Abstract
Levan, a type of β (2→6)-linked fructan, is a promising biopolymer with distinct properties and extensive applications in the fields of food, pharmaceutical, cosmetics, etc. However, the commercial availability of levan is still limited due to the relatively high production costs. Here, a new Paenibacillus sp. strain FP01 was isolated and identified as an efficient fructan producer with high yield (around 89.5 g/L fructan was obtained under 180 g/L sucrose) and conversation rate (49.7%). The fructan named Plev was structurally characterized as a linear levan-type fructan with a molecular mass of 3.11 × 106 Da. Aqueous solutions of Plev exhibited a non-Newtonian behavior at concentrations 3–5%. Heating and chilling had no obvious effects on apparent viscosities of Plev solutions. Plev also had good rheological stabilities toward pH (3–11) and metal salts (Na+, K+, Ca2+, Mg2+). Microbiome and metabolome analysis showed that Plev intervention increased the abundance of beneficial bacteria and elevated the levels of short-chain fatty acids (SCFAs) in feces of mice. Taken together, Plev could be considered a potential thickener and prebiotic supplement in food industry.
Key points
• Paenibacillus sp. strain FP01 was identified as a high-efficient levan producer.
• The levan Plev from FP01 exhibited good rheological properties and stabilities.
• The in vivo prebiotic activities of linear levan were revealed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fructans are a class of fructose-conjugated homopolysaccharides, widely distributed in plants as storage carbohydrates or synthesized by various environmental microorganisms as extracellular polysaccharide (EPS) or biofilm for defense (Benigar et al. 2014). Depending on the linkage of fructofuranosyl residues, fructans are classified into two main types, commonly known as inulin and levan. Inulin-type fructans are low-molecular-weight polymers linked by β (2→1) glycosidic bonds with degree of polymerization (DP) ranging from 3 to 60, traditionally extracted from tubers of Chicory and Jerusalem artichoke (Ahmed and Rashid 2019; Morreale et al. 2019). Levan is composed of β (2→6) linkages in the backbone with occasionally β (2→1) branches, produced by a variety of microorganisms and a few plants belonging to the tribe Poeae and Triticeae (Tamura et al. 2014). Bacterial levan is usually produced outside the cells by levansucrase-catalyzed transfructosylation with sucrose as the substrate. Compared with plant-derived levan which possesses small chains of DP 2-200, the size of bacterial levan-type polymers can be varied from low-molecular-weight oligosaccharides to thousands of fructose monomers (DP > 5 × 104), mainly depending on the producing strains, medium components, and cultivation conditions (Ortiz-Soto et al. 2019).
Compared with plant-derived fructan, bacterial levan confers more advantages in industrial applications because of ease of extraction and controllability of production, and has drawn extensive research attentions due to the extraordinary physicochemical and functional properties. Bacterial levan possesses high stability and solubility in water and oil (Ates 2015; Srikanth et al. 2015a), high water holding capacity (Haddar et al. 2020), low intrinsic viscosity (Arvidson et al. 2006), strong adhesiveness, favorable biocompatibility and biodegradability, good film-forming ability (Ağçeli and Cihangir 2020), etc., thus has been exploited as a stabilizer, emulsifier, food and feed additive, encapsulating agent, blood plasma extender, osmoregulator, and cryoprotector (Han 1990; Öner et al. 2016; Srikanth et al. 2015a). Besides, levan-type fructans have been reported to have antioxidant (Dahech et al. 2011b; Dahech et al. 2013), antidiabetic (Dahech et al. 2011a), antitumor (Abdel-Fattah et al. 2012), and immuno-stimulating (Dong et al. 2015) activities.
As a kind of non-digestible polysaccharide, fructan cannot be absorbed in the small intestine, but can be utilized by beneficial bacteria in the colon, and thus perform an excellent prebiotic action and have potential applications in the field of food, medical, and health care industries. Inulin and the related oligosaccharides have been regarded as efficient prebiotics for decades, whereas the prebiotic effects of bacterial levan still remain less investigated. Levan and levan-derived fructo-oligosaccharides have been reported as the substrates of several beneficial genera, including Bifidobacterium and some species of Bacteroides (Semjonovs and Zikmanis 2007) (Sonnenburg et al. 2010). The genes coding fructan-metabolizing enzymes (endo-levanase, fructofuranosidase, etc.) have also been identified in the genome of gut bacteria (Ávila-Fernández et al. 2016; Lammerts van Bueren et al. 2015; Mardo et al. 2017). In vitro model systems have been employed to assess the prebiotic potential of levan-type fructans (Kaarel et al. 2015). However, the results could be inconsistent, probably due to the model systems, different origins, chain length, and branching degree of levan (Liu et al. 2020; Szwengiel and Nkongha 2019). It is critical to understand the effects of levan on specific gut microbiome composition to gain an insight into the potential mechanisms of action.
Levan can be extracellularly produced from sucrose-based substrates by many bacterial species, including Zymomonas mobilis (Ferreira et al. 2019), Bacillus amyloliquefaciens (Cai et al. 2020), Bacillus subtilis (Bouallegue et al. 2020), Paenibacillus polymyxa (Grinev et al. 2020), Erwinia sp. (Liu et al. 2020), Halomonas smyrnensis (Tohme et al. 2018), Gluconobacter nephelii (Semjonovs et al. 2016), Acetobacter xylinum (Srikanth et al. 2015b), Leuconostoc citreum (Han et al. 2015), Brachybacterium sp. (Djurić et al. 2017), and Pseudomonas fluorescens (Jathore et al. 2012). The extracellular levansucrase from some bacteria could also be used for levan biosynthesis in vitro. Depending on fermentation conditions and producing strains, the bacterial levan may be of different chain length and branching degree, which determine its physiological functions and physicochemical characteristics.
Although many levan-producing microorganisms have been reported in recent years, levan is still a relatively cost-intensive polysaccharide due to high productive costs and limited commercial microbial systems, which hinder further study and applications of levan. Various strategies have been adopted to deal with these bottlenecks, including utilizing economically alternative substrates and nutrient sources (molasses, sugar cane syrups, chicken feather peptone) (de Oliveira et al. 2007; Veerapandian et al. 2020), optimizing medium and cultivation conditions (Erkorkmaz et al. 2018), immobilizing Zymomonas mobilis cells for continuous production of levan (Silbir et al. 2014), and modifying the metabolic pathway for levan biosynthesis (Feng et al. 2015a, b). Nevertheless, exploring new wild-type bacterial strains with high capacity of levan production still offers an efficient way. In this study, we intended to isolate and identify new bacterial strains which could produce levan-type fructans from sucrose-rich medium. The structure and rheological properties of the levan were characterized. Furthermore, the prebiotic activities of levan were evaluated based on intestinal microbiota and metabolism analysis. 16S rDNA high-throughput sequencing was employed to investigate the composition of the gut microbiota in mice, and the metabolic changes in the fecal metabolites were determined by 1H NMR-based metabolite analysis.
Materials and methods
Chemicals
Monosaccharide standards were purchased from Sigma-Aldrich Co. (St. Louis, USA). Carbon sources such as sucrose, starch, fructose, lactose, maltose, xylose, and nitrogen sources such as yeast extract and peptone were purchased from Solarbio Limited (Beijing, China). All other chemicals and solvents in this study were of analytical grade.
Microorganism and growth conditions
The syrup waste water from a local factory in Nanjing (Jiangsu, China) was used to screen fructan-producing bacteria. The samples were initially inoculated into PYS broth (containing in g/L: potato extract 10, yeast extract 5, sucrose 50, pH 7.0) and cultivated at 30 °C for 48 h. Dilution separation with sterile 0.9% saline water was used for bacteria isolation on PYS agar (containing in g/L: potato extract 10, yeast extract 5, sucrose 50, agar 15, pH 7.0). Mucous colonies were inoculated on new PYS agar plates by dilution spreading to obtain pure cultures. The isolates were identified based on colony morphological, physio-biochemical characteristics and 16S rDNA gene sequence analysis. Escherichia coli 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) were used for the 16S rDNA gene amplification. The procedure was set as 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 1.5 min; and one final extension at 72 °C for 10 min. The PCR product was purified and sequenced by Genscript (Nanjing, China). The sequence was compared by the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST) at the National Center for Biotechnology Information (NCBI, USA). The phylogenetic tree was conducted using the MEGA6.0 program (Tamura et al. 2013). Evolutionary distances were calculated according to the algorithm of the Kimura two-parameter model for the neighbor-joining method.
EPS preparation
The bacterial strain was inoculated into 50 ml MS medium (containing in g/L: sucrose 50, NaNO3 2.5, KH2PO4 0.5, CaCl2 0.1, MgSO4·7H2O 0.3, FeSO4·7H2O 0.01, MnSO4 0.004, pH 7.0) and cultivated on a shaker with a speed of 220 rpm at 30 °C. The culture broth was diluted with three volumes of deionized water and then the cell debris by centrifugation (16,000×g, 20 min). Protein was removed by the Sevag method (chloroform/1-butanol, v/v = 4:1). Double volumes of ethanol were added and the precipitate was collected by centrifugation at 7000×g for 10 min, re-dissolved in distilled water, dialyzed against several runs of distilled water for 24 h, and then lyophilized. The total sugar of EPS was determined by using phenol-sulfuric acid method with glucose as the standard. The protein content was measured using the bicinchoninic acid (BCA) reagent (Bio-Rad, Hercules, USA) with bovine serum albumin as a standard. All experiments were performed in triplicates.
Monosaccharide composition analysis
Monosaccharide compositions of the EPS were determined by gas chromatography (GC) according to Wang et al. (2019) with some modifications. Firstly, modified hydrolysis conditions were used for monosaccharide composition analysis of fructan-type polysaccharides. The purified EPS (10 mg) was dissolved in 2 ml of 1 M trifluoroacetic acid (TFA) solution and incubated at 70 °C for 30 min. Then the solution was dried under nitrogen, re-dissolved in methanol, and then dried under nitrogen to remove residual TFA. The dried residue was added to 1 ml pyridine, 0.2 ml trimethylchlorosilane, and 0.4 ml hexamethyldisilazane to react at 80 °C under nitrogen for 30 min. One milliliter of distilled water was added to terminate the reaction. The pyridine layer containing the trimethylsilylated derivatives was collected for gas chromatography (GC) with a Clarus 580 GC System (PerkinElmer, Waltham, USA) equipped with a Col Elite-5 column (30 m × 0.32 mm × 0.25 μm). Nitrogen flow rate was 1 ml/min. The injection volume was 1 μl with a split ratio of 10:1. The column temperature was firstly maintained at 100 °C for 2 min, then increased to 150 °C at 5 °C/min, held for 5 min, and finally raised to 210 °C at 3 °C/min. The same procedure was performed for standard monosaccharides (Fru, Rha, Glc, Gal, Xyl, and Man) and subjected to GC analysis.
Fourier transform infrared spectroscopy
Infrared spectra of the purified EPS were measured with a Nicolet Nexus 470 spectrometer (Thermo Nicolet, Madison, USA). The sample was prepared as a KBr pellet. All of the samples were scanned over a wavelength range of 4000–400 cm−1.
Nuclear magnetic resonance analysis of the EPS
Ten milligrams of the purified EPS was exchanged with D2O by three cycles of lyophilization and finally dissolved in 0.5 ml of D2O. 1H- and 13C-NMR spectra were recorded on an Avance 500-MHz spectrometer (Brucker, Billerica, USA) at 25 °C. 1H-NMR spectrum was referenced to residual HDO (δ = 4.79 ppm), and 13C-NMR spectrum was referenced to sodium-3-trimethylsilylpropionate (TMSP, Cambridge Isotope Laboratories, Andover, USA) as internal standards, respectively.
Molecular weight determination
The molecular weight distribution of EPS was determined by using a gel permeation chromatography (GPC) system (P230 GPC, Yilite Analytical Instruments, Dalian, China) equipped with a refractive index detector Shodex RID-20A (Shimadzu, Kyoto, Japan) and a PL aquagel-OH MIXED column (7.5 mm × 300 mm, Agilent, Santa Clara, USA). The EPS was dissolved in a 0.1 M NaNO3 and filtered through a nylon membrane (0.45 μm) before injection. NaNO3 (0.1 M) was used as the mobile phase with a flow rate of 1.0 ml/min. The column oven was at 40 °C. Dextran standards (100.6~3693 kDa, American Polymer Standards Corp., Ohio, USA) were used to calculate the molecular weight of EPS via a calibration curve.
Time course of EPS production
The MS medium supplied with varied sucrose concentrations (50–180 g/L) was examined for their effects on EPS production. The biomass, levan yield, sucrose, and glucose were monitored during 80-h fermentation. Cell growth was monitored by assaying the optical densities at 600 nm (OD600) with a Lambda35 UV/Vis spectrophotometer (PerkinElmer, Waltham, USA). After biomass removal by centrifugation, the supernatant was used for determination of EPS and glucose produced during fermentation, and residual sucrose in the fermentation broth. EPS was obtained by adding two volumes of ethanol to the supernatant, and the precipitations were collected and freeze-dried and weighted. Then the purified EPS was further used for GPC analysis to evaluate the molecular weight distribution of EPS under different sucrose concentrations. Glucose concentration in the supernatant was determined by using a D-Glucose Assay Kit (GOPOD Format, Megazyme, Wicklow, Ireland). Sucrose concentration was determined with a Agilent 1100 high-performance liquid chromatography (HPLC) system, equipped with a refractive index detector and a Zorbax Carbohydrate Analysis Column (4.6 × 250 mm, Agilent, Santa Clara, USA).
Rheological analysis
The purified EPSs were dissolved in distilled water to give a 0.5 to 5% (w/v) solution. The viscosity of the solutions was measured with a Physica MCR101 viscometer (Anton Paar GmbH, Graz, Austria) with a plate geometry sensor (PP-50, 1.000 mm gap). About 3 ml sample was carefully added to the platform and left to equilibrate for 5 min before each test. The apparent viscosity of the EPS solutions was measured with shear rate from 0.001 to 1000 s−1 at 20 °C. To study the stability after heating and freezing, Plev solutions were heated to 100 °C for 1 h, or cooled to − 20 °C for 2 h, and then both returned to 20 °C for the measurement of apparent viscosity. Effect of temperature (in range of 5–95 °C) on the viscosity of EPS solution (3–5%, w/v) was determined at a shear rate 0.1 s−1. Effect of pH (in range of 3.0–11.0) on the viscosity of EPS solution (5%, w/v) was measured with shear rate from 0.001 to 1000 s−1 at 20 °C. Effect of salts on the apparent viscosity was evaluated with EPS solutions containing 0.2 M of NaCl, KCl, CaCl2, and MgCl2, respectively.
Animals and treatments
Male C57BL/6J mice (weight 18–22 g, 4 weeks old) were obtained from the Model Animal Research Centre of Nanjing University (Nanjing, China). All mice were raised under a constant temperature (25 °C) and 12-h light/dark cycle (lights on at 7:00 am and off at 7:00 pm) with free access to food and water. After 7 days of adaptive feeding (week 0), the mice were randomly divided into three groups (n = 6 per group): (1) the control group (CK) fed with normal diet; (2) the low-dose Plev (LP) group fed with normal diet containing 5% (w/w) Plev; (3) the high-dose Plev (HP) group fed with normal diet containing 10% (w/w) Plev.
During the 30 days of feeding, the body weight and food intake of each mouse were measured every 2 days. On day 30, fresh feces samples were collected in individual sterilized cages, immediately frozen in liquid nitrogen and then stored at − 80 °C. The mice were anesthetized with 2–3% isoflurane (Forene®, Abbott, Queenborough, Kent, England), sacrificed by cervical dislocation. Colon and caecum were carefully dissected, weighted, and immersed in liquid nitrogen before storage at − 80 °C. All experimental schemes were approved by the Animal Care and Use Committee at Nanjing University of Science and Technology, China.
Histopathological evaluation
The small and large intestines were fixed in 10% formalin solution, dehydrated, embedded in paraffin, and sliced into 5-μm-thick sections. The sections were stained using hematoxylin and eosin (HE) solution for histopathological examination with a light microscope (Nikon, Tokyo, Japan).
454 pyrosequencing
The fecal DNA was extracted by using a TIANamp Stool DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufacturer’s protocols. DNA concentration was monitored by Nanodrop 3300 spectrophotometer (Thermo Scientific, Waltham, USA), assessed by agarose gel electrophoresis and stored at − 20 °C. Amplification and sequencing of the V3-V4 region were completed by Genewiz Bio-pharm Technology Co., Ltd. (Suzhou, China). Briefly, 16S rDNA sequence from fecal DNA was amplified by PCR with the primers 5′-CCTACGGRRBGCASCAGKVRVGAAT-3′ and 5′-GGACTACNVGGGTWTCTAATCC-3′ targeting the V3-V4 hypervariable regions of Bacteria and Archaea. Indexed adapters were added to the ends of the 16S rDNA amplicons to generate indexed libraries for downstream NGS sequencing on Illumina.
Bioinformatic analysis
Quality trimming of raw sequence data was performed with a QIIME program suite (Caporaso et al. 2010). The reads overlapping > 20 bp were assembled and the sequence length was retained longer than 200-bp sequence. Operational taxonomic unit (OTU) alignment was carried out using VSEARCH clustering (1.9.6, https://github.com/torognes/vsearch) (Rognes et al. 2016) against the Silva database 132 (http://www.arb-silva.de) (Quast et al. 2013). A 97% similarity cutoff was set for OTU clustering. Taxonomic information of the OTUs was annotated using the Ribosomal Database Project (RDP) classifier (http://rdp.cme.msu.edu/classifier/classifier.jsp) (Cole et al. 2014). Alpha diversities (ACE, Chao1, Simpson, and Shannon index) were calculated using QIIME (Caporaso et al. 2010). The comparison between the microbial communities was performed using weighted UniFrac analysis (Lozupone and Knight 2005). Beta diversities are displayed by principal co-ordinates analysis (PCoA) based on the distance between the matrix Brary-Curtis. Unweighted Pair Group Method with Arithmetic mean (UPGMA) tree was built from beta diversity distance matrix.
Quantitative RT-PCR
Quantification of Lactobacillus, Bifidobacterium, and Clostridium cluster XIVa was performed using quantitative RT-PCR (qRT-PCR) with an ABI 7300 real-time PCR system (Applied Biosystems, Foster City, USA). Bacterial DNA and primers (Supplemental Table S1) were mixed in a final volume of 20 μl containing 1 × SYBR Green PCR master mix (Applied Biosystems, Foster City, USA). The amplification procedure was set as one cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. The results were expressed as the ratio between the DNA levels of the species to the V3 region expression levels to correct the difference of the total DNA concentration between individual samples (Zhou et al. 2014).
Sample preparation for metabolomic analysis
Fecal samples were extracted according to a previously described procedure. Briefly, 50 mg of fecal samples was mixed with 500 μl of phosphate buffer (0.1 M K2HPO4/NaH2PO4 = 4/1, pH = 7.4) containing 99.9% D2O, 0.002% (w/v) of TMSP, and 0.03% of NaN3 (w/v). After vortex mixing for about 30 s, the samples were subjected to a freeze-thaw cycle 3 times in liquid nitrogen and subsequently homogenized with a tissue lyser (QIAGEN, Hilden, Germany) at 20 Hz, 90 s, followed by centrifugation at 16000g for 10 min at 4 °C. Supernatants were collected and transferred into 5-mm nuclear magnetic resonance (NMR) tubes for NMR experiment.
NMR spectroscopy and data analysis
1H-NMR spectra of fecal extracts were recorded on a Bruker Avance III 500-MHz NMR spectrometer (Bruker Biospin, Rheinstetten, Germany). A standard nuclear overhauser enhancement spectroscopy (NOESY) pulse sequence (relaxation delay-90-ms-90-tm-90-acquire-FID) was applied to suppress residual water signals. 1H NMR spectra were recorded with 128 scans into 32 K data points over a spectral width of 10 kHz, and then were Fourier transformed by an exponential window function with a line broadening of 0.5 Hz.
1H NMR spectra were manually phased and baseline corrected by using Bruker Topspin 3.0 software, and then automatically exported to ASCII files using MestRenova (version 8.0.1, Mestrelab Research SL, Santiago de Compostela, Spain). The ASCII format files were imported into R software (http://cran.r-project.org/) for multivariate data analysis with an R-script developed in-house (De Meyer et al. 2008). Each spectrum was binned between 0.5 and 9.5 ppm. After removal of residual water signals (4.5–5.45 ppm), the remaining regions were normalized to unity for sample comparison. Data were subsequently mean-centered and Pareto-scaled before multivariate analysis. Metabolites were identified and by using Chenomx NMR Suite 8.1 (Chenomx Inc., Edmonton, Canada), aided by statistical total correlation spectroscopy techniques.
The metabolic differences were disclosed by a supervised OPLS-DA (orthogonal partial least squares discriminant analysis). Repeated 2-fold cross-validation and a permutation test were used to validate the models. Model quality was assessed by the goodness-of-fit parameter R2 and cross-validation predictive ability Q2, respectively. Colored loadings plot and S plot were constructed to reveal the variables contributed to group separation. Fold-change (FC) values of metabolites and the associated P values corrected by Benjamini-Hochberg adjusted method were calculated and listed in colored tables (Benjamini and Hochberg 1995). Then, a heatmap drawn by TBtools software was used to identify clustered features (Chen et al. 2018).
Determination of lactate and short-chain fatty acids
Fecal samples (0.1 g) were suspended in 10 ml distilled water, vortexed for 10 min in ice bath, treated with sonication for 5 min, and then centrifuged at 10,000g for 10 min at 4 °C. The supernatants were filtered through a 0.2-μm nylon syringe filter and stored at − 20 °C. The concentration of organic acids was determined with a high-pressure ion chromatography system (ICS-2100, Dionex, USA) equipped with an IonPac AS11-HC column (250 × 4 mm, Thermo Scientific, Waltham, USA) and a CD-25 conductive detector (Zhou et al. 2014). The conductivity of the eluent was monitored and calibrated with organic acid standards (lactic acid, acetic acid, propionic acid, and butyric acid). Short-chain fatty acid (SCFA) concentrations were calculated as mean micromoles per gram wet weight of feces.
Statistical analysis
SPSS 20.0 software (IBM, Armonk, USA), R package (V3.5.1) (http://cran.r-project.org), and GraphPad Prism version 5.0 (GraphPad Software, San Diego, USA) were used for statistical analysis. Correlation coefficient between gut microbiota and metabolites was evaluated using Pearson correlation analysis. Data are expressed as means ± SEM or box-whisker plots. P < 0.05 was considered statistically significant.
Nucleotide sequence accession number
The 16S rDNA gene sequence of strain FP01 was submitted to NCBI with a GenBank number MK641394.1. The 16S rRNA sequencing data of the mice fecal microbiota was deposited in the Sequence Read Archive (SRA) database under accession number PRJNA649839.
Results
Isolation and characterization of strain FP01
Initial screening of fructan-producing bacteria was conducted on PYS medium with sucrose (5%, w/v) as the carbon source. A total of 15 strains exhibiting mucous appearance on PYS agar were isolated. One strain designated FP01 possessing the highest yield of EPS on PYS liquid medium was selected. Colonies of strain FP01 were raised, circular, cream white, and about 3 mm and 5 mm in diameter on PYS agar and MS agar, respectively, after 3 days of incubation at 30 °C (Fig. 1a). FP01 was identified as Gram-positive, rod-shaped, aerobic bacteria and can grow at temperature 15–35 °C and pH 4.0–9.0. The physiological and biochemical characteristics of FP01 are listed in Supplemental Table S2. FP01 could utilize monosaccharide (glucose), disaccharides (lactose, sucrose, and maltose), and many polysaccharides (starch, konjac glucomannan, carrageenan, guar gum, locust bean gum, and Arabic gum). The strain Paenibacillus sp. FP01 has been deposited in the China Center for Type Culture Collection (Wuhan, China) under CCTCC accession number M2019349.
Characterization of strain FP01 and structure analysis of the EPS from FP01. a FP01 on PYS and MS agar. b Neighbor-joining tree of FP01 based on 16S rDNA gene sequencing. The scale bar corresponds to 0.001 substitutions per nucleotide position. GenBank accession numbers are in parentheses. c GC profile of monosaccharide standards (rhamnose, fructose, galactose, glucose, mannose) and EPS hydrolysates. d Fourier transform-infrared spectrum of EPS. e 1H-NMR spectrum. f 13C-NMR spectrum. TMSP was used as the internal standard (0 ppm)
The molecular identification by 16S rDNA gene analysis indicated that strain FP01 possesses the highest similarity (99%) to the genus Paenibacillus, falling within the branch encompassing Paenibacillus jamilae and Paenibacillus polymyxa members (Fig. 1b). Accordingly, this isolate was identified as Paenibacillus sp. FP01 due to the unambiguous assignment on the species level. The 16S rDNA gene sequence of Paenibacillus sp. FP01 was submitted to GenBank under accession number MK641394.1.
Structural identification of the EPS
The EPS from Paenibacillus sp. FP01 was easily dissolved in water with a milky white, opalescent appearance, similar with that of most reported bacterial fructans. The monosaccharide composition of purified EPS was identified by GC analysis after acid hydrolysis. The GC spectrum of the trimethylsilyl-O-glycoside derivatives exhibited only one peak with retention time corresponding to that of fructose (Fig. 1c), indicating that the EPS derived from Paenibacillus sp. FP01 is a fructose polymer, named Plev. GPC analysis was performed to determine the molecular weight of Plev. The number average molecular weight (Mn), weight average molecular weight (Mw), and polydispersity index (Mw/Mn) of Plev were estimated to be 1.14 × 106 Da, 3.11 × 106 Da, and 2.73, respectively (Supplemental Fig. S1).
The Fourier transform infrared (FT-IR) spectrum of Plev displayed a strong, wide band at the wavenumber of 3327.9 cm−1, corresponding to the hydroxyl (OH) stretching vibration of polysaccharide (Fig. 1d). The band at 2937.1 cm−1 was assigned to the carbon-hydrogen (C–H) stretching absorption. The band at 1640.5 cm−1 indicated the existence of bound water. The absorption bands around 1125.7 cm−1 and 1020.8 cm−1 were due to C–O–C and C–O–H stretching vibrations in pyranose or furanose. Absorption band around 930.2, 874.7, and 815.8 cm−1 were typical signals of furanoid rings, being attributed to the symmetrical stretching vibration of furanose, transverse vibration of methyne, and bending vibration of D-type C–H of furanose, respectively. This result confirmed that Plev is fructan-type polysaccharide.
The 1H-NMR spectrum of Plev polysaccharide exhibited seven proton signals at 4.19 (H-3), 4.12 (H-4), 3.96 (H-5), 3.90 (H-6′), 3.77 (H-1′), 3.69 (H-1″), and 3.56 (H-6″) ppm (Fig. 1e). No peak in the anomeric region (5.3 to 4.3 ppm) was observed, confirming that this polymer is composed of only fructose residues. The 13C-NMR spectrum showed six intense signals at 107.05, 83.14, 79.20, 78.06, 66.25, and 62.81 ppm (Fig. 1f). Signals at 107.05 and 83.14 were assigned to the β-configuration of anomeric carbon atom (C-2) and furanose (C-5), respectively. Signals at 79.20 and 78.06 were attributed to the oxymethinic groups of fructosyl ring (C-3 and C-4), and those at 66.25 and 62.81 are due to methylene groups (C-6 and C-1). According to the reported data, levan-type fructan exhibits relatively closed chemical shifts values for C-3, C-4, and C-5 groups, whereas inulin exhibits closed peaks of C-1 and C-6 (Bouallegue et al. 2020; Hammer and Morgenlie 1990; Han 1990; Han and Clarke 1990). The comparison of Plev with the published chemical shifts of levan indicated that Plev is a (2→6)-β-d-fructofuranan or levan-type fructan (Supplemental Table S3). Furthermore, 13C-NMR spectroscopy has been used to distinguish linear or branched fructans (Hammer and Morgenlie 1990). Compared with the spectra of linear bacterial levans, those of slightly branched levans possess a minor C-3 signal, which is approximately 0.2 ppm downfield from the dominating C-3 peak (Seymour et al. 1979). There was no obvious weak signal companying the C-3 signal and other minor C signals in the 13C-NMR spectrum of Plev, demonstrating that Plev is a linear levan.
Fermentative curves of FP01
The fermentative process of FP01 under varying sucrose concentrations (50–180 g/L) was monitored and is depicted in Fig. 2. With 5%, 10%, and 15% sucrose, the maximum biomass of FP01 was observed by 48-h fermentation, at which time FP01 was still at the logarithmic phase under 18% sucrose concentration (Fig. 2a). Levan biosynthesis was growth-associated and reached the maximum at the pre-stationary phase (Fig. 2b). A stepwise increase in the EPS yield with increased sucrose concentration was observed. With 5%, 10%, and 15% sucrose, the maximum yields of levan (24.2, 49.5, and 74.6 g/L, respectively) were obtained after 48-h fermentation. Continuing growth up to 80 h had no obvious increase or decrease effect on EPS yields. For concentration of sucrose up to 180 g/L, the highest yield (89.5 g/L) was obtained with prolonged fermentation time (62 h). During fermentation, sucrose in the culture liquid was gradually utilized, coupled with release of glucose (Fig. 2c, d). No fructose was detected in the medium during the whole fermentative period, and about 49.5 ± 0.2% conversion rate was attained for all sucrose concentrations, indicating that almost all of the fructoside released from sucrose was employed to synthesize levan.
Flow and viscoelastic behavior of Plev solutions
The rheological properties of Plev aqueous solutions were investigated under different concentrations, pH levels, temperatures, and metal salts. The apparent viscosity of Plev increased gradually with increasing concentration (Fig. 3a), indicating good thickening properties. A non-Newtonian shear-thinning behavior was observed for Plev at concentrations ≥ 3% (w/v), while a Newtonian-like behavior is exhibited at concentrations ≤ 2%. According to Hundschell and Wagemans (2019), the transition of levan from Newtonian-like to shear-thinning behavior is between the concentrations of 1% and 4%, which lies on the macromolecular structure of levan. Shear thinning is a beneficial behavior to food processing and production as it makes liquid foods easier to pump and thinner to chew and swallow (Chen et al. 2014).
Rheological properties of the levan Plev produced by Paenibacillus sp. FP01. a Double logarithmic plot of shear rate vs apparent viscosity at different concentrations of Plev at 20 °C. b Effect of heating pretreatment on the apparent viscosity of Plev solutions with shear rate from 0.001 to 1000 s−1. c Effect of freezing pretreatment on the apparent viscosity of Plev solutions with shear rate from 0.001 to 1000 s−1. d Effect of temperature on the apparent viscosity of Plev at shear rate of 0.1 s−1. e Flow curves for 5% Plev solutions with varying pH at 20 °C. f Effect of various salts on the apparent viscosity of 5% Plev solutions at 20 °C
The effect of temperature on the apparent viscosity of Plev solutions (3–5%, w/v) was determined. After exposure to 100 °C or − 20 °C, the apparent viscosity of Plev was determined at 20 °C. No obvious change in flow behavior of Plev was observed, indicating that Plev has good thermostability and freeze-thaw stability, which is favorable in product processing that requires heating and freezing (Fig. 3b, c). In the temperature range of 5 to 95 °C, Plev solutions showed a gradual decline in the apparent viscosity in answer to the increasing temperature (Fig. 3d), which is similar with many polysaccharides, including pectin (Chen et al. 2014) and galactomannans (Kök et al. 1999).
Figure 3 e presents the effect of pH levels on the flow behavior of Plev solutions (5%, w/v). No obvious changes on viscosity were observed over a wide range of pH 3–11, indicating that Plev have good acid- and alkali-stable properties. The effect of common univalent and divalent ions on the viscosity of Plev was also determined. As shown in Fig. 3 f, the addition of NaCl, KCl, CaCl2, and MgCl2 (0.2 M) slightly increased the viscosity of Plev significantly at low shear rates (0.001~1 s−1), whereas it had no influence on viscosity at higher shear rates (1~1000 s−1). The results suggested that Plev can be potentially used to thicken foods containing salts.
Effect of Plev on body weight, food intake, cecal weight, and intestine histomorphology
The body weight of each mouse in all groups was determined every 2 days. Compared with the CK group, Plev feeding in both low and high doses exhibited higher food intake and resulted in slightly higher body weight gain (Fig. 4a, b). The cecum weight and cecal contents’ weight of mice in LP and HP groups were also significantly higher than those of the control group (Fig. 4c, d), implying a potential beneficial effects of levan on gut health. A similar phenomenon has been reported for the mice feed with the EPS salecan (Zhou et al. 2014). Histopathological observations after 30 days of feeding showed no abnormalities in the small and large intestines of the mice in all groups (Fig. 4e). Neither mucosal layer disruption nor infiltration of inflammatory cells was observed, indicating that Plev has little effect on intestine histomorphology.
Food intake (a), body weight gain (b), cecal wall weight (c), cecal contents weight (d), and photomicrographs of small- and large-intestine sections with HE staining (e) from mice fed with normal diet or diet containing Plev. CK, control group fed with normal diet; LP, low-dose Plev group (5%, w/w); HP, high-dose Plev group (10%, w/w)
Structural changes of the fecal microbiota
The fecal samples from the CK and HP groups were used to evaluate the prebiotic activity of Plev. The effect of Plev on the gut microbiota was determined by 16S rDNA sequencing. Based on 97% similarity cutout, averages of 270 and 281 OTUs for bacterial diversity were obtained for the CK and HP groups, respectively, and 266 OTUs coexisted in both groups. No obvious differences were observed between two groups by common microbial alpha diversity analysis, indicating that the diversity and richness of the gut microbiota were not significantly changed by Plev intervention (Supplemental Fig. S2a). PCoA score plots based on the Unweight UniFrac algorithm exhibited the relationships between the gut microbiota communities of groups. A palpable clustering of the gut microbiota constitution for each group was observed, suggesting that supplementation of Plev induces substantial changes in gut microbiota composition (Supplemental Fig. S2b). Hierarchical clustering also showed the clustering of each group (Supplemental Fig. S2c), being consistent with the result of PCoA analysis.
Gut microbiota composition at the phylum and genus levels
At the phylum level, the dominant components of the gut microbiota were Bacteroidetes and Firmicutes. In Plev-fed mice, the relative abundance of Bacteroidetes showed a significant increase, while that of Firmicutes significantly decreased in comparison with that of control group (Fig. 5a). The Firmicutes/Bacteroidetes (F/B) ratio was notably decreased due to the supplementation of Plev in the diet (Fig. 5b).
Relative abundance of the microbiota communities. a Phyla level. b Firmicutes/Bacteroidetes (F/B) ratio. c Statistical charts of bacteria with significant difference at the genus level. d The relative abundance of Lactobacillus, Bifidobacterium, and Clostridium cluster XIVa by qRT-PCR. CK, control group (n = 6); HP, high-dose Plev feeding group (n = 6). Unpaired t test was used to assess the statistical significance. *P < 0.05; **P < 0.01
At the genus level, the relative abundance of several beneficial bacteria, including Ruminococcaceae UCG-014, [Eubacterium] coprostanoligenes_group, Prevotellaceae_NK3B31_group, and Anaerostipes was significantly increased. Contrarily, the abundance of potentially harmful bacteria, including Odoribacter, Enterorhabdus, Turicibacter, and Ruminiclostridium, was significantly downregulated after supplementation of Plev (P < 0.05) (Fig. 5c). Besides, slight increases were detected for Muribaculaceae_Unclassified, Alloprevotella, Faecalibaculum, Parasutterella, Eggerthellaceae_Unclassified, Bifidobacterium, and Ruminococcaceae UCG-003. No obvious change was observed for Lactobacillus, Roseburia, Ruminococcaceae UCG-013, and Ruminococcaceae UCG-009. Quantitative RT-PCR was performed to further confirm the abundance of probiotic bacteria Bifidobacterium, Lactobacillus, and butyrate-producing Clostridium cluster XIVa (Van den Abbeele et al. 2013). As shown in Fig. 5 d, a slight increase was detected for the levels of Bifidobacterium and Clostridium cluster XIVa, in accordance with the results of 16S rDNA metagenomic analysis. No obvious change was observed for the Lactobacillus level.
Feces metabolic profile
The metabolic changes in the fecal samples were assessed using 1H-NMR-based metabolomics. A total of 30 specific metabolites, predominantly organic acids (formate, lactate, acetate, propionate, butyrate, isobutyrate, valerate, isovalerate, succinate) and amino acids (alanine, arginine, glycine, leucine, tyrosine, aspartate), were identified (Table 1 and Supplemental Fig. S3). To obtain more details about the metabonomic shifts introduced by Plev supplementation, the 1H-NMR data of feces extracts were further analyzed using multivariate statistical analysis. Two distinct clusters were observed in the OPLS-DA score plots (Fig. 6a), in which the HP group was located in the right region, far away from the CK group, suggesting notably altered fecal metabolites between Plev-fed mice and normal mice. The S-plot exhibited contributions of divergent metabolites to the group separation. Further away from the center, more contributions provided (Fig. 6b). The color-coded loading plots based on the correlation coefficients visualized the changed metabolites in the HP group compared with the control group (Fig. 6c, d). In comparison with the CK group, significant increases (P < 0.05) of SCFAs (valerate, propionate, butyrate, and acetate) and slight increases of lactate and branched-chain fatty acids (isobutyrate, isovalerate) were observed in the HP group. The levels of some amino acids (arginine, glycine) and organic acids (phenylacetate, malonate, hippurate, formate) were decreased due to Plev intervention. The discriminating metabolites between CK and HP groups were further visualized by heatmap analysis and an apparent hierarchical clustering of CK and HP groups was observed (Fig. 6e).
OPLS-DA analysis of metabolic profiles between control and Plev-fed groups for feces. a Score plot. b S-plot points represent different variables (metabolites). c, d Color-coded coefficient loadings plots. Red and blue color represent metabolites that statistically significant or no statistically significant contributions to the separation of groups, respectively. Peaks in positive or negative status indicated a relatively decreased or increased metabolites level in HP group
Correlation analysis
The correlation analysis was conducted by calculating the Pearson correlation coefficient to investigate the interactions between gut microbiota and metabolic changes introduced by Plev supplementation. An apparent correlation between specific genus of gut bacteria and fecal metabolites is illustrated in Fig. 7. The abundance of Anaerostipes, Clostridiales vadinBB60 group, Eggerthellaceae_Unclassified, Muribaculaceae, Parasutterella Ruminococcaceae UCG-014, and [Eubacterium]_coprostanoligenes_group showed a positive correlation with SCFAs (acetate, propionate, butyrate, valerate) and branched-chain fatty acids (BCFAs) (isobutyrate, isovalerate). On the contrary, some other genera were negatively related to SCFAs (Turicibacter, Dubosiella, Enterorhabdus, Odoribacter, Desulfovibrio, and Ruminiclostridium). Besides, possible positive correlations between Odoribacter and arginine, and Desulfovibrio and formate, were also revealed.
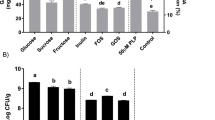
Quantitative analysis of lactate and SCFAs in feces
SCFAs in gut are one of the most critical microbial metabolites which have been acknowledged to be beneficial to host health, especially in host with an abnormal status. To confirm the effect of Plev on the concentrations of SCFAs, the lactate and SCFAs levels in feces were further quantified by ion chromatography. As shown in Fig. 8, the levels of lactate, acetate, propionate, and butyrate remarkably elevated in the HP group than those in the CK group, implying the beneficial effects of levan metabolism. The molar ratio between lactate, acetate, propionate, and butyrate was about 2.65:43.35:6.79:8.68 in fecal samples of the Plev-fed mice, and 1.78: 20.92:5.19:2.08 in the control group. The main fatty acid in fecal samples donated was acetate, followed by butyrate and propionate. These results confirmed that supplementation of Plev can elevate fecal SCFAs effectively.
Discussion
As a functional biopolymer with distinct physicochemical properties and multiple activities, levan has drawn extensive research attentions in recent decades. Due to the high production cost and limited levan producers, levan has only been explored in small quantities. In this study, a new Paenibacillus strain FP01 was isolated and identified as high-level levan producer. Compared with most reported bacterial levan producers (Table 2), Paenibacillus sp. FP01 was outstanding for its exceptionally high conversion rate of sucrose and high yield of levan. Besides, FP01 fermentation was performed in chemically defined medium with relatively low-cost inorganic nitrogen source (NaNO3) rather than commonly used organic nitrogen sources (yeast extract, peptone, etc.), thus could be employed as a potential strain for levan production in industrial scale.
The levan Plev produced by Paenibacillus sp. FP01 was structurally characterized as a linear levan with a high molecular weight (Mw) of 3.11 × 106 Da. Microbial levans from different bacteria have Mw varying from 104 to 1010 Da. Extremely high Mw of 2.466 × 109 Da was reported for the levan from Kozakia baliensis (Jakob et al. 2013). Most reported Bacillus and Paenibacillus species produce levan with Mw in the range of 105–107 Da (Gojgic-Cvijovic et al. 2019), except for the levan (2.826 × 104 Da) from Bacillus licheniformis 8-37-0-1 (Liu et al. 2010a). Plev solutions showed lower intrinsic viscosity than commonly used polysaccharide thickeners (xanthan, gellan gum, pectin, etc.) (Mahmood et al. 2017; Zhang et al. 2020). Nevertheless, the apparent viscosity of Plev is still higher than that of most plant-derived fructans. It should be noted that the viscosity of Plev could be improved by adding common salts including NaCl, KCl, CaCl2, and MgCl2, which was rarely reported for other levan-type fructans. Hundschell et al. (2020) reported that NaCl concentration plays a minor role for the intrinsic viscosity of levan from Gluconobacter albidus TMW 2.1191. The viscosity of levan from Zymomonas mobilis strain 113 “S” strain was only improved by high concentrations of NaCl (15–30%), whereas it may even be reduced by KCl (Vina et al. 1998). Good acid and alkali stability, thermostability, and freeze-thaw stability were also observed for Plev solutions. All these properties make Plev a potential fructan-type thickening agent in the food industry.
The prebiotic activities of inulin-type fructans have been comprehensively investigated, whereas that of levan-type fructans are rarely reported, except for some in vitro studies. Here, we evaluated the prebiotic activities of Plev with an in vivo mice model by using metagenomic sequencing. Significant changes of gut microbiota were observed at the phylum and genus levels. The Firmicutes/Bacteroidetes (F/B) ratio was notably decreased due to Plev intervention. The F/B ratio has been proposed as an eventual biomarker, and a higher F/B ratio in gut microbiota has been associated with obesity, type 2 diabetes, and cardiovascular diseases (Magne et al. 2020). At the genus level, a remarkable upregulation of beneficial bacteria (Ruminococcaceae UCG-014, [Eubacterium]_coprostanoligenes_group, Prevotellaceae_NK3B31_group, and Anaerostipes) and downregulation of potential harmful bacteria (Odoribacter, Enterorhabdus, Turicibacter, and Ruminiclostridium) were detected. It should be noted that no obvious change was observed for the Lactobacillus level. Kaarel et al. (2015) reported that in vitro fermentation of levan by fecal samples from a healthy human containing levan could not enhance the growth of Lactobacillus. A high-molecular-weight (HMW) levan-type fructan produced by Lactobacillus sanfranciscensis TMW1.392 also could not enhance the growth of Lactobacilli by in vitro cultivation (Bello et al. 2001). These results indicated that HMW levan could not stimulate the proliferation of Lactobacillus in guts.
SCFAs are one of the most critical microbial metabolites which have been acknowledged to be beneficial to host health, especially in hosts with an abnormal status. The gut beneficial effects of Plev were determined by 1H-NMR-based metabolomics and quantitative determination of fecal SCFAs. The levels of SCFAs were significantly increased in Plev-fed mice, confirming the prebiotic activities of Plev. Correlation analysis revealed a positive relevance between SCFA levels and potential SCFA-producing bacteria (Anaerostipes, Clostridiales vadinBB60 group, Parasutterella Ruminococcaceae UCG-014, [Eubacterium]_coprostanoligenes_group, etc.) (Allen-Vercoe et al. 2012; Bas-Bellver et al. 2020; Duncan et al. 2004). The effect of Turicibacter on SCFA production has not been revealed, but it has been reported that the abundance of Turicibacter was significantly upregulated in mice with dextran sulfate sodium–induced colitis (Wang et al. 2020). As a H2S producing bacterium, Desulfovibrio has been related with hypoplasia and hyperpermeability of intestinal epithelial cells (Rohr et al. 2019). Odoribacter was revealed to be responsible for production sulfonolipids which was notably increased in high-fat-fed mice (Walker et al. 2017). These findings indicated that the gut microbiota acts as a complex and sophisticated modulator of intestinal metabolite metabolism.
In conclusion, the present study established the potential of the new Paenibacillus isolate FP01 for cost-effective production of levan. Maximum production of levan was obtained at 30 °C after 62-h incubation with 180 g/L sucrose in chemically defined medium. The levan Plev from Paenibacillus sp. FP01 was a linear HMW levan with good rheological properties. Plev solutions were quite steady and exhibited relatively low intrinsic viscosities and specific non-Newtonian flow behavior, which are favorable characteristics for food processing and production. More importantly, Plev has outstanding prebiotic activities by regulating gut microbiota, stimulating SCFA production and thus improving the intestinal metabolism. Based on these results, Plev could be proposed as a promising biopolymer for large-scale applications in food, feed, and pharmaceutical industries.
Data availability
Sequence data that support the findings of this study have been deposited in GenBank with the primary accession code MK641394.1 and in the Sequence Read Archive (SRA) with the primary accession code PRJNA649839.
References
Abdel-Fattah AM, Gamal-Eldeen AM, Helmy WA, Esawy MA (2012) Antitumor and antioxidant activities of levan and its derivative from the isolate Bacillus subtilis NRC1aza. Carbohydr Polym 89(2):314–322. https://doi.org/10.1016/j.carbpol.2012.02.041
Abou-Taleb K, Abdel-Monem M, Yassin M, Draz A (2015) Production, purification and characterization of levan polymer from Bacillus lentus V8 strain. Br Microbiol Res J 5:22–32. https://doi.org/10.9734/BMRJ/2015/12448
Ağçeli GK, Cihangir N (2020) Synthesis, characterization and antimicrobial performance of novel nanostructured biopolymer film based on levan/clay/LL-37 antimicrobial peptide. Biocatal Agric Biotechnol 23:101421. https://doi.org/10.1016/j.bcab.2019.101421
Ahmed W, Rashid S (2019) Functional and therapeutic potential of inulin: a comprehensive review. Crit Rev Food Sci Nutr 59(1):1–13. https://doi.org/10.1080/10408398.2017.1355775
Allen-Vercoe E, Daigneault M, White A, Panaccione R, Duncan SH, Flint HJ, O'Neal L, Lawson PA (2012) Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976. Anaerobe 18(5):523–529. https://doi.org/10.1016/j.anaerobe.2012.09.002
Aramsangtienchai P, Kongmon T, Pechroj S, Srisook K (2020) Enhanced production and immunomodulatory activity of levan from the acetic acid bacterium, Tanticharoenia sakaeratensis. Int J Biol Macromol 163:574–581. https://doi.org/10.1016/j.ijbiomac.2020.07.001
Arvidson SA, Rinehart BT, Gadala-Maria F (2006) Concentration regimes of solutions of levan polysaccharide from Bacillus sp. Carbohydr Polym 65(2):144–149. https://doi.org/10.1016/j.carbpol.2005.12.039
Ates O (2015) Systems biology of microbial exopolysaccharides production. Front Bioeng Biotechnol 3:200. https://doi.org/10.3389/fbioe.2015.00200
Ávila-Fernández Á, Cuevas-Juárez E, Rodríguez-Alegría ME, Olvera C, López-Munguía A (2016) Functional characterization of a novel β-fructofuranosidase from Bifidobacterium longum subsp. infantis ATCC 15697 on structurally diverse fructans. J Appl Microbiol 121(1):263–276. https://doi.org/10.1111/jam.13154
Bas-Bellver C, Andrés C, Segui L, Barrera C, Jimenéz-Hernandéz N, Artacho A, Betoret N, Gosalbes MJ (2020) Valorisation of persimmon and blueberry by-products to obtain functional powders: in vitro digestion and fermentation by gut microbiota. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.0c02088
Bello FD, Walter J, Hertel C, Hammes WP (2001) In vitro study of prebiotic properties of levan-type exopolysaccharides from Lactobacilli and non-digestible carbohydrates using denaturing gradient gel electrophoresis. Syst Appl Microbiol 24(2):232–237. https://doi.org/10.1078/0723-2020-00033
Benigar E, Dogsa I, Stopar D, Jamnik A, Cigić IK, Tomšič M (2014) Structure and dynamics of a polysaccharide matrix: aqueous solutions of bacterial levan. Langmuir 30(14):4172–4182. https://doi.org/10.1021/la500830j
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Bouallegue A, Casillo A, Chaari F, La Gatta A, Lanzetta R, Corsaro MM, Bachoual R, Ellouz-Chaabouni S (2020) Levan from a new isolated Bacillus subtilis AF17: purification, structural analysis and antioxidant activities. Int J Biol Macromol 144:316–324. https://doi.org/10.1016/j.ijbiomac.2019.12.108
Cai G, Wu D, Li X, Lu J (2020) Levan from Bacillus amyloliquefaciens JN4 acts as a prebiotic for enhancing the intestinal adhesion capacity of Lactobacillus reuteri JN101. Int J Biol Macromol 146:482–487. https://doi.org/10.1016/j.ijbiomac.2019.12.212
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Chen C, Xia R, Chen H, He Y (2018) TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv:289660. https://doi.org/10.1101/289660
Chen Y, Zhang J-G, Sun H-J, Wei Z-J (2014) Pectin from Abelmoschus esculentus: optimization of extraction and rheological properties. Int J Biol Macromol 70:498–505. https://doi.org/10.1016/j.ijbiomac.2014.07.024
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(D1):D633–D642. https://doi.org/10.1093/nar/gki038
Dahech I, Belghith KS, Hamden K, Feki A, Belghith H, Mejdoub H (2011a) Antidiabetic activity of levan polysaccharide in alloxan-induced diabetic rats. Int J Biol Macromol 49(4):742–746. https://doi.org/10.1016/j.ijbiomac.2011.07.007
Dahech I, Belghith KS, Hamden K, Feki A, Belghith H, Mejdoub H (2011b) Oral administration of levan polysaccharide reduces the alloxan-induced oxidative stress in rats. Int J Biol Macromol 49(5):942–947. https://doi.org/10.1016/j.ijbiomac.2011.08.011
Dahech I, Harrabi B, Hamden K, Feki A, Mejdoub H, Belghith H, Belghith KS (2013) Antioxidant effect of nondigestible levan and its impact on cardiovascular disease and atherosclerosis. Int J Biol Macromol 58:281–286. https://doi.org/10.1016/j.ijbiomac.2013.04.058
De Meyer T, Sinnaeve D, Van Gasse B, Tsiporkova E, Rietzschel ER, De Buyzere ML, Gillebert TC, Bekaert S, Martins JC, Van Criekinge W (2008) NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem 80(10):3783–3790. https://doi.org/10.1021/ac7025964
de Oliveira MR, da Silva RSSF, Buzato JB, Celligoi MAPC (2007) Study of levan production by Zymomonas mobilis using regional low-cost carbohydrate sources. Biochem Eng J 37(2):177–183. https://doi.org/10.1016/j.bej.2007.04.009
Djurić A, Gojgić-Cvijović G, Jakovljević D, Kekez B, Kojić JS, Mattinen ML, Harju IE, Vrvić MM, Beškoski VP (2017) Brachybacterium sp. CH-KOV3 isolated from an oil-polluted environment-a new producer of levan. Int J Biol Macromol 104(Pt A):311–321. https://doi.org/10.1016/j.ijbiomac.2017.06.034
Dong C-X, Zhang L-J, Xu R, Zhang G, Zhou Y-B, Han X-Q, Zhang Y, Sun Y-X (2015) Structural characterization and immunostimulating activity of a levan-type fructan from Curcuma kwangsiensis. Int J Biol Macromol 77:99–104. https://doi.org/10.1016/j.ijbiomac.2015.03.009
Dos Santos LF, De Melo FBC, Paiva WM, Borsato D, Da Silva MCC, Celligoi MPC (2013) Characterization and optimization of levan production by Bacillus subtilis NATTO. Rom Biotechnol Lett 18(4):8413–8422. https://doi.org/10.3103/S0891416813030026
Duncan SH, Louis P, Flint HJ (2004) Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70(10):5810–5817. https://doi.org/10.1128/aem.70.10.5810-5817.2004
Erkorkmaz BA, Kırtel O, Ateş Duru Ö, Toksoy Öner E (2018) Development of a cost-effective production process for Halomonas levan. Bioprocess Biosyst Eng 41(9):1247–1259. https://doi.org/10.1007/s00449-018-1952-x
Feng J, Gu Y, Han L, Bi K, Quan Y, Yang C, Zhang W, Cao M, Wang S, Gao W, Sun Y, Song C (2015a) Construction of a Bacillus amyloliquefaciens strain for high purity levan production. FEMS Microbiol Lett 362(11):fnv079. https://doi.org/10.1093/femsle/fnv079
Feng J, Gu Y, Quan Y, Zhang W, Cao M, Gao W, Song C, Yang C, Wang S (2015b) Recruiting a new strategy to improve levan production in Bacillus amyloliquefaciens. Sci Rep 5(1):13814. https://doi.org/10.1038/srep13814
Ferreira J, Santos VAQ, Calegari GC, Cruz CHG (2019) Assessment of levan production from sucrose by Zymomonas mobilis CCT 4494. Orbital: Electron J Chem 11(6):394–398. https://doi.org/10.17807/orbital.v11i6.1229
Gojgic-Cvijovic GD, Jakovljevic DM, Loncarevic BD, Todorovic NM, Pergal MV, Ciric J, Loos K, Beskoski VP, Vrvic MM (2019) Production of levan by Bacillus licheniformis NS032 in sugar beet molasses-based medium. Int J Biol Macromol 121:142–151. https://doi.org/10.1016/j.ijbiomac.2018.10.019
González-Garcinuño Á, Tabernero A, Sánchez-Álvarez JM, Galán MA, del Valle EMM (2017) Effect of bacteria type and sucrose concentration on levan yield and its molecular weight. Microb Cell Factories 16(1):91. https://doi.org/10.1186/s12934-017-0703-z
Grinev VS, Tregubova KV, Anis’kov AA, Sigida EN, Shirokov AA, Fedonenko YP, Yegorenkova IV (2020) Isolation, structure, and potential biotechnological applications of the exopolysaccharide from Paenibacillus polymyxa 92. Carbohydr Polym 232:115780. https://doi.org/10.1016/j.carbpol.2019.115780
Haddar A, Hamed M, Bouallegue A, Bastos R, Coelho E, Coimbra MA (2020) Structural elucidation and interfacial properties of a levan isolated from Bacillus mojavensis. Food Chem 128456. https://doi.org/10.1016/j.foodchem.2020.128456
Hammer H, Morgenlie S (1990) Classification of grass fructans by 13C NMR spectroscopy. Acta Chem Scand 44:158–160. https://doi.org/10.3891/acta.chem.scand.44-0158
Han J, Xu X, Gao C, Liu Z, Wu Z (2015) Levan-producing Leuconostoc citreum strain BD1707 and its growth in tomato juice supplemented with sucrose. Appl Environ Microbiol 82(5):1383–1390. https://doi.org/10.1128/aem.02944-15
Han YW (1990) Microbial levan. Adv Appl Microbiol 35:171–194. https://doi.org/10.1016/s0065-2164(08)70244-2
Han YW, Clarke MA (1990) Production and characterization of microbial levan. J Agric Food Chem 38(2):393–396. https://doi.org/10.1021/jf00092a011
Hundschell CS, Bäther S, Drusch S, Wagemans AM (2020) Osmometric and viscometric study of levan, β-lactoglobulin and their mixtures. Food Hydrocoll 101:105580. https://doi.org/10.1016/j.foodhyd.2019.10558
Hundschell CS, Wagemans AM (2019) Rheology of common uncharged exopolysaccharides for food applications. Curr Opin Food Sci 27:1–7. https://doi.org/10.1016/j.cofs.2019.02.011
Jakob F, Pfaff A, Novoa-Carballal R, Rübsam H, Becker T, Vogel RF (2013) Structural analysis of fructans produced by acetic acid bacteria reveals a relation to hydrocolloid function. Carbohydr Polym 92(2):1234–1242. https://doi.org/10.1016/j.carbpol.2012.10.054
Jathore NR, Bule MV, Tilay AV, Annapure US (2012) Microbial levan from Pseudomonas fluorescens: characterization and medium optimization for enhanced production. Food Sci Biotechnol 21(4):1045–1053. https://doi.org/10.1007/s10068-012-0136-8
Kök MS, Hill SE, Mitchell JR (1999) Viscosity of galactomannans during high temperature processing: influence of degradation and solubilisation. Food Hydrocoll 13(6):535–542. https://doi.org/10.1016/S0268-005X(99)00040-5
Kaarel A, Katrin T, Tiina T, Ksenia P, Marju P, Triinu V, Tiina AE, Signe A, Dipshikha C (2015) Levan enhances associated growth of Bacteroides, Escherichia, Streptococcus and Faecalibacterium in fecal microbiota. PLoS One 10(12):e0144042. https://doi.org/10.1371/journal.pone.0144042
Kazak Sarilmiser H, Ates O, Ozdemir G, Arga KY, Toksoy Oner E (2015) Effective stimulating factors for microbial levan production by Halomonas smyrnensis AAD6T. J Biosci Bioeng 119(4):455–463. https://doi.org/10.1016/j.jbiosc.2014.09.019
Kekez B, Gojgic-Cvijovic G, Jakovljevic D, Kojic JS, Markovic M, Beskoski V, Vrvic M (2015) High levan production by Bacillus licheniformis NS032 using ammonium chloride as the sole nitrogen source. Appl Biochem Biotechnol 175(6):3068–3083. https://doi.org/10.1007/s12010-015-1475-8
Lammerts van Bueren A, Saraf A, Martens EC, Dijkhuizen L (2015) Differential metabolism of exopolysaccharides from probiotic Lactobacilli by the human gut symbiont Bacteroides thetaiotaomicron. Appl Environ Microbiol 81(12):3973–3983. https://doi.org/10.1128/aem.00149-15
Liu C, Kolida S, Charalampopoulos D, Rastall RA (2020) An evaluation of the prebiotic potential of microbial levans from Erwinia sp. 10119. J Funct Foods 64:103668. https://doi.org/10.1016/j.jff.2019.103668
Liu C, Lu J, Lu L, Liu Y, Wang F, Xiao M (2010a) Isolation, structural characterization and immunological activity of an exopolysaccharide produced by Bacillus licheniformis 8–37–0-1. Bioresour Technol 101(14):5528–5533. https://doi.org/10.1016/j.biortech.2010.01.151
Liu J, Luo J, Ye H, Sun Y, Lu Z, Zeng X (2010b) Medium optimization and structural characterization of exopolysaccharides from endophytic bacterium Paenibacillus polymyxa EJS-3. Carbohydr Polym 79(1):206–213. https://doi.org/10.1016/j.carbpol.2009.07.055
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235. https://doi.org/10.1128/AEM.71.12.8228-8235.2005
Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R (2020) The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12(5):1474. https://doi.org/10.3390/nu12051474
Mahmood K, Kamilah H, Shang PL, Sulaiman S, Ariffin F, Alias AK (2017) A review: interaction of starch/non-starch hydrocolloid blending and the recent food applications. Food Biosci 19:110–120. https://doi.org/10.1016/j.fbio.2017.05.006
Mardo K, Visnapuu T, Vija H, Aasamets A, Viigand K, Alamäe T (2017) A highly active endo-levanase BT1760 of a dominant mammalian gut commensal Bacteroides thetaiotaomicron cleaves not only various bacterial levans, but also levan of timothy grass. PLoS One 12(1):e0169989. https://doi.org/10.1371/journal.pone.0169989
Molinari ML, Boiardi JL (2013) Levans production by Gluconacetobacter diazotrophicus. Electron J Biotechnol 16:7–7. https://doi.org/10.2225/vol16-issue3-fulltext-9
Morreale F, Benavent-Gil Y, Rosell CM (2019) Inulin enrichment of gluten free breads: interaction between inulin and yeast. Food Chem 278:545–551. https://doi.org/10.1016/j.foodchem.2018.11.066
Nasir A, Sattar F, Ashfaq I, Lindemann SR, Chen M-H, Van den Ende W, Ӧner ET, Kirtel O, Khaliq S, Ghauri MA, Anwar MA (2020) Production and characterization of a high molecular weight levan and fructooligosaccharides from a rhizospheric isolate of Bacillus aryabhattai. LWT 123:109093. https://doi.org/10.1016/j.lwt.2020.109093
Öner ET, Hernández L, Combie J (2016) Review of levan polysaccharide: from a century of past experiences to future prospects. Biotechnol Adv 34(5):827–844. https://doi.org/10.1016/j.biotechadv.2016.05.002
Ortiz-Soto ME, Porras-Domínguez JR, Seibel J, López-Munguía A (2019) A close look at the structural features and reaction conditions that modulate the synthesis of low and high molecular weight fructans by levansucrases. Carbohydr Polym 219:130–142. https://doi.org/10.1016/j.carbpol.2019.05.014
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596. https://doi.org/10.1093/nar/gks1219
Rütering M, Schmid J, Rühmann B, Schilling M, Sieber V (2016) Controlled production of polysaccharides-exploiting nutrient supply for levan and heteropolysaccharide formation in Paenibacillus sp. Carbohydr Polym 148:326–334. https://doi.org/10.1016/j.carbpol.2016.04.074
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, Parthasarathy S (2019) Negative effects of a high-fat diet on intestinal permeability: a review. Adv Nutr 11(1):77–91. https://doi.org/10.1093/advances/nmz061
Semjonovs P, Shakirova L, Treimane R, Shvirksts K, Auzina L, Cleenwerck I, Zikmanis P (2016) Production of extracellular fructans by Gluconobacter nephelii P1464. Lett Appl Microbiol 62(2):145–152. https://doi.org/10.1111/lam.12521
Semjonovs P, Zikmanis P (2007) An influence of levan on the fermentation of milk by a probiotic ABT-type starter. J Food Technol 5(2):123–130
Seymour FR, Knapp RD, Zweig JE, Bishop SH (1979) 13C-nuclear magnetic resonance spectra of compounds containing β-D-fructofuranosyl groups or residues. Carbohydr Res 72:57–69. https://doi.org/10.1016/S0008-6215(00)83923-3
Silbir S, Dagbagli S, Yegin S, Baysal T, Goksungur Y (2014) Levan production by Zymomonas mobilis in batch and continuous fermentation systems. Carbohydr Polym 99:454–461. https://doi.org/10.1016/j.carbpol.2013.08.031
Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL (2010) Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141(7):1241–1252. https://doi.org/10.1016/j.cell.2010.05.005
Srikanth R, Reddy CH, Siddartha G, Ramaiah MJ, Uppuluri KB (2015a) Review on production, characterization and applications of microbial levan. Carbohydr Polym 120:102–114. https://doi.org/10.1016/j.carbpol.2014.12.003
Srikanth R, Siddartha G, Sundhar Reddy CH, Harish BS, Janaki Ramaiah M, Uppuluri KB (2015b) Antioxidant and anti-inflammatory levan produced from Acetobacter xylinum NCIM2526 and its statistical optimization. Carbohydr Polym 123:8–16. https://doi.org/10.1016/j.carbpol.2014.12.079
Szwengiel A, Nkongha GL (2019) Influence of acid depolymerization parameters on levan molar mass distribution and its utilization by bacteria. Carbohydr Polym 206:371–379. https://doi.org/10.1016/j.carbpol.2018.11.029
Tamura K-i, Sanada Y, Tase K, Kawakami A, Yoshida M, Yamada T (2014) Comparative study of transgenic Brachypodium distachyon expressing sucrose: fructan 6-fructosyltransferases from wheat and timothy grass with different enzymatic properties. Planta 239(4):783–792. https://doi.org/10.1007/s00425-013-2016-8
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tohme S, Hacıosmanoğlu GG, Eroğlu MS, Kasavi C, Genç S, Can ZS, Toksoy Oner E (2018) Halomonas smyrnensis as a cell factory for co-production of PHB and levan. Int J Biol Macromol 118(Pt A):1238–1246. https://doi.org/10.1016/j.ijbiomac.2018.06.197
Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM, Van de Wiele T (2013) Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7(5):949–961. https://doi.org/10.1038/ismej.2012.158
Veerapandian B, Shanmugam SR, Varadhan S, Sarwareddy KK, Mani KP, Ponnusami V (2020) Levan production from sucrose using chicken feather peptone as a low cost supplemental nutrient source. Carbohydr Polym 227:115361. https://doi.org/10.1016/j.carbpol.2019.115361
Vina I, Karsakevich A, Gonta S, Linde R, Bekers M (1998) Influence of some physicochemical factors on the viscosity of aqueous levan solutions of Zymomonas mobilis. Acta Biotechnol 18(2):167–174. https://doi.org/10.1002/abio.370180214
Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell WZ, Haller D, Schmid M, Hartmann A, Schmitt-Kopplin P (2017) Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci Rep 7(1):11047. https://doi.org/10.1038/s41598-017-10369-z
Wang Y, Zhang N, Kan J, Zhang X, Wu X, Sun R, Tang S, Liu J, Qian C, Jin C (2019) Structural characterization of water-soluble polysaccharide from Arctium lappa and its effects on colitis mice. Carbohydr Polym 213:89–99. https://doi.org/10.1016/j.carbpol.2019.02.090
Wang Z, Liang Y, Yu J, Zhang D, Ren L, Zhang Z, Liu Y, Wu X, Liu L, Tang Z (2020) Guchang Zhixie Wan protects mice against dextran sulfate sodium-induced colitis through modulating the gut microbiota in colon. J Ethnopharmacol 260:112991. https://doi.org/10.1016/j.jep.2020.112991
Wu F-C, Chou S-Z, Shih I-L (2013) Factors affecting the production and molecular weight of levan of Bacillus subtilis natto in batch and fed-batch culture in fermenter. J Taiwan Inst Chem Eng 44(6):846–853. https://doi.org/10.1016/j.jtice.2013.03.009
Xu X, Gao C, Liu Z, Wu J, Han J, Yan M, Wu Z (2016) Characterization of the levan produced by Paenibacillus bovis sp. nov BD3526 and its immunological activity. Carbohydr Polym 144:178–186. https://doi.org/10.1016/j.carbpol.2016.02.049
Zhang H, Zhang F, Yuan R (2020) Chapter 13-Applications of natural polymer-based hydrogels in the food industry. In: Chen Y (ed) Hydrogels Based on Natural Polymers, 1st edn. Elsevier, Amsterdam, pp 357–410
Zhou M, Pu C, Xia L, Yu X, Zhu B, Cheng R, Xu L, Zhang J (2014) Salecan diet increases short chain fatty acids and enriches beneficial microbiota in the mouse cecum. Carbohydr Polym 102:772–779. https://doi.org/10.1016/j.carbpol.2013.10.091
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31600043) and the China Postdoctoral Science Foundation (grant number 2015M581803).
Author information
Authors and Affiliations
Contributions
RC and JFZ conceived and designed the project. LC and LW conducted the experiments. YZ and RC analyzed the data. RC wrote the original manuscript. JFZ and SMW critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical statement
This study was conducted in strict accordance with the recommendations in the guide for the Animal Care and Use Committee at Nanjing University of Science and Technology (NJUST-AC-05-1217). All efforts were made to minimize suffering of the mice.
Consent to participate
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 493 kb)
Rights and permissions
About this article
Cite this article
Cheng, R., Cheng, L., Zhao, Y. et al. Biosynthesis and prebiotic activity of a linear levan from a new Paenibacillus isolate. Appl Microbiol Biotechnol 105, 769–787 (2021). https://doi.org/10.1007/s00253-020-11088-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11088-8