Abstract
Streptococcus agalactiae is a major pathogen causing streptococcosis. To prevent and control this bacterial disease, antagonistic bacteria have become a new research hotspot. This study evaluated the probiotic potential of Bacillus velezensis LF01 strain, which is antagonistic to S. agalactiae. The active compounds produced by LF01 showed antimicrobial activity against a broad spectrum of fish pathogens, including S. agalactiae, Streptococcus iniae, Aeromonas hydrophila, Edwardsiella tarda, Edwardsiella ictaluri, Aeromonas schubertii, Aeromonas veronii, Aeromonas jandaei, and Vibrio harveyi. The antimicrobial compounds were heat stable, pH stable, UV stable, resistant to proteases, and could be stored for a long time. To evaluate the probiotic function of LF01 in Nile tilapia, juveniles were divided into three treatment groups: a control group, an interval feeding group, and a continuous feeding group. Tilapia fed with LF01-supplemented diets (1.0 × 109 CFU/g) showed significantly better growth performances than those of the control group (P < 0.05). Tilapia fed with LF01-supplemented diets significantly increased lysozyme (LZY) and superoxide dismutase (SOD) activities. The expression of three immune-related genes (C3, lyzc, and MHC-IIβ) was higher in the intestine, head kidney, and gill of tilapia from the continuous feeding group than in those from the control group (P < 0.05). Tilapia fed with LF01-supplemented diets showed remarkably improved survival rates after S. agalactiae infection, and analysis of their intestinal tract pathogens revealed that the abundance of Edwardsiella and Plesiomonas had significantly decreased compared with the control group. Our findings demonstrate that LF01 is an effective antagonist against various fish pathogens and has potential for controlling infections by Streptococcus spp. and other pathogens in tilapia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tilapia is one of the main farmed fish in tropical and subtropical regions worldwide. It has the advantages of palatable fish meat, fast growth, strong reproductive capacity, and wide adaptability (Watanabe et al. 2002). China has the largest tilapia production worldwide, reaching 1.62 million tons in 2018 (Xu et al 2019). With the increase of tilapia culture density and the consequent culture environment deterioration, healthy development of tilapia aquaculture has been plagued by a series of diseases. Tilapia streptococcosis caused by Streptococcus agalactiae has become one of the most important factors limiting the sustainable development of tilapia in the past decade, especially in China (Li et al. 2013). Additionally, bacterial diseases in tilapia infected with pathogens such as Aeromonas sp. (Dong et al. 2017), Aeromonas schubertii (Liu et al. 2018a), and Edwardsiella tarda (Nagy et al. 2018) can also cause a large number of tilapia deaths, resulting in huge economic losses.
At present, there are no safe and effective control measures for bacterial diseases in tilapia. The use of chemical drugs can easily lead to resistance of pathogenic bacteria, making the related diseases more difficult to control (Currie et al. 2014; Dunstan et al. 2013). Therefore, safe and efficient methods to prevent and control tilapia bacterial diseases have become key to the sustainable development of tilapia aquaculture. Vaccine immunization is an important and effective method to prevent and control fish diseases. However, there is no commercialized vaccine in China to control tilapia streptococcosis and other bacterial diseases.
Probiotics are defined as live microbial adjuncts that have a beneficial effect on the host. They can improve water quality, enhance the host’s resistance to pathogens, and inhibit the growth/reproduction of harmful bacteria by decomposing organic matter in the water, activating the host’s humoral and cellular immunity, and secreting antagonistic substances to inhibit pathogens (Martínez et al. 2012; Zorriehzahra et al. 2016). Their mechanisms of biological control include triggering induced systemic resistance, competition for nutrients and space, and the production of active compounds (Chen et al. 2009; Islam et al. 2012; Torres et al. 2016). The use of probiotics in the prevention and treatment of aquatic animal diseases can effectively overcome the shortcomings of antibiotics such as drug residues and drug resistance. Recently, screening antagonistic bacteria with secretion of antagonistic substances has become a new research hotspot for prevention and control of aquatic animal bacterial diseases.
Probiotics, including Bacillus and lactic acid bacteria, are widely applied in aquaculture (Afrilasari et al. 2016; Sanlar et al. 2017; Wang et al. 2017a). Bacillus velezensis is an important biological control agent that is widely used in plant and animal disease control (Chen et al. 2018; Jiang et al. 2018; Lim et al. 2017; Yi et al. 2018). However, limited information is available on the use of B. velezensis to prevent and control fish diseases in aquaculture. The probiotic strain LF01 (herein LF01) was isolated from Nile tilapia (Oreochromis niloticus) and was identified as B. velezensis based on physiological and biochemical characteristics and phylogenetic analysis in our previous study (Gao et al. 2019). In the present study, we (i) measured the in vitro antagonistic activity of LF01 against several fish pathogenic bacteria; (ii) extracted and determined the antimicrobial compounds from LF01, and investigated their temperature, pH, UV, protease, and storage stability; (iii) evaluated the probiotic function of LF01 in Nile tilapia; (iv) investigated streptococcosis resistance of tilapia after feeding with LF01.
Materials and methods
Strains and culture condition
The B. velezensis strain LF01 was isolated from tilapia and stored at our laboratory. This strain has been deposited in the Guangdong Microbial Culture Center (GDMCC) with deposit number 60344. Nine fish pathogenic bacteria including S. agalactiae (WC1535, GenBank accession No. CP016501.2) (Zhang et al. 2019), Streptococcus iniae (Sn03), Aeromonas hydrophila (Ca1701), Edwardsiella tarda (GD1701), Edwardsiella ictaluri (Pef1401), Aeromonas schubertii (WL1707), Aeromonas veronii (Ci1361), Aeromonas jandaei (Ip121), and Vibrio harveyi (JZL1401) were isolated from diseased fish. Strains WC1535, Sn03, and GD1701 were isolated from diseased tilapia, while WL1707, Ca1701, Pef1401, Ci1361, Ip121, and JZL1401 were isolated from diseased snakehead fish (Channa argus), crucian carp (Carassius auratus), Pelteobagrus fulvidraco, grass carp (Ctenopharyngodon idellus), Ictalurus punetaus, and Micropterus salmoides, respectively. LF01 and fish pathogens were grown in brain heart infusion (BHI) liquid/agar medium at 30 °C.
Extraction of antimicrobial compounds
As described by Chen et al. (2010), LF01 was cultured in BHI broth at 30 °C for 48 h, and then cells were separated by centrifugation (7500 r/min) for 20 min at 4 °C. The antimicrobial compounds were precipitated by adding HCl (1 mol/L) to the supernatant with a final pH of 2. The antimicrobial compounds in precipitates were collected by centrifugation (7500 r/min for 20 min), dissolved in 100 mL sterile phosphate-buffered saline (PBS, pH 7.4), and filtered through a 0.22-μm micro-filter (Life, USA) to obtain cell-free extracts. The extracts were divided into 1.5-mL centrifuge tubes and stored at − 80 °C until use.
Stability and activity of the antimicrobial compounds
Well diffusion agar assay (WDAA) was used to determine the antibacterial activity of active compounds produced by LF01 as the previously described (Ambas et al. 2015; Didinen et al. 2018). Briefly, nine fish pathogens were cultured in BHI broth at 30 °C, and then each of them was adjusted to 107 CFU/mL in BHI broth, and 100 μL of each bacterium were streaked on BHI agar plates. Plates were dried for 10 min and a well (6 mm in diameter) was made in the center of each plate. Finally, 60 μL cell-free extracts were added into each well, and the plates were incubated overnight at 30 °C. The extracts’ antibacterial activity was determined based on the inhibition zone after 24 h of incubation at 30 °C. The experiments were performed at least three times.
Thermal, acid-base, proteinase, UV (ultraviolet rays), and storage stability were also determined. The extracts were treated at temperatures of 30, 40, 50, 60, 70, 80, 90, 100, and 121 °C for 30 min in triplicate to determine the thermal stability; adjusted to pH 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, and 12.0 with 1 mol/L HCl or 1 mol/L NaOH in triplicate, stored at 4 °C overnight, and re-adjusted to pH 7.0 to determine the acid-base stability; added the final concentration of 1 mg/mL proteinase K/pepsin/trypsin, and put in a 37 °C water bath for 1 h (the control with the same volume of PBS) to determine proteinase stability; treated at UV for 5, 10, 15, and 20 min in triplicate to estimate UV stability; and stored at different temperatures (25, 4, − 20, and − 80 °C) for 1, 2, 4, 6, and 8 weeks to assess storage stability. Afterward, antibacterial activities against S. agalactiae and A. hydrophila of the crude extracts treated under different conditions were determined by WDAA method.
Time-kill assay
To further evaluate the antimicrobial characteristics of the active compounds, time-kill experiments were performed as previously described (Sopirala et al. 2010). The LF01 was fermented in 1 L BHI liquid and the supernatant was obtained by centrifugation. The crude extracts were precipitated by adding HCl and then centrifuged to collect them according to “Extraction of antimicrobial compounds.” The crude extracts were dissolved in 100 mL sterile BHI broth and filtered through 0.22-μm micro-filter (Life, USA) to obtain 10-fold concentration of cell-free extracts. The logarithmic-phase S. agalactiae WC1535 strain was added (in triplicate) into the 10-fold concentrated cell-free extract medium and BHI broth (as control) to adjust the final concentrations of WC1535 strain to 2.5 × 105 CFU/mL. Finally, the medium was incubated for 12 h with shaking at 30 °C. Aliquots were removed at 0, 1, 2, 3, 4, 6, 8, 10, and 12 h post incubation, serially diluted and plated on BHI agar for enumeration of viable colonies. The cell concentration of the control group was monitored by measuring the optical density of cultures at a wavelength of 600 nm (OD600).
Fish and experimental set-up
Juvenile tilapias (average weight approximately 4.0 g) were obtained from the National Tilapia Seed Farm (Guangzhou City, Guangdong Province, China). After being transferred to the laboratory, fish were acclimated to the experimental conditions (30 °C) in 600-L aquaria (1.3 × 0.8 × 0.6 m, L × W × H) equipped with an air pump and fed with the basal diet for 2 weeks before the experiment started.
Juveniles were randomly divided into 6 aquaria with water flowing evenly (600 L volume with 300 fish per aquarium) and assigned to three treatments: two aquaria with nonsupplemented control diets, four aquaria with LF01-supplemented diets at 1.0 × 109 CFU/g (Yi et al. 2018), including interval (two aquaria, fed with LF01 at 3-week intervals) and continuous (two aquaria, continuously fed with LF01) feeding groups. Fish were fed to apparent satiation twice daily (09:00 and 18:00) with floating micro-pellets at a rate of 3% body weight for 9 weeks. During the experimental period, the fish culturing conditions were as follows: water temperature 28.5 ± 1.5 °C, pH 7.0 ± 0.3, dissolved oxygen = 6.1 ± 0.5 mg/L, NH4+-N < 0.5 mg/L, and NO2-N < 0.05 mg/L. In order to maintain the water quality, approximately 20% of the water volume in each aquarium was exchanged with flowing water every day (Srisapoome and Areechon 2017). The control, interval, and continuous groups were defined as CK, JG, and LX, respectively.
Growth performance
At the end of the 9th week of feeding trial, fish were starved for 24 h before sampling. Forty fish in each group were individually weighed to evaluate the weight gain and specific growth rate. The spleen index (9 fish per group) assay was performed in triplicate. The growth parameters, spleen index, and survival rate (on the 6th and 9th week) were calculated with the following formulae:
Challenge test
On the 6th and 9th week of the experiment, 45 fish from each group were equally and randomly divided into three 200-L water glass tanks (N = 15, each group). Each fish in each group was intraperitoneally injected with WC1535 strain at a dose of 3.0 × 108 CFU/mL (Mian et al. 2009). Each group was performed in triplicate. The mortality and clinical signs such as swimming, appetite, and fish surface color were monitored daily in each tank for up to 7 days. The biocontrol ability of LF01 against S. agalactiae was evaluated by the relative percent survival (RPS) using the formula:
Nonspecific immune-related enzymes activities and biochemical indices
Six fish were taken randomly from each group at the end of the experiment (9 weeks) and anesthetized with MS-222 (ethyl 3-aminobenzoate methanesulfonate, Shanghai Macklin Biochemical Co., Ltd.) at the dose of 40 mg/L. Blood samples were collected by caudal venipuncture using syringes. The serum was isolated by centrifugation, and it was used for determining the lysozyme (LZY), superoxide dismutase (SOD), alkaline phosphatase (AKP), total proteins (TP), albumin (ALB), and globulin (GLO). The measurements of LZY and SOD were performed with commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. The other indices were assayed by enzymic procedures utilizing an automatic biochemical analyzer (Hitachi 7600, Tokyo, Japan).
RNA extraction and real-time quantitative PCR
The intestine, head kidney, and gill were sampled on the 6th and 9th week to analyze gene expression, including the immune-related genes such as lyzc (C-type lysozyme gene), complement C3, and MHC-IIβ (major histocompatibility complex class IIβ gene). Total RNA was extracted from the intestine, head kidney, and gill using TRIzol reagent (Life, USA). Total RNA purity and concentration were measured using electrophoresis on 1.0% agarose gels and a micro-volume spectrophotometer (OSE-260, Tiangen Biotech, Beijing, China) at an absorbance ratio of 260/280 nm. The total RNA was treated with DNase-I (Takara, Japan) to remove genomic DNA. Then, the extracted total RNA was reverse-transcribed to cDNA using the PrimeScript™ RT reagent Kit (Perfect Real Time, Takara) according to the manufacturer’s instructions.
Real-time qPCR was performed with TransStart Top Green qPCR SuperMix (TransGen Biotech, AQ141, Beijing, China) in a LightCycler® 96 Real Time PCR System (Roche, Basel, Switzerland). Primers are listed in Table 1 and the β-actin gene was used as an internal reference gene. The reaction was conducted in a total volume of 20 μL, containing 0.4 μL of forward and reverse primers, 1.0 μL cDNA template, 10 μL TransStart Top Green qPCR SuperMix (2×), and 8.2 μL RNase/DNase-free water. The relative mRNA expression levels of three target genes were calculated by using the 2-ΔΔCT method (Livak and Schmittgen 2001) and normalized with β-actin gene.
Illumina high-throughput sequencing of barcoded 16S rRNA genes and bioinformatics statistical analyses
After 9 weeks of experiment, 9 fish were collected from each group (i.e., CK, JG, and LX) and euthanized with overdose of MS-222. The foregut, midgut, and hindgut from the 9 fish in each group were dissected and pooled together (3 fish per pool). Bacterial DNA was extracted from the gut using the Hipure Soil DNA Mini Kit (Magen, Guangzhou, China) according to the manufacturer’s instructions, and stored at − 80 °C until use.
Next-generation sequencing library preparations and Illumina MiSeq sequencing were conducted at IGEbio, Inc. (Guangzhou, China). V4 hypervariable regions of prokaryotic 16S rDNA were selected for generating amplicons and subsequent taxonomy analysis (Wang et al. 2017b). The V4 hypervariable regions were PCR-amplified from the microbial genomic DNA using primers 515F (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTG(C/T)CAGC(A/C)GCCGCGGTAA-3′) and 806R (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGG(A/G/C/T)(A/C/G)
GGGT(A/T)TCTAA-3′). The PCR conditions were 94 °C for 4 min, followed by 21 cycles of 94 °C for 30 s, 58 °C for 30 s (annealing) and 72 °C for 30 s (elongation), and then 72 °C for 5 min (extension). Thirty nanograms of each purified PCR product were subjected to Illumina-based high-throughput sequencing (IGEbio, Inc, Guangzhou, China). The Illumina MiSeq raw data has been submitted to GenBank, and the BioSample accessions: KDDM00000000.
Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using QIIME bioinformatics pipeline, and chimeric sequences were identified and removed using UCHIME. Reads that were observed less than 2 times and reads with length < 150 bp were discarded. The remaining sequences were used in the final analysis. The phylogenetic affiliation of each 16S rDNA gene sequence was analyzed by RDP Classifier (version 2.2) against the SILVA database using a confidence threshold of 0.8. The taxonomic richness and diversity estimators were determined in each library. Rarefaction curves were created to determine whether sequencing depth was sufficient to cover the expected number of OTUs. Principal component analysis (PCA) and nonmetric multidimensional scaling (NMDS) were used to distinguish different microbial communities among different groups.
Statistical analysis
Experiments including antimicrobial activity, growth performance, immune parameters, expression levels of immune-related genes, and diversity of intestinal microbiota were performed in triplicate. Data were expressed as means ± SD (standard deviation) of three replicates. One-way analysis of variance (ANOVA) and Duncan’s multiple range tests were conducted using SPSS version 21.0 statistical software (SPSS, Inc., Chicago, IL, USA) to determine the significant differences (P < 0.05).
Results
Antimicrobial activity and stability
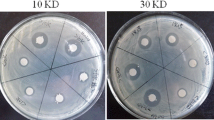
The crude extracts produced by LF01 excellently inhibited the growth of nine species of fish pathogens, and the inhibition effect showed the following decreasing order: A. hydrophila > S. iniae > E. tarda and A. jandae > A. schubertii > S. agalactiae, E. ictaluri, and A. veronii > V. harveyi (Fig. 1). Overall, the crude extracts inhibited Gram-negative fish pathogens better than Gram-positive pathogens. Moreover, the crude extracts significantly inhibited four common pathogens of tilapia in China (i.e., A. hydrophila, S. agalactiae, S. iniae, and E. tarda).
Antibacterial activity of the active compounds extracted from LF01 strain. Antagonistic effect of the active compounds (a). The antimicrobial ability of the active compounds against fish pathogens (b). WC1535, S. agalactiae ; Sn03, S. iniae; Ca1701, A. hydrophila; GD1701, E. tarda; Pef1401, E. ictaluri; WL1707, A. schubertii; Ci1361, A. veronii; Ip121, A. jandaei; JZL1401, V. harveyi
The antimicrobial compounds showed high thermal stability. The inhibition zone diameter was smaller (13.5 ± 0.3 mm) after treatment at 121 °C for 30 min than in the positive control (19.5 ± 0.5 mm). The extracts were resistant to enzyme digestion and UV radiation, as their activity showed no significant differences after treatment with proteases and decreased only slightly when exposed to the UV radiation for 10 min at a distance of 30 cm (Table 2). Additionally, the extracts still maintained high inhibition activity at pH 2 or 12, the relative antimicrobial activities were still as high as 84.16–88.12% and 85.58–88.46% for S. agalactiae and A. hydrophila, respectively.
In addition, the inhibitory activities of antimicrobial compounds at 25 °C and 4 °C showed a decrease with storage time, and the decrease was significantly larger at 25 °C than at 4 °C (P < 0.05). By the 8th week, the activity of antimicrobial compounds stored at 25 °C nearly disappeared (data not shown), and the inhibition zone diameter was less than 10 mm. However, high inhibitory activity still remained at the same time when stored at 4 °C. Furthermore, microbial compounds maintained high activity at − 20 and − 80 °C for a long time (> 8 weeks), indicating that they are resistant to low-temperature storage. Briefly, the antagonist substances tolerated well the temperature, UV, and acid-base stress, and were not sensitive to proteases, confirming that they were not proteins.
Time-kill assays
The time-kill curve of antimicrobial compounds against S. agalactiae was opposite to that in BHI broth (Fig. 2). In the case of S. agalactiae WC1535, the amount of viable WC1535 strain in antimicrobial compounds decreased by at least 2 log decades during the first 3 h of exposure, and no viable bacteria could be detected after 6-h incubation in the active compounds group. In BHI broth, after incubation for 0–6 h, S. agalactiae was in the growth lag phase and entered the log phase after 6 h with rapid proliferation. The result revealed that LF01 antimicrobial compounds could inhibit or kill this S. agalactiae strain.
Growth curves of S. agalactiae WC1535 strain cultured in BHI broth with and without the crude extracts. The cell-free extracts group was monitored according to the enumeration of viable colonies, whereas the control group was measured by the OD600 using the spectrophotometer. Time-kill assays were independently performed 3 times. Mean values of the triplicate measurements from a single experiment are plotted
Growth performance and survival rate
Effects of different feeding times on growth and survival rate of tilapia are presented in Table S1. The final weight, weight gain, spleen index, and specific growth rate of individuals fed with the probiotics diet in LX were significantly higher than those in CK after 9 weeks of feeding (P < 0.05), whereas no significant differences were observed between JG and LX at the same period (P > 0.05). Additionally, there was no significant difference in the survival rate between control and the treatment groups during the experimental period.
Resistance against S. agalactiae
The challenge test indicated that LF01-supplemented diet significantly enhanced tilapia resistance against S. agalactiae infection compared with the control diet (Fig. 3). After 6 weeks, tilapia exhibited a survival rate of 11.11%, 59.88%, and 64.44% in CK, JG, and LX groups, respectively. The RPSs of JG and LX were 54.87% and 60.71%, respectively, and significantly different between them. After 9 weeks, the fish fed with diets containing LF01 exhibited a survival rate of 70.83% and 79.17% in JG and LX groups, respectively, with an RPS of 58.82% and 70.59%, respectively. More importantly, the RPS of tilapia in LX was significantly higher than that of tilapia in JG (P < 0.05), suggesting that continuous feeding with LF01 could significantly improve disease resistance of tilapia compared with interval feeding. These results revealed that tilapia fed with an LF01 dietary supplement showed an improved resistance to streptococcosis.
Nonspecific immune-related enzymes activities and biochemical indices
The nonspecific immune-related enzyme activities and biochemical indices of tilapia are shown in Fig. 4. There was no significant difference in AKP, TP, ALB, and GLO among different groups at the end of trial (P > 0.05). However, there were significant differences in LZY and SOD between probiotic-fed group and the control group (P < 0.05). In addition, SOD in LX was significantly higher than that in JG (P < 0.05), whereas no significant differences in LZY were observed between JG and LX groups (P > 0.05).
The expression patterns of immune-related genes
The expression levels of immune-related genes including lyzc, C3, and MHC-IIβ in intestine, head kidney, and gill of tilapia are shown in Fig. 5. In the intestine, the relative expression levels of the three genes were significantly higher in JG and LX groups than in CK (P < 0.05). In the head kidney, the expression levels of C3 and lyzc were significantly higher in JG and LX than in CK (P < 0.05). After 6 weeks, there was no significant difference in the expression level of MHC-IIβ between LX and CK groups, whereas the expression level of MHC-IIβ was significantly higher in JG than in CK. In the gill, the expression levels of C3, lyzc, and MHC-IIβ were significantly higher in JG and LX than in CK. These results revealed that feeding LF01 significantly increased the expression of immune-related genes in tilapia.
Gene expression levels of lyzc (a–c), complement C3 (d–f), and MHC-IIβ (g–i) in the intestine, head kidney, and gill of tilapia after 6-week and 9-week treatments. Data are expressed as the mean fold changes (means ± SD, n = 3) from the 0-day group. Different superscript lowercase letters represent significant differences among the groups at the same time (P < 0.05)
Diversities and changes of microbial communities
Good’s coverage estimators for all groups were > 0.99, indicating that sufficient sampling depth was achieved for each sample. According to the species richness of OTUs in the sample, diversity, richness, and coverage estimations were calculated for all data sets. The Chao index and number of OTUs are estimators of phylotype richness, and the Shannon and Simpson indices of diversity reflect both the richness and community evenness. After 6 weeks, JG showed a higher richness (OTUs, 8649 and Chao1, 15,808) and diversity (Shannon = 6.117; Simpson = 0.963) than LX and CK (P < 0.05), suggesting that interval feeding could give rise to a higher level of biodiversity than the other treatments (Table S2). PCA showed that the samples in each group were closely clustered but there was a long distance among the three groups, especially at the end of the experiment, indicating that the species composition was different among groups. These results revealed that feeding LF01 could obviously affect the intestinal flora structure of tilapia (Fig. S1).
The information on classification and abundance of the OTU list per intestine sample was sorted at the phylum level (Fig. S2). After 6 weeks of feeding, CK group was dominated by Planctomycetes (35.42 %), while JG and LX were dominated by Fusobacteria, with 41.41% and 38.88% abundance, respectively (Table S3). After 9 weeks of feeding, Fusobacteria dominated in CK (64.97%) and JG (29.71%), whereas LX was dominated by Verrucomicrobia (30.47 %) and the abundance of Fusobacteria was lower than in the other groups (17.37 %) (Table S3). These results revealed that continuous feeding with LF01 could reduce the proportion of Fusobacteria in the intestinal tract. At the genus level, Cetobacterium were significantly less abundant in JG and LX than in CK after 9 weeks, and Candidatus Xiphinematobacter were extremely more abundant in LX than in CK (Fig. 6).
In addition, the proportions of Edwardsiella in the intestinal tracts of tilapia in JG (0.01%) and LX (0.22%) were significantly lower than that of tilapia in CK (2.33%) (P < 0.05) (Fig. 6). Plesiomonas abundance in the intestinal tracts of tilapia was also significantly lower in LX (0.70%) than in CK (2.69%) and JG (2.74%), whereas no significant difference was observed between CK and JG (P > 0.05) (Fig. 6). The results indicated that feeding a diet containing B. velezensis LF01 could lead to the accumulation of more beneficial microorganisms in the intestinal tract of tilapia, while inhibiting the growth of potentially pathogenic Edwardsiella and Plesiomonas.
Discussion
The use of antibiotics as growth promoters has been restricted in the European Union since 2006 (Castanon 2007). Thus, natural and safe feed additives have been considered as substitutes for antibiotics due to their positive effects on animal growth and their disease prevention and control abilities (Dimitroglou et al. 2011; Millet and Maertens 2011). Many bacterial antagonists to microbial pathogens belong to the genus Bacillus, and the number of important antagonists is increasing rapidly (Grady et al. 2019). Bacillus spp. offer several advantages for protection from pathogen infections owing to their ability to form endospores and the broad spectrum activity of their antibiotics. B. velezensis is a newly species of Bacillus named by Ruiz-García in 2005 (Ruiz-García et al. 2005). As an antagonistic probiotic bacterium, it has been reported in the prevention and control of bacterial and fungal diseases in plants, mammals, and aquatic animals. For example, B. velezensis JW strain was isolated from common carp with antimicrobial activity against a broad range of fish pathogenic bacteria (Yi et al. 2018). Additionally, tilapia fed with B. velezensis H3.1 showed beneficial effects on innate immunity, protection against infection, and growth performance (Doan et al. 2018). In this study, LF01 had a high capacity to inhibit nine species of fish pathogens. Subsequently, we determined the stability of LF01 active compounds and evaluated its potential value as a feed additive.
The antimicrobial compounds of LF01 purified by hydrochloric acid treatment had the ability to antagonize several fish pathogens and, based on the time-kill assays, the number of S. agalactiae strains decreased significantly after adding antimicrobial compounds compared with the control group. In addition, they have the ability to tolerate the acid, alkali, UV radiation, and proteases digestion, as well as being resistant to high temperature. Similar to results of this study, the active peptide produced by Bacillus amyloliquefaciens 6256 could tolerate the treatment of temperature, pH, protease K, and trypsin, and caused a sharp decline in the viable count for both Gram-positive and Gram-negative bacteria within a short time at a concentration of 2× minimum inhibitory concentration (MIC) (Prasad et al. 2018; Zhang et al. 2018). Our results suggested that LF01 inhibited the growth of fish pathogenic bacteria (or killed them) by producing antagonistic substances.
Bacillus spp. have previously been reported to improve growth and control disease in farmed fish (Gao et al. 2018; Liu et al. 2018b; Meidong et al. 2018). In this study, tilapia fed with added LF01 at a dose of 1.0 × 109 CFU/g in feed had significantly higher final weight, weight gain rate, spleen index, and specific growth rate than the control group at the end of the experiment. Furthermore, they could resist S. agalactiae infection according to the challenge test. The results were consistent with previous reports about probiotic dietary supplementation enhancing fish growth performance and disease resistance (Abarike et al. 2018; Hoseinifar et al. 2015; Tan et al. 2019; Wang et al. 2017a; Xia et al. 2018) and suggested that B. velezensis LF01 could be effective to promote the growth of tilapia, improve feed utilization rate, and increase resistance to streptococcosis.
Humoral components including serum and mucus-related immune substances (i.e., LZY, SOD, AKP, TP, GLO, proteases, and antiproteases) play an important role in host defense against pathogenic bacteria (Munir et al. 2016). LZY is an important defense molecule of the fish innate immune system because it can degrade peptidoglycan of the bacterial cell wall leading to rapid killing of Gram-positive organisms (Saurabh and Sahoo 2008). AKP is one of the most important enzymes for the growth and survival of organisms. It participates in the transmembrane transport of substances, ion secrection, protein synthesis, cartilage calcification, and immune defense (Sharma et al. 2014). Additionally, SOD, as an antioxidant enzyme, protects the host from oxidative stress. In this study, the activity of LZY and SOD in serum of tilapia was significantly increased by dietary supplementation with LF01, while AKP activity showed no significant change. According to previous reports, the activities of LZY and SOD in tilapia were enhanced by feeding with probiotics, such as Bacillus subtilis HAINUP40 (Liu et al. 2017), Bacillus licheniformis Dahb1 (Gobi et al. 2018), Rummeliibacillus stabekisii (Tan et al. 2019), and probiotics containing B. subtilis and B. licheniformis (Abarike et al. 2018). Additionally, serum proteins, albumin, and globulin play important roles in immune responses. Increases in serum protein, albumin, and globulin levels are thought to be associated with a stronger innate immune response in fish (Wiegertjes et al. 1996). However, there was no significant difference in total proteins, albumin, and globulin among treatments during the experiment. The serum LZY and SOD activities of tilapia increased significantly after 9-week dietary supplementation with LF01, which may be associated with the stronger microbial killing capacity of its macrophages (Giri et al. 2013) and higher Bacillus antigen-stimulated secretion of antioxidant enzymes and antioxidants (Zhang et al. 2013). These results showed that dietary supplementation with LF01 could improve the humoral immune factors of tilapia, such as LZY and SOD activity.
In this study, the anti-S. agalactiae effect of LF01 was remarkable. More importantly, tilapia fed with LF01 supplement showed obvious resistance to streptococcosis and RPSs ranging from 54.87 to 70.59%. Probiotics are able to modulate host immunity by stimulating the expression of innate immunity genes. Our study suggested that the expression levels of C3, lyzc, and MHC-IIβ were significantly upregulated in tilapia fed with LF01 compared with the control group. Similar results have been observed in tilapia, with a significant increase in lyzc expression in the intestines and the head kidney of tilapia fed with diets including B. subtilis and B. licheniformis (Abarike et al. 2018). According to previous reports, LZY and C3 play important roles in protecting fish from microbial invasion and other environmental stressors, and are widely distributed in many tissues (Bao et al. 2005; Saurabh and Sahoo 2008). Furthermore, major histocompatibility complex (MHC) class II molecules play important roles in the immune system of vertebrates (Zhou et al. 2013). Therefore, our results revealed that dietary LF01 significantly enhanced the expression of innate immunity genes, which may be attributable to the activation of nonspecific immune defense system in tilapia.
The intestinal tract is a complex system that plays an important role not only in digestion, absorption, and osmoregulation, but also in the defense against pathogens. Bacillus can affect the richness and diversity of commensal microbiota (Luis-Villaseñor et al. 2011). The dominant intestinal microbial community of sea cucumber can be modulated by B. subtilis 2-1, which can decrease the abundance and number of species of genus Vibrio, while increasing Psychrobacter and Bacillus genera abundance (Zhao et al. 2018). Our study revealed no significant difference in intestinal flora diversity between tilapia fed with or without LF01 after 9 weeks. However, tilapia in the interval feeding group, fed with LF01-supplement diet, did show the highest increase in diversity index at week 6. Interestingly, the number of fish pathogens Edwardsiella and Plesiomonas in the intestinal tract of tilapia fed with LF01 was significantly reduced, indicating that the opportunistic pathogens of the indigenous intestinal microbiota may be reduced by the probiotics supplement. There have been reports of Edwardsiella in O. niloticus (Nagy et al. 2018) and Scophthalmus maximus (Xiao et al. 2010), and Plesiomonas in tilapia (Yilmaz 2019) that confirmed Edwardsiella and Plesiomonas as pathogens. Although adding LF01 did not increase the abundance of Bacillus in tilapia intestine, putative pathogens such as Edwardsiella and Plesiomonas were significantly reduced. These results indicated that feeding tilapia with LF01 did not change the diversity of intestinal flora, but could reduce the abundance of certain pathogenic bacteria.
In conclusion, B. velezensis LF01 strain can antagonize various fish pathogenic bacteria by producing antimicrobial compounds and its active compounds were heat stable, pH stable, UV stable, and not sensitive to protease. Feeding tilapia with LF01 supplement diet significantly improved their growth and caused immune response stimulation, resistance to S. agalactiae infection, and a decrease of opportunistic pathogens in the intestinal tract. Therefore, LF01 can be used as a biocontrol agent to improve growth performance and diseases control in tilapia aquaculture.
References
Abarike ED, Cai J, Lu YS, Yu H, Chen LH, Jian JC, Tang JF, Jun L, Kuebutornye FKA (2018) Effects of a commercial probiotic BS containing Bacillus subtilis and Bacillus licheniformis on growth, immune response and disease resistance in Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol 82:229–238. https://doi.org/10.1016/j.fsi.2018.08.037
Afrilasari W, Widanarni, Meryandini A (2016) Effect of probiotic Bacillus megaterium PTB 1.4 on the population of intestinal microflora, digestive enzyme activity and the growth of Catfish (Clarias sp.). HAYATI J Biosci 23(4):168–172. https://doi.org/10.1016/j.hjb.2016.12.005
Ambas I, Buller N, Fotedar R (2015) Isolation and screening of probiotic candidates from marron, Cherax cainii(Austin, 2002) gastrointestinal tract (GIT) and commercial probiotic products for the use in marron culture. J Fish Dis 38(5):467–476. https://doi.org/10.1111/jfd.12257
Bao B, Peatman E, Li P, He CB, Liu ZJ (2005) Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specifc upregulation after bacterial infection. Dev Comp Immunol 29(11):939–950. https://doi.org/10.1016/j.dci.2005.03.006
Castanon JIR (2007) History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci 86(11):2466–2471. https://doi.org/10.3382/ps.2007-00249
Chen XH, Koumoutsi A, Scholz R, Borriss R (2009) More than anticipated-production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J Mol Microbiol Biotechnol 16(1-2):14–24. https://doi.org/10.1159/000142891
Chen H, Wang L, Su CX, Gong GH, Wang P, Yu ZL (2010) Isolation and characterization of lipopeptide antibiotics produced by Bacillus subtilis. Lett Appl Microbiol 47(3):180–186. https://doi.org/10.1111/j.1472-765X.2008.02412.x
Chen L, Heng JY, Qin SY, Bian K (2018) A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS One 13(6):e0198560. https://doi.org/10.1371/journal.pone.0198560
Currie J, Lin WC, Meng JJ (2014) Addressing antibiotic abuse in China: an experimental audit study. J Dev Econ 110:39–51. https://doi.org/10.1016/j.jdeveco.2014.05.006
Didinen BI, Onuk EE, Metin S, Cayli O (2018) Identification and characterization of lactic acid bacteria isolated from rainbow trout (Oncorhynchus mykiss, Walbaum 1792), with inhibitory activity against Vagococcus salmoninarum and Lactococcus garvieae. Aquac Nutr 24(1):400–407. https://doi.org/10.1111/anu.12571
Dimitroglou A, Merrifield DL, Carnevali O, Picchietti S, Avella M, Daniels C, Guroy D, Davies SJ (2011) Microbial manipulations to improve fish health and production-A mediterranean perspective. Fish Shellfish Immunol 30(1):1–16. https://doi.org/10.1016/j.fsi.2010.08.009
Doan HV, Hoseinifar SH, Khanongnuch C, Kanpiengjai A, Unban K, Kim VV, Srichaiyo S (2018) Host-associated probiotics boosted mucosal and serum immunity, disease resistance and growth performance of Nile tilapia (Oreochromis niloticus). Aquaculture 491:94–100. https://doi.org/10.1016/j.aquaculture.2018.03.019
Dong HT, Techatanakitarnan C, Jindakittikul P, Thaiprayoon A, Taengphu S, Charoensapsri W, Khunrae P, Rattanarojpong T, Senapin S (2017) Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J Fish Dis 40(10):1395–1403. https://doi.org/10.1111/jfd.12617
Dunstan RA, Heinz E, Wijeyewickrema LC, Pike RN, Purcell AW, Evans TJ, Praszkier J, Robins-Browne RM, Strugnell RA, Korotkov KV, Lithgow T (2013) Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel Pilotin AspS. PLoS Pathog 9(1):303–309. https://doi.org/10.1371/journal.ppat.1003117
Gao X, Zhang M, Li X, Han Y, Wu F, Liu Y (2018) Effects of a probiotic (Bacillus licheniformis) on the growth, immunity, and disease resistance of Haliotis discus hannai Ino. Fish Shellfish Immunol 76:143–152. https://doi.org/10.1016/j.fsi.2018.02.028
Gao YX, Zhang DF, Ke XL, Liu ZG, Yi MM, Wang M, Han XQ, Lu MX (2019) Selection and characteristics of intestinal Bacillus strain antagonistic against pathogenic Streptococcus agalactiae of tilapia. Acta Microbiol Sin 59(5):926–938. (In Chinese). https://doi.org/10.13343/j.cnki.wsxb.20180368
Giri SS, Sukumaran V, Oviya M (2013) Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish Immunol 34(2):660–666. https://doi.org/10.1016/j.fsi.2012.12.008
Gobi N, Vaseeharan B, Chen JC, Rekha R, Vijayakumar S, Anjugam M, Iswarya A (2018) Dietary supplementation of probiotic Bacillus licheniformis Dahb1 improves growth performance, mucus and serum immune parameters, antioxidant enzyme activity as well as resistance against Aeromonas hydrophila in tilapia Oreochromis mossambicus. Fish Shellfish Immunol 74:501–508. https://doi.org/10.1016/j.fsi.2017.12.066
Grady EN, MacDonald J, Ho MT, Weselowski B, Mcdowell T, Solomon O, Renaud J, Yuan ZC (2019) Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol 19(1):5. https://doi.org/10.1186/s12866-018-1380-8
Hoseinifar SH, Roosta Z, Hajimoradloo A, Vakili F (2015) The effects of Lactobacillus acidophilus as feed supplement on skin mucosal immune parameters, intestinal microbiota, stress resistance and growth performance of black swordtail (Xiphophorus helleri). Fish Shellfish Immunol 42(2):533–538. https://doi.org/10.1016/j.fsi.2014.12.003
Islam MR, Jeong YT, Lee YS, Song CH (2012) Isolation and identification of antifungal compounds from Bacillus subtilis C9 inhibiting the growth of plant pathogenic fungi. Mycobiology 40(1):59–65. https://doi.org/10.5941/MYCO.2012.40.1.059
Jiang CH, Liao MJ, Wang HK, Zheng MZ, Xu JJ, Guo JH (2018) Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol Control 126:147–157. https://doi.org/10.1016/j.biocontrol.2018.07.017
Li LP, Wang R, Liang WW, Gan X, Huang T, Huang Y, Li J, Shi YL, Chen M, Luo HL (2013) Rare serotype occurrence and PFGE genotypic diversity of Streptococcus agalactiae isolated from tilapia in China. Vet Microbiol 167(3-4):719–724. https://doi.org/10.1016/j.vetmic.2013.09.001
Lim SM, Yoon MY, Choi GJ, Choi YH, Jang KS, Shin TS, Park HW, Yu NH, Kim YH, Kim JC (2017) Diffusible and volatile antifungal compounds produced by an antagonistic Bacillus velezensis G341 against various phytopathogenic fungi. Plant Pathol J 33(5):488–498. https://doi.org/10.5423/ppj.oa.04.2017.0073
Liu H, Wang S, Cai Y, Guo X, Cao Z, Zhang Y, Liu S, Yuan W, Zhu W, Zheng Y, Xie Z, Guo W, Zhou Y (2017) Dietary administration of Bacillus subtilis HAINUP40 enhances growth, digestive enzyme activities, innate immune responses and disease resistance of tilapia, Oreochromis niloticus. Fish Shellfish Immunol 60:326–333. https://doi.org/10.1016/j.fsi.2016.12.003
Liu C, Chang OQ, Zhang DF, Li KB, Wang F, Lin MH, Shi CB, Jiang L, Wang Q, Bergmann SM (2018a) Aeromonas shuberti as a cause of multi-organ necrosis in internal organs of Nile tilapia, Oreochromis niloticus. J Fish Dis 41:1529–1538. https://doi.org/10.1111/jfd.12848
Liu CH, Wu K, Chu TW, Wu TM (2018b) Dietary supplementation of probiotic, Bacillus subtilis E20, enhances the growth performance and disease resistance against Vibrio alginolyticus in parrot fish (Oplegnathus fasciatus). Aquac Int 26(1):63–74. https://doi.org/10.1007/s10499-017-0189-z
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Luis-Villaseñor IE, Macías-Rodríguez ME, Gómez-Gil B, Ascencio-Valle F, Campa-Córdov AI (2011) Beneficial effects of four Bacillus strains on the larval cultivation of Litopenaeus vannamei. Aquaculture 321(1-2):136–144. https://doi.org/10.1016/j.aquaculture.2011.08.036
Martínez CP, Ibáñez AL, Monroy Hermosillo OA, Ramirez SHC (2012) Use of probiotics in aquaculture. ISRN Microbiol 2012:1–13. https://doi.org/10.5402/2012/916845
Meidong R, Khotchanalekha K, Doolgindachbaporn S, Nagasawa T, Nakao M, Sakai K, Tongpim S (2018) Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellfish Immunol 73:1–10. https://doi.org/10.1016/j.fsi.2017.11.032
Mian GF, Godoy DT, Leal CAG, Yuhara TY, Costa GM, Figueiredo HCP (2009) Aspects of the natural history and virulence of S. agalactiae infection in Nile tilapia. Vet Microbiol 136(1):180–183. https://doi.org/10.1016/j.vetmic.2008.10.016
Millet S, Maertens L (2011) The European ban on antibiotic growth promoters in animal feed: from challenges to opportunities. Vet J 187(2):143–144. https://doi.org/10.1016/j.tvjl.2010.05.001
Munir MB, Hashim R, Chai YH, Marsh TL, Nor SAM (2016) Dietary prebiotics and probiotics influence growth performance, nutrient digestibility and the expression of immune regulatory genes in snakehead (Channa striata) fingerlings. Aquaculture 460:59–68. https://doi.org/10.1016/j.aquaculture.2016.03.041
Nagy E, Fadel A, Almoghny FA, Ibrahim MS (2018) Isolation, Identification and pathogenicity characterization of Edwardsiella tarda isolated from Oreochromis niloticus fish farms in Kafr-Elshiekh, Egypt. Alex J Vet Sci 57(1):171–179. https://doi.org/10.5455/ajvs.294237
Prasad S, Saleshier MF, Krishnan S, Bharathi P (2018) Synthesis, spectroscopic studies, antibacterial activity, and colorimetric evaluation of the time-kill assay for newly synthesized chalcones using resazurin. Pharm Chem J 52(6):518–525. https://doi.org/10.1007/s11094-018-1852-z
Qiang J, He J, Yang H, Xu P, Habte-Tsion HM, Ma XY, Zhu ZX (2016) The changes in cortisol and expression of immune genes of GIFT tilapia Oreochromis niloticus (L.) at different rearing densities under Streptococcus iniae infection. Aquac Int 24(5):1365–1378. https://doi.org/10.1007/s10499-016-9995-y
Ruiz-García C, Béjar V, Martínez-Checa F, Llamas I, Quesada E (2005) Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int J Syst Evol Microbiol 55(1):191–195. https://doi.org/10.1099/ijs.0.63310-0
Sanlar H, Philip B, Philip R, Singh ISB (2017) Effect of probiotics on digestive enzyme activities and growth of cichlids, Etroplus suratensis (Pearl spot) and Oreochromis mossambicus (Tilapia). Aquac Nutr 23(4):852–864. https://doi.org/10.1111/anu.12452
Saurabh S, Sahoo PK (2008) Lysozyme: an important defence molecule of fish innate immune system. Aquac Res 39(3):223–239. https://doi.org/10.1111/j.1365-2109.2007.01883.x
Sharma U, Pal D, Prasad R (2014) Alkaline phosphatase: an overview. Indian J Clin Biochem 29(3):269–278. https://doi.org/10.1007/s12291-013-0408-y
Sopirala MM, Mangino JE, Gebreyes WA, Biller B, Bannerman T, Balada-Llasat JM, Pancholi P (2010) Synergy testing by Etest, microdilution checkerboard, and time-kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 54(11):4678–4683. https://doi.org/10.1128/AAC.00497-10
Srisapoome P, Areechon N (2017) Efficacy of viable Bacillus pumilus isolated from farmed fish on immune responses and increased disease resistance in Nile tilapia (Oreochromis niloticus): Laboratory and on-farm trials. Fish Shellfish Immunol 67:199–210. https://doi.org/10.1016/j.fsi.2017.06.018
Tan HY, Chen SW, Hu SY (2019) Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol 92:265–275. https://doi.org/10.1016/j.fsi.2019.06.027
Torres MJ, Brandan CP, Petroselli G, Erra-Balsells R, Audisio MC (2016) Antagonistic effects of Bacillus subtilis subsp. subtilis and B. amyloliquefaciens against Macrophomina phaseolina: SEM study of fungal changes and UV-MALDI-TOF MS analysis of their bioactive compounds. Microbiol Res 182:31–39. https://doi.org/10.1016/j.micres.2015.09.005
Wang M, Liu GB, Lu MX, Ke XL, Liu ZG, Gao FY, Cao JM, Zhu HP, Yi MM, Yu DG (2017a) Effect of Bacillus cereus as a water or feed additive on the gut microbiota and immunological parameters of Nile tilapia. Aquac Res 48(6):3163–3173. https://doi.org/10.1111/are.13146
Wang XT, Sun YX, Wang LL, Li XY, Qu KL, Xu YP (2017b) Synbiotic dietary supplement affects growth, immune responses and intestinal microbiota of Apostichopus japonicas. Fish Shellfish Immunol 68:232–242. https://doi.org/10.1016/j.fsi.2017.07.027
Watanabe WO, Losordo TM, Fitzsimmons K, Hanley F (2002) Tilapia production systems in the Americas: technological advances, trends, and challenges. Rev Fish Sci 10(3-4):465–498. https://doi.org/10.1080/20026491051758
Wiegertjes GF, Stet RJM, Parmentier HK, Muiswinkel WBV (1996) Immunogenetics of disease resistance in fish: a comparative approach. Dev Comp Immunol 20(6):365–381. https://doi.org/10.1016/S0145-305X(96)00032-8
Xia Y, Lu M, Chen G, Cao JM, Gao FY, Wang M, Liu ZG, Zhang DF, Zhu HP, Yi MM (2018) Effects of dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp. lactis JCM5805 on the growth, intestinal microbiota, morphology, immune response and disease resistance of juvenile Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol 76:368–379. https://doi.org/10.1016/j.fsi.2018.03.020
Xiao JF, Wang QY, Liu Q, Wang X, Liu H, Zhang YX (2010) Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquac Res 40(1):13–17. https://doi.org/10.1111/j.1365-2109.2008.02101.x
Xu LJ, Wu FX, Guo Y, Gao HQ, Yu HS, Wang Q, Wang JX, Wang SH, Wang XH, Zhan Y, Yu Y, Zhu M, Zhu AF, Zhu JX, Liu X, Liu W, Liu XW, Liu LM, An YY, Zhu ZH, Li J, Li SB, Li YP, Li XT, Yang YM, Yang CJ, Yang GS, Yang JB, Wu SS, Wu XQ, He Y, Zou GH, Sha S, Zhang L, Zhang SY, Zhang XA, Chen Y, Chen ZY, Ou HQ, Pang FM, Meng H, Zhao XY, Jiang ZY, Xia Y, Qian YL, Cao Y, Cao ZH, Wen B, Lei X (2019) China Fishery Statistics Yearbook. China Agriculture Press, Beijing. (In Chinese)
Yi Y, Zhang Z, Zhao F, Liu H, Yu LJ, Zha JW, Wang GX (2018) Probiotic potential of Bacillus velezensis JW: antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol 78:322–330. https://doi.org/10.1016/j.fsi.2018.04.055
Yilmaz S (2019) Effects of dietary blackberry syrup supplement on growth performance, antioxidant, and immunological responses, and resistance of Nile tilapia, Oreochromis niloticus to Plesiomonas shigelloides. Fish Shellfish Immunol 84:1125–1133. https://doi.org/10.1016/j.fsi.2018.11.012
Zhang CN, Li XF, Xu WN, Jiang GZ, Lu KL, Wang LN, Liu WB (2013) Combined effects of dietary fructooligosaccharide and Bacillus licheniformis on innate immunity, antioxidant capability and disease resistance of triangular bream (Megalobrama terminalis). Fish Shellfish Immunol 35(5):1380–1386. https://doi.org/10.1016/j.fsi.2013.07.047
Zhang QX, Zhang Y, He LL, Ji ZL, Tong YH (2018) Identification of a small antimycotic peptide produced by Bacillus amyloliquefaciens 6256. Pestic Biochem Physiol 150:78–82. https://doi.org/10.1016/j.pestbp.2018.07.006
Zhang D, Ke X, Liu Z, Cao J, Su Y, Lu M, Gao F, Wang M, Yi M, Qin F (2019) Capsular polysaccharide of Streptococcus agalactiae is an essential virulence factor for infection in Nile tilapia (Oreochromis niloticus Linn.). J Fish Dis 42:293–302. https://doi.org/10.1111/jfd.12935
Zhao YC, Yuan L, Wan JL, Sun HS, Wang YY, Zhang Q (2018) Effects of a potential autochthonous probiotic Bacillus subtilis 2-1 on the growth and intestinal microbiota of juvenile sea cucumber, Apostichopus japonicus Selenka. J Ocean U China 17(2):363–370. https://doi.org/10.1007/s11802-018-3402-4
Zhou FN, Dong ZD, Fu Y, Li TM, Zeng YQ, Ji XS, Chen WY, Zhang J, Wang H (2013) Molecular cloning, genomic structure, polymorphism and expression analysis of major histocompatibility complex class II B gene of Nile tilapia (Oreochromis niloticus). Aquaculture 372(1):149–157. https://doi.org/10.1016/j.aquaculture.2012.10.032
Zorriehzahra MJ, Delshad ST, Adel M, Tiwari R, Karthik K, Dhama K, Lazado CC (2016) Probiotics as beneficial microbes in aquaculture: an update on their multiple modes of action: a review. Vet Q 36(4):228–241. https://doi.org/10.1080/01652176.2016.1172132
Funding
This work was supported by the Guangzhou Science and Technology Plan Project (201904020004), China Agriculture Research System (CARS-46), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2019XN-003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All experimental procedures involving aquatic animals followed the animal welfare standards and were approved by the Ethical Committee for Animal Experiments of Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, China. All animal experiments complied with the guidelines of the Animal Welfare Council of China.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 648 kb)
Rights and permissions
About this article
Cite this article
Zhang, D., Gao, Y., Ke, X. et al. Bacillus velezensis LF01: in vitro antimicrobial activity against fish pathogens, growth performance enhancement, and disease resistance against streptococcosis in Nile tilapia (Oreochromis niloticus). Appl Microbiol Biotechnol 103, 9023–9035 (2019). https://doi.org/10.1007/s00253-019-10176-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10176-8