Abstract
Techniques from the field of artificial intelligence, and more specifically machine (deep) learning methods, have been core components of most recent developments in the field of medical imaging. They are already being exploited or are being considered to tackle most tasks, including image reconstruction, processing (denoising, segmentation), analysis and predictive modelling. In this review we introduce and define these key concepts and discuss how the techniques from this field can be applied to nuclear medicine imaging applications with a particular focus on radio(geno)mics.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nuclear medicine and radiology have evolved greatly over the last 20 years thanks mainly to hardware developments, such as the deployment of multimodality imaging devices in the 2000s [1], and the development of fast detector technologies [1, 2]. Software innovations have also led to substantial improvements in the spatial resolution and the signal-to-noise ratio of reconstructed images, thanks for example to point-spread function and time-of-flight (ToF) information being incorporated into PET image reconstruction [3]. Despite being quantitative by nature, nuclear medicine images are, in most clinical publications, clinical trials and obviously routine clinical practice, exploited in a very restrictive manner (i.e. analysed mostly visually or semiquantitatively). There seems to be growing interest in more automatic analysis of medical images coupled to extraction of multiple features, including some that may not be accessible to the naked eye, even the expert trained eye [4, 5]. The main objective of this evolution and change in paradigm is to better harness the information provided by imaging studies in terms of influencing patient management workflow within the context of precision medicine. As a result, within this new framework, medical imaging should play an enhanced role, and become essential beyond diagnosis to cover therapy planning, as well as therapy monitoring and assessment, and predictive modelling and stratification, to become overall an integral part of future clinical decision-making systems.
The aim of the present paper is to provide definitions of artificial intelligence (AI, machine/deep learning) and radio(geno)mics, as well as some insights into their potential applications in nuclear medicine imaging.
Definitions of artificial intelligence, machine (deep) learning and radio(geno)mics
Artificial intelligence
The term artificial intelligence is a ‘fuzzy concept’ with a number of possible definitions depending on context, time and applications. As an academic discipline, it is considered to have been founded in 1956 at the Dartmouth conference [6]. A rather general definition is “intelligence demonstrated by machines, in contrast to the natural intelligence displayed by humans and other animals”. However in the present context of medical imaging, a more specific definition may be more appropriate: “a system’s ability to correctly interpret external data, to learn from such data, and to use those learnings to achieve specific goals and tasks through flexible adaptation” [7]. As algorithms tackle increasingly complex tasks, those considered to require ‘intelligence’ are sometimes removed from the field of AI, leading to the assertion that “AI is whatever has not been done yet” [8]. An example of this is character recognition, which may no longer be considered as ‘artificial intelligence’, because it is now a standard routinely used technology, for example by postal services. Well-known capabilities of today’s algorithms usually considered as AI include speech recognition and more importantly understanding, language translation, mastering complex games such as Go [9] and more recently even more complex strategy video gamesFootnote 1, or autonomously driving cars.
AI systems can be classified as analytical, human-inspired or humanized AI [7]. Analytical systems possess only characteristics related to cognitive intelligence, using learning based on past experience to make predictions. Human-inspired AI systems possess emotional intelligence and understanding in addition to cognitive elements. Humanized AI systems are able to demonstrate cognitive, emotional and social intelligence, with self-consciousness and self-awareness in interactions with others. In the twenty-first century, AI techniques have benefited from improved theoretical understanding (e.g. in neural network mathematics), advances in computer power (e.g. graphical processing units, GPU), wider availability of always larger quantities of data for learning (e.g. through the development of social networks and other platforms, cloud storage/computing, etc.), and availability of the algorithms and libraries themselves. As a result, older concepts and theories can now be actually applied to real-life problems and tasks, even by nonexperts on commercially available systems.
Regarding medical imaging, a number of tasks that clinicians perform that rely on images could therefore theoretically be carried out by AI, including but not limited to, lesion detection, disease classification, diagnosis and staging, quantification, treatment planning (delineation of targets and organs at risk, and dosimetry optimization), assessing response to treatment and prognosis [10]. Automation is expected to allow these tasks to be achieved with much higher robustness and reproducibility, and potentially with fewer errors, in a much shorter time. Obviously, there are other aspects of medical imaging in which AI could provide solutions to improve practice, such as improving operational workflow, finance management and quality improvement, amongst others [11]. Most, if not all, AI systems developed for medical image analysis tasks belong to the class of analytical systems, and can therefore be classified as machine/deep-learning techniques.
Machine (deep) learning
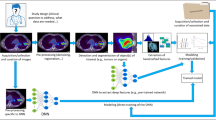
Machine learning is the study of algorithms that learn and improve through experience, and as such is a fundamental concept of AI. Learning is usually considered to be unsupervised or (semi)supervised. Unsupervised learning consists of finding patterns in unlabelled data [12], whereas supervised learning uses labels to infer classification or regression, and semisupervised learning is typically carried out with a small amount of labelled data and a large amount of unlabelled data [13]. In the context of medical imaging, the standard workflow or machine learning pipeline is usually directly applied to address most tasks (Fig. 1).
Data (e.g. images or parts of images) are fed into a feature extractor. The goal of the feature extractor is to compute from the input data a number of ‘handcrafted’ (or ‘engineered’) features. These can be based partly on expert knowledge, can be more or less relevant to the task at hand, more or less complex, and in small or large numbers. These features are then fed into a classifier (or a regression) algorithm whose goal is to map the features with the classification task, for example differentiating between two types of tumour, or stratifying patients based on clinical outcome. This part is often divided into two steps: feature selection and actual modelling. Feature selection consists of identifying a smaller relevant subset amongst the features calculated by the extractor. This is usually done to facilitate the subsequent modelling step by reducing training times, avoid dimensionality issues and reduce overfitting, as well as to simplify the resulting models and improve their interpretability. There are several techniques to perform feature selection, that can be either linked to or independent of the chosen classifier that will subsequently combine these selected features into some sort of multiparametric model. These techniques can be categorized into three main approaches: filter, wrapper and embedded [14]. Filter methods identify variables independently of the model that will be combining them, based only on metrics such as the correlation between each variable and the endpoint, suppressing the least valuable ones. Despite not being prone to overfitting, filter methods tend to retain redundant features, as they do not take into consideration correlations between them [15]. Wrapper approaches evaluate subsets of features, which allows relationships between features to be taken into account, in contrast to filter methods. Their main associated drawbacks are the higher computation time and risk of overfitting with small samples. Embedded methods combine the advantages of wrapper and filter approaches, by ‘embedding’ the feature selection process within a learning algorithm, performing feature selection and classification simultaneously. Actual modelling (i.e. mapping a combination of features with the endpoint) can be performed thanks to a classifier or a regression algorithm. A large number of techniques have been developed in the field of machine learning. Popular examples include random forest, support vector machines and artificial neural networks [16].
Deep learning (as opposed to ‘shallow’ learning methods, as described above) is a category of methods belonging to the machine learning field, that are mostly based on the use of specific types of artificial neural networks, sometimes with a very large number of layers and nodes. Thus deep learning is a specific type of machine learning, which is itself part of AI (see Fig. 2) [17].
These techniques rely on a cascade of multiple layers of nonlinear processing units for feature extraction and transformation, where the input to each successive layer is the output from the previous layer, so that multiple levels of representation that correspond to different levels of abstraction are learned [18]. Even though neural networks were designed much earlier, ‘deep’ networks, with their ability to learn efficiently through a general purpose procedure, are more recent. Furthermore, the main impact of convolutional neural networks (CNN) on computer vision and imaging applications was considered to be a real breakthrough in the years 2011 and 2012. CNNs trained using the backpropagation algorithm had existed for decades, and GPU implementations for years. However in 2012, Cireşan et al. showed how max-pooling CNN implemented on a GPU could provide improved results in a number of vision benchmarks. The same year, Krizhevsky et al. won the ImageNet competition with much better performance than shallow machine learning methods, using a similar CNN design [19]. Cireşan et al. also won both the ICPR and the MICCAI Grand Challenge on mitosis detection [20]. Over the next years, the performance obtained by challengers in the ImageNet competition steadily improved thanks to new CNN designs and techniques [21]. Deep learning methods, especially CNN but also other types of network (e.g. recurrent neural networks and generative adversarial networks) have since been exploited to address existing challenges in a number of medical imaging tasks, including but not limited to, image registration, reconstruction, classification, pattern recognition, segmentation, denoising and super resolution. In most of these tasks, they often achieved unprecedented performance (in terms of computational efficiency and/or performance in the specific task), becoming de facto the new standard and benchmark to beat [22].
One of the major differences between these techniques and the ‘older’ machine learning approaches described above is that the aim of these networks is to learn specific patterns relevant to a given task (e.g. segmentation or endpoint prediction) from the data (i.e. images) themselves, instead of relying on ‘engineered’ or ‘handcrafted’ features (including expert knowledge) [22, 23]. In that regard, these methods can be considered a paradigm shift, as they may provide an ‘end-to-end’ workflow, relying on a general purpose learning procedure (Fig. 1).
User intervention, for example detection and selection of objects of interest for further characterization, could therefore be significantly simplified or even deemed unnecessary. On the other hand, a number of challenges need to be well understood when considering the use of these techniques. Deep neural networks have a large number of hyper parameters, and exploring the parameter space to identify optimal ones is usually not feasible due to limitations in computational resources and time. Tricks such as computing the gradient on several examples at once (batch processing) can help speed up computation. The large processing capabilities of GPUs have enabled significant improvements in training speed. Deep neural networks are also prone to overfitting. This is due in part to their ability to model rare dependencies observed in the training data, thanks to their numerous layers. Various approaches such as regularization and dropout are usually implemented to limit overfitting [24]. Data can also be augmented by methods such as zooming and rotating to increase the size of the required training sets [25]. Finally, transfer learning is an important component in which networks pretrained on different, although much larger datasets, are fine-tuned using smaller datasets more specific to the task at hand [26, 27].
Radio(geno)mics
In parallel to the improvements in PET/CT hardware and reconstruction software over the last two decades, several developments have been made in the field of PET/CT image processing and analysis: noise filtering [28, 29] and partial volume effect correction [30] methods can further improve both the visual quality and quantitative accuracy of PET images. In addition, (semi)automated image analysis algorithms can detect lesions of interest [31] and also delineate them with similar accuracy and higher reproducibility and robustness than human experts [32,32,34]. This has opened the way to a more comprehensive characterization of organs and tumours, by extracting quantitative metrics (‘handcrafted’ or ‘engineered’ image features) from preprocessed and segmented PET/CT images. In this context, most of the current work on PET/CT imaging has concentrated on the radiotracer most commonly used clinically, namely 18F-fluorodeoxyglucose (18F-FDG), with very few studies considering other tracers [35]. The four methodological components shown in Fig. 1 (preprocessing, segmentation, feature extraction, modelling) are the key building blocks of the scientific field known today as radiomics. The term ‘radiomics’ first appeared in 2010 [35] and the fully formalized framework of radiomics was described in 2012 [36]. As can be understood from the previous sections, radiomics is simply a translation of the standard machine learning pipeline (Fig. 1) applied to medical images. The rationale behind the development of the radiomics field of research is that medical images contain features of tumour phenotypes that can reflect at least part of the underlying pathophysiological processes at smaller scales, including down to the genetic level. This is why the term radiomics is often associated with genomics, in the term ‘radiogenomics’. Radiogenomics actually has two different meanings. The first, older one is related to radiobiology and is not relevant in the present context. The second concerns the association/combination of radiomics and genomics, which can be categorized into two different methodological approaches. The first investigates the links between the two, i.e. what part of the genomics information can be explained or ‘decoded’ by radiomics, which has been described as ‘imaging genomics’ [37, 38] and investigated in a number of studies [39, 40]. The alternative approach concerns the development of methodology that combines two sources of information making use of their complementary value in order to build more efficient predictive models.
Nuclear medicine imaging applications of artificial intelligence, deep learning and radio(geno)mics
Applications of AI in nuclear medicine are extremely wide and promising and may impact different aspects [41]. The first step concerns the use of AI for data processing at the detector level for image reconstruction, including corrections for the different physics processes associated with the detection process (e.g. attenuation, scatter). Beyond the image reconstruction step, AI may be useful for different image processing steps including denoising, segmentation and fusion. Finally, AI can be used in the construction of models based on information extracted from images that would help achieve predictive, personalized medicine relying on images.
At the detection level, recent efforts include the use of CNN to enhance PET image resolution and improve the noise properties of PET scanners with large pixelated crystals [42] and to estimate ToF directly from pairs of coincident digitized detector waveforms [43]. Integrating a deep neural network into the iterative image reconstruction process may improve final image quality [44, 45]. Deep learning methods have already been proposed for attenuation correction and registration in PET/CT and PET/MR, and have been shown to be able to generate attenuation maps with high accuracy [46,47,48,49,50]. In the same context, deep learning has been used for improving maximum-likelihood reconstruction of activity and attenuation (MLAA) with ToF PET data [51]. Denoising is one of the most popular image processing applications for which deep learning techniques have been successfully used, for example to generate full-dose PET images from low-dose images [52] or to directly filter reconstructed PET images [29].
Automated detection, counting and segmentation/characterization of lesions in images may have wide applications in diagnosis, as well as in planning treatment and monitoring response, but more broadly, also for all radio(geno)mics applications. For a long time, methods relying on older, shallow machine learning frameworks were not able to reach the required levels of automation and accuracy to be fully transferred to clinical practice or to allow the fast processing of hundreds of patients in radiomics analysis. Some recent developments still involve the use of ‘older’ machine learning techniques [53], but a growing number are relying on deep learning methods with the hope of greatly improving both automation and performance. Indeed, CNNs have been especially successful for medical image segmentation tasks [22]. This is explained by the fact that, contrary to classification tasks (one label per image), segmentation learning occurs at the voxel level (one label per voxel). The amount of learning data thus allows efficient training of the network parameters. For example, despite having only a few training examples available in the recent PET functional volume segmentation MICCAI challenge, the method based on a pretrained CNN achieved the highest score (although not significantly higher than the scores of some of the more conventional techniques) [32]. CNNs have also been applied to the problem of multimodal PET/CT cosegmentation [34, 54, 55]. Pipelines for tumour detection and segmentation based on a deep learning framework [31, 55, 56] are likely to provide fully automated solutions for this step of the radiomics pipeline, thereby eliminating this important bottleneck.
Predictive modelling and radio(geno)mics studies already heavily rely on machine learning methods [16, 57,58,59], although most of them are in the field of radiology and not nuclear medicine. Evaluation of machine learning and deep learning methods have shown improved feature selection, more robust model building and harmonization of radiomic PET features [59,60,61,62,63]. However, only a limited number of studies have explored the potential of deep network CNNs to reach a higher levels of automation by using them as an end-to-end methodology, and most of them have been in the field of CT or MR imaging [64,65,66,67,68,69,70], with only a few examples of their use in nuclear medicine imaging such as FDG PET [71,72,73] and SPECT [74].
Discussion
Although most currently available studies on the use of deep features, and their combination with usual radiomic features, have been carried out in the fields of CT and MRI, the same concepts can be applied to nuclear medicine imaging. Replacing the usual machine learning/radiomics pipeline by one based on end-to-end deep learning (Fig. 1) may be an attractive solution to solving some of the issues and limitations of radiomics. In this approach, all steps performed separately and sequentially (segmentation, feature extraction, modelling) are performed by one (or several) neural network(s). However, this approach actually replaces previous challenges by others more specific to the use of deep learning. First, these methods are data-hungry and therefore datasets much larger than those usually available in radiomics studies are needed for efficient training. Thus, techniques and tricks such as transfer learning and data augmentation, or reliance on segmentation networks to build classifiers [75], become crucially important. Second, the requirement to provide interpretable models is also important in clinical applications. Therefore there is a need to provide feedback and explanation to the end-users regarding a network decision, using for example network visualization techniques [76] to generate heat maps in the input images highlighting the areas in the image, or even within the tumour, that were the most relevant in reaching the decision. This is also important in understanding and correcting the remaining errors that the algorithms make, and in trying to address other issues including regulatory, legal and accountability issues [77].
A major paradigm shift is occurring in the design of most computer approaches designed for use in the clinic. It is unclear how much time is needed for deep learning methods to be integrated into clinical nuclear medicine practice, and to achieve full automation of most clinical tasks. Currently, these developments have focused on tackling the most common clinical problems for which sufficient data are available.
The aim of most developed methods is to solve one problem within one specific task. While they may excel in interpreting image and contextual information, they are usually not able to make associations the way a human brain does, and cannot replace clinicians for all tasks they perform. In addition, they may not yet have reached the same level of performance as the expert in all situations, and therefore a full artificial nuclear medicine physician still belongs to the domain of science fiction. On the other hand, the role of nuclear medicine physicians is likely to evolve as these new techniques are integrated into their practice, and it is therefore important that the acquisition of a basic understanding of these methods and concepts is part of their training. They will themselves also probably contribute to the training of AI, providing additional expert knowledge and experience to the tools they will then use.
The availability of data remains a crucial bottleneck in AI system learning, because curated data (ensuring the training data conforms to a number of quality criteria, which usually involves experts and is time-expensive) is simply not yet available for all tasks and in sufficient amount. On the other hand, the deep learning software platforms are open-source and because of this, experimentation and sharing of innovations has been fast and on a massive scale, and this may eventually also help in terms of data processing and availability. An additional concern for proper training of machine and deep learning models is the lack of standardization in both image acquisition and reconstruction (despite long-term efforts by societies such as the EANM, SNMMI and RSNA, and others) and in machine (deep) learning techniques themselves (including but not limited to, radiomics definitions, nomenclature, implementation, software, machine learning methodology and implementation, and optimization). The strong heterogeneity and variability in scanner models, vendors, acquisition protocols and reconstruction settings is a considerable challenge for training generalizable models that remains to be fully addressed. Efforts already ongoing, for example the Image Biomarker Standardization Initiative (IBSI) for radiomics standardization [78,79,80] and harmonization/normalization techniques [63, 81], should clearly be emphasized and supported to further improve these aspects in the future.
References
Beyer T, Townsend DW, Brun T, Kinahan PE, Charron M, Roddy R, et al. A combined PET/CT scanner for clinical oncology. J Nucl Med. 2000;41:1369–79.
Lecoq P. Pushing the limits in time-of-flight PET imaging. IEEE Trans Radiat Plasma Med Sci. 2017;1(6):473–85.
Berg E, Cherry SR. Innovations in instrumentation for positron emission tomography. Semin Nucl Med. 2018;48:311–31.
Aerts H. Radiomics: there is more than meets the eye in medical imaging. Plenary presentation from SPIE Medical Imaging 2016: computer-aided diagnosis. https://doi.org/10.1117/12.2214251.
Hatt M, Tixier F, Visvikis D, Cheze Le Rest C. Radiomics in PET/CT: more than meets the eye? J Nucl Med. 2017;58:365–6.
Moor J. The Dartmouth College Artificial Intelligence Conference: the next fifty years. Al Mag. 2006;27:87–91.
Kaplan A, Haenlein M. Siri, Siri, in my hand: Who’s the fairest in the land? On the interpretations, illustrations, and implications of artificial intelligence. Bus Horiz. 2019;62:15–25.
Maloof M. Artificial intelligence: an introduction. Washington DC: Georgetown University. 2015. http://people.cs.georgetown.edu/~maloof/cosc270.f17/cosc270-intro-handout.pdf.
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484–9.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10.
Palmer WJ. Artificial intelligence in radiology: friend or foe? Diagnostic Imaging. 2019. http://www.diagnosticimaging.com/di-executive/artificial-intelligence-radiology-friend-or-foe.
Duda RO, Hart PE, Stork DG. Pattern classification. New York: Wiley; 2012.
Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge, MA: MIT Press; 2012.
Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–82.
Phuong TM, Lin Z, Altman RB. Choosing SNPs using feature selection. Proceedings of the 2005 IEEE Computational Systems Bioinformatics Conference. 2005; p. 301–9.
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117.
Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems, vol. 1; 2012. p. 1097–105.
Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Berlin: Springer; 2013. p. 411–8.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115:211–52.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ. Quantitative imaging of cancer in the postgenomic era: radio(geno)mics, deep learning, and habitats. Cancer. 2018;124:4633–49.
Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. 2013. p. 8609–13.
Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops: CVPRW 2014. Washington, DC, USA: IEEE Computer Society; 2014. p. 512–519. https://doi.org/10.1109/CVPRW.2014.131.
Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–59.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–98.
Le Pogam A, Hanzouli H, Hatt M, Cheze Le Rest C, Visvikis D. Denoising of PET images by combining wavelets and curvelets for improved preservation of resolution and quantitation. Med Image Anal. 2013;17:877–91.
Gong K, Guan J, Liu C, Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):153–61.
Le Pogam A, Hatt M, Descourt P, Boussion N, Tsoumpas C, Turkheimer FE, et al. Evaluation of a 3D local multiresolution algorithm for the correction of partial volume effects in positron emission tomography. Med Phys. 2011;38:4920–3.
Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-net convolutional neural network study. PLoS One. 2018;13:e0195798.
Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177–95.
Hatt M, Lee JA, Schmidtlein CR, Naqa IE, Caldwell C, De Bernardi E, et al. Classification and evaluation strategies of auto-segmentation approaches for PET: report of AAPM task group no. 211. Med Phys. 2017;44:e1–42.
Zhao X, Li L, Lu W, Tan S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys Med Biol. 2018;64:015011.
Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol. 2010;65:517–21.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6.
Bai HX, Lee AM, Yang L, Zhang P, Davatzikos C, Maris JM, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89:20151030.
Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–41.
Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–80.
Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
Hatt M, Parmar C, Qi J, Naqa IE. Machine (deep) learning methods for image processing and radiomics. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):104–8.
Hong X, Zan Y, Weng F, Tao W, Peng Q, Huang Q. Enhancing the image quality via transferred deep residual learning of coarse PET sinograms. IEEE Trans Med Imaging. 2018;37:2322–32.
Berg E, Cherry SR. Using convolutional neural networks to estimate time-of-flight from PET detector waveforms. Phys Med Biol. 2018;63:02LT01.
Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2018;38:675–85.
Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Trans Med Imaging. 2018;37:1478–87.
Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. In: Deep Learning and Data Labeling for Medical Applications – First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, held in conjunction with MICCAI 2016, Athens, Greece, 21 October 2016; 2016. p. 170–8.
Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med. 2019;60(3):429–35.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019. https://doi.org/10.2967/jnumed.118.219493.
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286(2):676–84.
Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Zero-echo-time and Dixon deep pseudo-CT (ZeDD CT): direct generation of pseudo-CT images for pelvic PET/MRI attenuation correction using deep convolutional neural networks with multiparametric MRI. J Nucl Med. 2018;59:852–8.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9.
Kaplan S, Zhu Y-M. Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging. 2018. https://doi.org/10.1007/s10278-018-0150-3.
Perk T, Bradshaw T, Chen S, Im H-J, Cho S, Perlman S, et al. Automated classification of benign and malignant lesions in 18F-NaF PET/CT images using machine learning. Phys Med Biol. 2018;63:225019.
Zhong Z, Kim Y, Plichta K, Allen BG, Zhou L, Buatti J, et al. Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med Phys. 2018;46(2):619–33.
Guo Z, Li X, Huang H, Guo N, Li Q. Deep learning-based image segmentation on multi-modal medical imaging. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):162–9.
Schwyzer M, Ferraro DA, Muehlematter UJ, Curioni-Fontecedro A, Huellner MW, von Schulthess GK, et al. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks – initial results. Lung Cancer. 2018;126:170–3.
Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zöphel K, et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7:13206.
Deist TM, Dankers FJWM, Valdes G, Wijsman R, Hsu I-C, Oberije C, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med Phys. 2018;45:3449–59.
Upadhaya T, Vallières M, Chatterjee A, Lucia F, Bonaffini PA, Masson I, et al. Comparison of radiomics models built through machine learning in a multicentric context with independent testing: identical data, similar algorithms, different methodologies. IEEE Trans Radiat Plasma Med Sci. 2018;3(2):192–200.
Wu J, Lian C, Ruan S, Mazur TR, Mutic S, Anastasio MA, et al. Treatment outcome prediction for cancer patients based on radiomics and belief function theory. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):216–24.
Klyuzhin IS, Fu JF, Shenkov N, Rahmim A, Sossi V. Use of generative disease models for analysis and selection of radiomic features in PET. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):178–91.
Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Bist V, et al. An empirical approach for avoiding false discoveries when applying high-dimensional radiomics to small datasets. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):201–9.
Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Saif S, et al. Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):210–5.
Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711.
Paul R, Hall L, Goldgof D, Schabath M, Gillies R. Predicting nodule malignancy using a CNN ensemble approach. Proc Int Jt Conf Neural Netw. 2018. https://doi.org/10.1109/IJCNN.2018.8489345.
Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290:290–7.
Bibault J-E, Giraud P, Housset M, Durdux C, Taieb J, Berger A, et al. Deep learning and radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611.
Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7:5467.
Cha KH, Hadjiiski L, Chan H-P, Weizer AZ, Alva A, Cohan RH, et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep. 2017;7:8738.
Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, et al. Convolutional neural network using a breast MRI tumor dataset can predict oncotype Dx recurrence score. J Magn Reson Imaging. 2019;49:518–24.
Ypsilantis P-P, Siddique M, Sohn H-M, Davies A, Cook G, Goh V, et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS One. 2015;10:e0137036.
Amyar A, Ruan S, Gardin I, Chatelain C, Decazes P, Modzelewski R. 3D RPET-NET: development of a 3D PET imaging convolutional neural network for radiomics analysis and outcome prediction. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):225–31.
Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, et al. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2019;290:456–64.
Mabrouk R, Chikhaoui B, Bentabet L. Machine learning based classification using clinical and DaTSCAN SPECT imaging features: a study on Parkinson’s disease and SWEDD. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):170–7.
Wong KCL, Syeda-Mahmood T, Moradi M. Building medical image classifiers with very limited data using segmentation networks. Med Image Anal. 2018;49:105–16.
Yosinski J, Clune J, Nguyen A, Fuchs T, Lipson H. Understanding neural networks through deep visualization. arXiv:1506.06579 [cs.CV]. http://arxiv.org/abs/1506.06579.
Ford R, Price W. Privacy and accountability in black-box medicine. Mich Telecommun Technol Law Rev. 2016;23:1–43.
Vallières M, Zwanenburg A, Badic B, Cheze-Le Rest C, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. 2018;59(2):189–93.
Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv:1612.07003 [cs.CV]. http://arxiv.org/abs/1612.07003.
Hatt M, Vallières M, Visvikis D, Zwanenburg A. IBSI: an international community radiomics standardization initiative. J Nucl Med. 2018;59:287.
Lucia F, Visvikis D, Desseroit M-C, Miranda O, Malhaire J-P, Robin P, et al. External validation of a combined PET and MRI radiomics model for prediction of distance recurrence and locoregional control in locally advanced cervical cancer patients treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2019;46(4):864–77.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
N/A.
Conflict of interest
None.
Ethical approval
This is a review article for which ethical approval is not required.
Informed consent
This is a review article for which informed consent is not relevant.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Rights and permissions
About this article
Cite this article
Visvikis, D., Cheze Le Rest, C., Jaouen, V. et al. Artificial intelligence, machine (deep) learning and radio(geno)mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging 46, 2630–2637 (2019). https://doi.org/10.1007/s00259-019-04373-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04373-w