method for predicting EGFR mutation status, which can be very useful in the clinical practice to identify candidates for targeted therapy.
References
Hsu WH, Yang JC, Mok TS, Loong HH. Overview of current systemic management of EGFR-mutant NSCLC. Ann Oncol. 2018;29:i3–9.
An N, Zhang Y, Niu H, Li Z, Cai J, Zhao Q, et al. EGFR-TKIs versus taxanes agents in therapy for nonsmall-cell lung cancer patients: a PRISMA-compliant systematic review with meta-analysis and meta-regression. Medicine (Baltimore). 2016;95:e5601.
Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:1800986.
Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics─guiding principles and technical considerations. Radiology. 2014;270(2):320–5.
Ozkan E, West A, Dedelow JA, Chu BF, Zhao W, Yildiz VO, et al. CT gray-level texture analysis as a quantitative imaging biomarker of epidermal growth factor receptor mutation status in adenocarcinoma of the lung. AJR Am J Roentgenol. 2015;205:1016–25.
Bianconi F, Fravolini ML, Bello-Cerezo R, Minestrini M, Scialpi M, Palumbo B. Evaluation of shape and textural features from CT as prognostic biomarkers in non-small cell lung cancer. Anticancer Res. 2018;38:2155–60.
Digumarthy SR, Padole AM, Gullo RL, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore). 2019;98:e13963.
Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018;102:1083–9.
Lee SM, Bae SK, Jung SJ, Kim CK. FDG uptake in non-small cell lung cancer is not an independent predictor of EGFR or KRAS mutation status: a retrospective analysis of 206 patients. Clin Nucl Med. 2015;40:950–8.
Takamochi K, Mogushi K, Kawaji H, Imashimizu K, Fukui M, Oh S, et al. Correlation of EGFR or KRAS mutation status with 18F-FDG uptake on PET-CT scan in lung adenocarcinoma. PLoS One. 2017;12:e0175622.
Zwanenburg A, Leger S, Vallières M, Löck S, for the Image Biomarker Standardisation Initiative (IBSI). Image biomarker standardisation initiative — feature definitions. 2018 [Current version V10 2019]. https://arxiv.org/abs/1612.07003.
Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42:328–54.
Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78:4786–9.
Kirienko M, Cozzi L, Rossi A, Voulaz E, Antunovic L, Fogliata A, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur J Nucl Med Mol Imaging. 2018;45:1649–60.
She Y, Zhang L, Zhu H, Dai C, Xie D, Xie H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol. 2018;28:5121–8.
Goldman JW, Noor ZS, Remon J, Besse B, Rosenfeld N. Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol. 2018;29:i38–46.
Kim TO, Oh IJ, Kho BG, Park HY, Chang JS, Park CK, et al. Feasibility of re-biopsy and EGFR mutation analysis in patients with non-small cell lung cancer. Thorac Cancer. 2018;9:856–64.
Li W, Qiu T, Ling Y, Gao S, Ying J. Subjecting appropriate lung adenocarcinoma samples to next-generation sequencing-based molecular testing: challenges and possible solutions. Mol Oncol. 2018;12:677–89.
Kim YI, Paeng JC, Park YS, Cheon GJ, Lee DS, Chung JK, et al. Relation of EGFR mutation status to metabolic activity in localized lung adenocarcinoma and its influence on the use of FDG PET/CT parameters in prognosis. AJR Am J Roentgenol. 2018;210:1346–51.
Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–93.
Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol. 2016;26:32–42.
Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y, et al. CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology. 2016;280:271–80.
Shi Z, Zheng X, Shi R, Song C, Yang R, Zhang Q, et al. Radiological and clinical features associated with epidermal growth factor receptor mutation status of Exon 19 and 21 in lung adenocarcinoma. Sci Rep. 2017;7:364.
Sacconi B, Anzidei M, Leonardi A, Boni F, Saba L, Scipione R, et al. Analysis of CT features and quantitative texture analysis in patients with lung adenocarcinoma: a correlation with EGFR mutations and survival rates. Clin Radiol. 2017;72:443–50.
Desseroit MC. Tixier F, Weber WA, Siegel BA, Cheze Le Rest C, Visvikis D, et al. Reliability of PET/CT shape and heterogeneity features in functional and morphologic components of non-small cell lung cancer tumors: a repeatability analysis in a prospective multicenter cohort. J Nucl Med. 2017;58:406–11.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
Yip SS, Kim J, Coroller TP, Parmar C, Velazquez ER, Huynh E, et al. Associations between somatic mutations and metabolic imaging phenotypes in non-small cell lung cancer. J Nucl Med. 2017;58:569–76.
Rios Velazquez E, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77:3922–30.
Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–77.
Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–62.
Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer. 2016;17:441–8.e6.
Zhang L, Chen B, Liu X, Song J, Fang M, Hu C, et al. Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non-small cell lung cancer. Transl Oncol. 2018;11:94–101.
Acknowledgments
The authors thank all their coworkers involved in the study for their support and assistance.
Funding
This study was financially supported by the Foundation of Science and Technology Department of Hebei Province, China (grant No.15277776D).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in the study and involving human participants were carried out in accordance with the ethical standards of the institutional and/or national research committee and with the principles of the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
This retrospective analysis was approved by the Institutional Review Board of the Fourth Hospital of Hebei Medical University (Approval No. 2019MEC031), and the requirement of informed consent was waived.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Zhao, X., Zhao, Y. et al. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging 47, 1137–1146 (2020). https://doi.org/10.1007/s00259-019-04592-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04592-1