Abstract
Microaerobic bioreactor treatment for enriched sulfide and nitrate has been demonstrated as an effective strategy to improve the efficiencies of elemental sulfur (S0) generation, sulfide oxidation, and nitrate reduction. However, there is little detailed information for the effect and mechanism of dissolved oxygen (DO) on the variations of microbial community in sulfur generation, sulfide oxidation, and nitrate reduction systems. Polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) was employed to evaluate the variations of microbial community structures in a sulfide oxidation and nitrate reduction reactor under different DO conditions (DO 0–0.7 mg · L−1). Experimental results revealed that the activity of sulfide-oxidizing bacteria (SOB) and nitrate-reducing bacteria (NRB) could be greatly stimulated in 0.1–0.3 mg-DO · L−1. However, when the DO concentration was further elevated to more than 0.5 mg · L−1, the abundance of NRB was markedly decreased, while the heterotrophic microorganisms, especially carbon degradation species, were enriched. The reaction pathways for sulfide and nitrate removal under microaerobic conditions were also deduced by combining batch experiments with functional species analysis. It was likely that the oxidation of sulfide to sulfur could be performed by both aerobic heterotrophic SOB and sulfur-based autotrophic denitrification bacteria with oxygen and nitrate as terminal electron acceptor, respectively. The nitrate could be reduced to nitrite by both autotrophic and heterotrophic denitrification, and then the generated nitrite could be completely converted to nitrogen gas via heterotrophic denitrification. This study provides new insights into the impacts of microaerobic conditions on the microbial community functional structures of sulfide-oxidizing, nitrate-reducing, and sulfur-producing bioreactors, which revealing the potential linkage between functional microbial communities and reactor performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
SO2 and NOx released from the process of coal, fuel oils, and waste combustion result in serious environmental pollution and impact significantly on the ecosystem and human life. Utilizing microorganisms to treat SO2 and NOx seems to be one of the most promising alternatives to conventional air pollution control techniques, because biological process is cost-effective with no secondary pollutant production (Ding et al. 2014; Wang et al. 2015).
A novel method for simultaneous flue gas desulfurization and denitrification was investigated recently (Fig. S1). The novel method composed of two processes was demonstrated performing well with respect to sulfide and nitrate treatment in microaerobic conditions (Wang et al. 2015). When sulfide, nitrate, and carbon source were present simultaneously, the absence of oxygen makes the activities of heterotrophic denitrifiers significantly depressed by sulfide of high concentrations, resulting in the accumulation of abundant nitrite. And then the accumulated nitrite could inhibit the activities of autotrophic denitrifiers, which followed by the collapse of the denitrifying sulfide removal process (Chen et al. 2010a, c). Nevertheless, the addition of limited oxygen could greatly relieve the inhibition of sulfide to denitrification by oxidizing sulfide to sulfur or sulfate via the following oxidation-reduction reaction equation (Eqs. 1–5) (Chen et al. 2010a; Wang et al. 2016).
Furthermore, under microaerobic conditions, the glucose could be degraded by fermenting bacteria to small molecule organic acids (lactate, alanine, valine, acetate, and α-ketoglutarate) (Huang et al. 1998), which were more easily utilized by microorganism (NRB) and could enhance the performance of bioreactor with respect to denitrification. Therefore, the nitrate removal efficiency in the simultaneous sulfide, nitrate, and carbon source removal system could be improved with limited oxygen (Chen et al. 2010a, c). However, the abundance of carbon degradation bacteria (fermenting bacteria) would continuously increase with the raising of DO levels (Xu et al. 2014b), and superfluous fermenting bacteria would compete with denitrification bacteria for carbon source (Bernat and Wojnowska-Baryła 2007). On the other hand, oxygen appeared to be as an alternate and energetically preferable electron acceptor for facultative NRB (Xu et al. 2014b). So, oxygen of excess may compete with nitrate for the same enzymes (or electron donor), resulting in a lower nitrate reduction rate (Bernat and Wojnowska-Baryła 2007; Knowles 1982). According to the above analysis, the DO is a key factor to determine the coexistence and performance of SOB, carbon degradation bacteria, and NRB and then influences the efficiencies of sulfide oxidation, nitrate reduction, and S0 generation (Chen et al. 2010a; Wang et al. 2015; Xu et al. 2014b).
So, understanding the relationship among SOB, carbon degradation bacteria and NRB under different DO in the bioreactor are meaningful to the improvement of sulfide and nitrate removal efficiencies. However, previous studies were focused on sulfide and nitrate removal efficiencies in microaerobic conditions (Wang et al. 2015) or the diversity of microbial communities in anoxic sulfide and nitrate removal bioreactors (Chen et al. 2008; Huang et al. 2015a, b; Mora et al. 2015). The bacterial community structures of microaerobic bioreactors need to be explored urgently. The aims of this work are to investigate the changes of the diversity, structure, and abundances of functional species at different DO levels with the use of polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) technique and to reveal the linkage between the abundances of functional species and the performance of reactor. The reaction pathways for sulfide and nitrate removal under microaerobic conditions were also deduced by combining batch experiments with functional species analysis to better understand the interaction between sulfide-oxidizing and denitrification, which was helpful for better reactor performance and control.
Materials and methods
Bioreactor design
A polymethyl methacrylate cylinder (diameter 100 mm and height 800 mm) with 3.7 L active volume was used for this research. The microaerobic activated sludge reactor was covered by a water jacket to keep the operational temperature at 30.0 ± 0.5 °C (Fig. S2). Influent was introduced into the bottom of the bioreactor with a peristaltic pump. Air stream is introduced into the bioreactor by a sparger fixed at the bottom. DO was adjusted by changing gas flow rates (20–600 mL · min−1).
Activated sludge obtained from the secondary sedimentation tank of a municipal wastewater treatment plant in Chunliu Wastewater Treatment Plant, Dalian, China was incubated into the bioreactor. SOB and NRB cultures were enriched by sulfide and nitrate-laden medium. Table S1 shows the reactor startup stages operation parameters. The sulfide and nitrate-laden synthetic wastewater were prepared according to the following composition: first with 800 ± 30 mg · L−1 S2− (Na2S · 9H2O), 220 ± 10 mg-N · L−1 (NaNO3), 2200 ± 105 mg-COD · L−1 (glucose), 500 ± 20 mg · L−1 yeast extract, 2000 ± 98 mg · L−1 NaHCO3, 46 ± 2 mg-P · L−1 (KH2PO4), and other trace metals at hydraulic retention time (HRT) of 8.8 h, pH 8.0 ± 0.2. After the domestication of activated sludge in microaerobic system for sulfide and nitrate removal was finished, the loading rate increased stepwise in 67 days to 8.72 ± 0.04 (S), 2.49 ± 0.04 (N), and 23.52 ± 0.55 (COD) kg · m−3day−1, respectively, at HRT of 2.2 h.
Effects of DO on S0 generation efficiency and sulfide and nitrate removal efficiencies
When the performance of the bioreactor was stable, the DO was stepwise changed in order to investigate the effects of DO on S0 generation efficiency and sulfide and nitrate removal efficiencies. The degradation period consisted of four stages, I (DO < 0.1 mg · L−1), II (0.1 < DO < 0.3 mg · L−1), III (0.3 < DO < 0.5 mg · L−1), and IV (0.5 < DO < 0.7 mg · L−1). During each changing period, influents and effluents were collected every day to measure concentrations of sulfide, sulfate, thiosulfate, nitrate, nitrite, and COD.
Batch cultures
Batch tests were conducted in order to explore the substrate consumption mechanism. Two media used in this study were prepared as follows: organic carbon source (S + N + C) fed medium, containing 263.8 ± 4.1 (8.2 ± 0.1 mM) mg-S2− · L−1 (Na2S · 9H2O), 144.0 ± 1.7 (10.3 ± 0.1 mM) mg NO3 −-N · L−1 (NaNO3), 500 ± 20 mg · L−1 yeast extract, 1500 ± 83 mg-COD · L−1 (glucose) (8.06 ± 0.11 mM) and other trace metals, and organic-free medium (S + N), which was identical to the one above, except that it contained 0 mM organic carbon. Mature sulfide and nitrate removal granules were formed after cultivation for 129 days in microaerobic conditions. Activated sludge was collected by centrifugation at 10,000 rpm for 5 min, washed twice with required medium, and resuspended in the same medium to achieve initial VSS of 4000 ± 196 mg · L−1. One hundred milliliter aliquots of required culture were transferred into 125-mL serum bottles. Serum bottles were kept open and were not subjected to helium gas stripping. The bottles then were inoculated at 30 ± 1.0 °C with no shaking (Chen et al. 2010a), while the pH was adjusted to 8.0 ± 0.2. In each batch tests experimental condition, three serum bottles with identical feed were prepared to confirm data reproducibility, and an abiotic control was also performed. Liquid samples were periodically taken with sterile needle and syringe.
Analysis procedures
An ion chromatography (Dionex ICS-1100, USA) was employed to monitor the concentrations of sulfate, thiosulfate, nitrate, and nitrite in the collected liquor samples which had been filtrated through 0.45-μm filters. Sample separation and elution were performed in an IonPac AG22 AS22 4-mm analytical column with carbonate/bicarbonate as eluent (Chen et al. 2009). The dissolved sulfide concentration was measured by methylene blue method (Trüper and Schlegel 1964). A pH/ORP meter (pHS-30, China) was employed to determine the pH and oxidation-reduction potential (ORP) of liquid samples. Concentrations of DO were measured by a DO meter (YSI 550A, USA). Quantities of COD, SS, and VSS were determined according to the standard methods (APHA 2005). Elemental sulfur production was calculated by theoretic equation according to (Eq. 6) (de Graaff et al. 2012):
DNA extraction, 16S rDNA amplification, and DGGE analysis
Microaerobic sludge samples were collected from stages I (DO < 0.1 mg · L−1), II (0.1 < DO < 0.3 mg · L−1), III (0.3 < DO < 0.5 mg · L−1), and IV (0.5 < DO < 0.7 mg · L−1) to analyze the bacterial community. In each period, once the sulfide and nitrate removal efficiencies were stable, activated sludge was sampled from the bioreactors for 3 days and the samples were fully mixed. All of the samples were stored at −80 °C before the extraction of total DNA.
After 0.25 g (wet weight) mixed sludge of each stage was weighed, the total DNA extraction was performed according to the manufacturer’s instructions with a PowerSoil DNA Isolation Kit (MO BIO, USA). The extracted DNA samples were stored in a −20 °C freezer before use. For DGGE analysis, the bacterial 16S ribosomal RNA (rRNA) gene was amplified by bacterial primer sets GC-341F (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′) and 907R (5′-CCT ACG GGA GGC AGC AG-3′). Polymerase chain reaction (PCR) amplification was conducted by a thermal cycler (Veriti 96-Well Fast Thermal Cycler, Applied Biosystems, USA) with a touchdown PCR method as following program: initial reaction mixtures were held at 95 °C for 5 min, followed by 30 cycles of 94 °C for 30 sec, 50 °C for 1 min, and 72 °C for 2 min per cycle. Finally, an extension step of 10 min at 72 °C was performed. PCR products of the proper size were confirmed by electrophoresis through a 1 % agarose gel stained with GoldView (Sangon Biotech Co., Ltd., Shanghai, China) in 0.5 × TAE buffer, and DGGE was performed at 60 °C with a D-CODE System Universal Mutation (BioRad Co., Ltd., USA), which was conducted according to the manufacturer’s instruction. The PCR products were applied directly on a DGGE gel of 8 % polyacrylamide with a linear denaturing gradient ranging from 30 to 70 % (100 % denaturant was a mixture of 7 mM urea and 40 % [v/v] formamide). Electrophoresis was run at a constant voltage of 180 V for 6 h in 1 × TAE buffer. Subsequently, the gels were stained with GoldView (Sangon Biotech Co., Ltd., Shanghai, China) in 1 × TAE buffer for 40 min, and gel digital images were obtained by the Gel Doc 2000 System (BioRad Co., Ltd., USA).
Each gel slice containing an obvious electrophoretic band was excised and placed in a 1.5-mL Eppendorf tube incubated with TE buffer. Then, the Eppendorf tubes were stored at 4 °C for 12 h. DGGE profiles were analyzed using the Quantity One software. The Shannon-Wiener index was calculated using the Shannon method. The richness (S) of the microbial population was examined using the number of bands in each lane. Selected DGGE bands were excised and re-amplified by PCR using the primers described above without the GC clamp. The PCR products were sequenced at Sangon Biotech Co., Ltd. (Shanghai, China). The sequenced 16S rDNA gene was analyzed using the NCBI search tool to identify the closest matching sequence. The sequences were compared with the GenBank databases using the Basic Local Alignment Search Tool algorithm to determine the approximate phylogenetic affiliations. Representatives from the different clone types were submitted to the ribosome database (http://rdp.cme.msu.edu/index.jsp).
The sequences obtained were aligned in the GenBank DNA database with the BLAST algorithm (http://www.ncbi.nlm.nih.gov/genbank/). Sequences obtained from band 1 to 11 were deposited in the GenBank with the accession number of KT819574, KT825876∼KT825885.
Results
Effects of DO on S0 generation and sulfide and nitrate removal
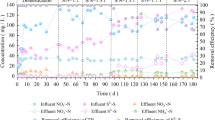
After start-up (66 days), the operation consisting of four stages continued for 63 days, totaling running for 129 days (Table S1). Figures 1 and 2 show the time course of S2− loading rate, S0 loading rate, TN loading rate, COD loading rate, effluent thiosulfate, effluent sulfate, effluent nitrite, effluent nitrate, influent and effluent COD, S2− removal (S2− Re), S0 generation (S0 Ge), TN removal (TNRe), nitrate removal (NRe), and COD removal (CODRe) at four different DO levels during 63 days of continuous operation of the lab-scale system. As DO levels were shifted stepwise from <0.1 mg · L−1 to 0.1 mg · L−1 < DO < 0.3 mg · L−1, 0.3 mg · L−1 < DO < 0.5 mg · L−1, and then to 0.5 mg · L−1 < DO < 0.7 mg · L−1, there were significant increases and subsequent decreases in the S0 Ge (from 83.1 ± 3.1 to 98.3 ± 2.6 %, 67.1 ± 1.9 and then to 42.9 ± 1.2 %), TNRe (from 75.9 ± 8.2 to 91.5 ± 3.6 %, 74.8 ± 3.3 and then to 22.5 ± 3.7 %), CODRe (from 79.9 ± 5.3 to 81.0 ± 7.3 %, 75.4 ± 3.4 and then to 53.7 ± 2.6 %), while S2− Re increased significantly with DO concentration improved to 0.1–0.3 mg · L−1 and then kept stable with the further increase of DO levels (from 83.3 ± 3.1 to 97.8 ± 3.8 %, 99.7 ± 0.7 and then to 100 ± 0.1 %). Overall, the results showed that S0 generation and sulfide, nitrate, and COD removal performance was significantly affected by DO concentrations.
Effects of DO on loading rates and removal efficiency. I (DO < 0.1 mg · L−1), II (0.1 < DO < 0.3 mg · L−1), III (0.3 < DO < 0.5 mg · L−1), IV (0.5 < DO < 0.7 mg · L−1). a Effects of DO on sulfide removal efficiency and S0 generation loading rate. b The effects of DO on TN influent and effluent loading rate, TN removal efficiency. c The effects of DO on COD influent and effluent loading rate, COD removal efficiency
The effects of DO on conversion efficiency and effluent concentrations. I (DO < 0.1 mg · L−1), II (0.1 < DO < 0.3 mg · L−1), III (0.3 < DO < 0.5 mg · L−1), IV (0.5 < DO < 0.7 mg · L−1). a Effects of DO on S0 generation efficiency and effluent concentrations of sulfate and thiosulfate. b Effects of DO on TN removal efficiency and effluent concentrations of nitrate and nitrite. c Effects of DO on COD removal efficiency and concentrations of influent and effluent
As shown in Figs. 1 and 2, the sulfide, nitrate, and COD removal simultaneously reached the highest efficiencies in the 0.1–0.3 mg-DO · L−1, and at same time, the S0 generation efficiency also reached a peak. A similar behavior was also observed in microaerobic bioreactor (Wang et al. 2015). It was suggested by previous research that the optimal DO concentration for nitrogen removal in CANON reactor was 0.2 mg-DO · L−1, in which 75.4 % total nitrogen was removed (Varas et al. 2015). S0 generation efficiency decreased with DO increasing to more than 0.3 mg · L−1, because partial sulfide was further oxidized into thiosulfate and sulfate by excessive DO. The values of oxidized sulfide (sulfate or thiosulfate) in effluent were indeed improved in higher DO levels when the influent sulfide concentration remained unchanged (Fig. 2a). A similar phenomenon was also observed in previous report that the increase of DO resulted in significant increases in S0 generation efficiency and subsequent decreases with the presence of a certain amount of sulfide (Roosta et al. 2011). During the degradation period (I–II), the consumed sulfide was mainly oxidized to elemental sulfur with the sulfur generation efficiency gradually increased from 83.1 ± 3.1 to 98.3 ± 2.6 % (Figs. 1 and 2). The results of the present work were in agreement with those reported earlier, which implied that under limited oxygen conditions, almost all of sulfide was oxidized to elemental sulfur instead of sulfate in fluidized loop reactor (Krishnakumar et al. 2005).
Effect of DO concentration on microbial community
Relative abundances (%) of bacterial species based on DGGE profile and Shannon diversity analysis were shown in Fig. 3 and Table 1. While the most similar sequences of predominant DGGE bands were listed in Table 2. Through phylogenetic analyses, the dominant populations in the reactors were mainly divided into five groups, including Bacilli, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Clostridia (Fig. 3c). According to DGGE profile and phylogenetic analysis, the microbial population structure and the microbial population diversity of microaerobic reactor were both varied among different stages of the sulfide and nitrate removal reactor. In the first stage of the operation (DO < 0.1 mg · L−1), the stripe intensity of sulfide and nitrate removal reactor was weaker, while microbial species (e.g., bands 3, 4, 6, 8, and 10) similar to species Clostridium sp., Facklamia sp., Methanogenic prokaryote, Paracoccus denitrificans, Thauera aromatica, respectively, were detected. In the second stage of the operation (0.1 mg · L−1 < DO < 0.3 mg · L−1), the variation in the number of DGGE bands and the enhanced stripe intensity indicated that the microbial population structure of sulfide and nitrate removal reactor was greatly altered with a sharp increase in microbial populations. Some new species appeared in the sulfide and nitrate removal reactor with stronger brightness, such as bands 1, 7, 9, and 11, the most similar species of which were Streptococcus sp., Pantoea vagans, Paracoccus sp., and Azoarcus sp., respectively. In the third stage of the continuous operation (0.3 mg · L−1 < DO < 0.5 mg · L−1), the microbial population diversity in the sulfide and nitrate removal reactor decreased significantly, and the stripe intensity became weaker. Some species disappeared in the sulfide and nitrate removal reactor, such as bands 1, 7, and 9, whose maximum similarity bacteria were Streptococcus sp., Pantoea vagans, and Paracoccus sp., respectively. In the fourth stage of the continuous operation (0.5 mg · L−1 < DO < 0.7 mg · L−1), the microbial community was altered distinctly. The carbon degradation bacteria (Bacilli and Clostridia) was obviously increased, while the NRB (Azoarcus and Thauera) was decreased dramatically. Some new species appeared in the sulfide and nitrate removal reactor, such as band 5, whose maximum similar species was Fusibacter tunisiensis. On the other hand, some species disappeared in the sulfide and nitrate removal reactor, such as bands 7 and 11, whose maximum similar species were Pantoea vagans and Azoarcus sp. According to the changes of microbial populations in sulfide and nitrate removal reactor, it could be concluded that the diversity and the number of microbial populations of sulfide and nitrate removal reactor had a closed relationship to DO concentration.
To obtain more mechanistic insights into the effect of increased DO concentrations on functional species, several key species involved in sulfur, nitrate, and carbon cycling were analyzed. The detected species involved in sulfide oxidation, denitrification, and carbon degradation were highly diverse, and the relative abundance of these genes varied considerably among the four stages (Fig. 3 and Table 1).
Carbon degradation bacteria including Streptococcus sp. (band 1) (Mickelson 1972), Clostridium sp. (band 3) (Monot et al. 1982), Facklamia sp. (band 4) (Di Giacomo et al. 2007), Methanogenic prokaryote (band 6) (Ito et al. 2012), and Fusibacter tunisiensis (band 5) (Ben Hania et al. 2012) were detected in the reactor. The DGGE identified five denitrifiers in the bioreactor, namely Mesorhizobium sp. (band 2) (Costa et al. 2000), Pantoea vagans (band 7) (Feng et al. 2012), Paracoccus denitrificans (band 8) (Shi et al. 2013), Paracoccus sp. (band 9) (Shi et al. 2013), Thauera aromatica (band 10) (Zhao et al. 2013), and Azoarcus sp. (band 11) (Lee et al. 2014) (Fig. 3). Additionally, the sulfide-oxidizing bacteria in the microaerobic reactor were noted to be Paracoccus (bands 8 and 9) (Quentmeier et al. 2003).
Mesorhizobium sp. was reported to have the capability for denitrification with methane as external carbon source in oxygen-limited conditions (Costa et al. 2000). Pantoea vagans was associated with denitrification and refractory organic degradation under low DO concentration (Feng et al. 2012). Paracoccus sp. was confirmed to perform denitrification reactions no matter under anaerobic (Vackova et al. 2011) or aerobic conditions (Shi et al. 2013). Azoarcus sp. could reduce nitrate through autotrophic pathways (Lee et al. 2014). In detail, nitrate could be reduced by the reducing equivalent formed in the process of sulfide oxidation (Lee and Wong 2014). Meanwhile, Azoarcus sp. was also reported to have the capability for heterotrophic nitrate-reducing in both anaerobic and aerobic conditions (Rhee et al. 1997). Thauera aromatica was reported to be able to reduce nitrate with organics as substrate and was also demonstrated that CO2 and N2 were the end products in both anaerobic (Hubert and Voordouw 2007) and aerobic conditions (Luo et al. 2014; Zhao et al. 2013). Paracoccus sp. is an SOB that can oxidized sulfide with O2 as electron acceptors whether in lithotrophic or mixotrophic conditions (Quentmeier et al. 2003), and it also has been proven to have the sulfide-oxidizing ability under denitrifying conditions (Chen et al. 2010b).
Batch cultures
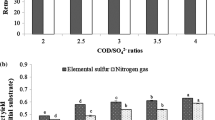
Batch cultures were conducted to further investigate the mechanism of the sulfide and nitrate removal process under microaerobic conditions. The removal performances of batch tests are presented in Fig. 4. It could be observed that the sulfide and nitrate removal sludge in S + N + C medium oxidized sulfide and reduced nitrate at higher rates than that in S + N medium (Fig. 4).
Effects of carbon source on sulfide, nitrate removal in microaerobic conditions, and the suggested pathway involved in the degradation process. a Effects of carbon source on sulfide removal in microaerobic conditions. b. Effects of carbon source on nitrate removal in microaerobic conditions. c. Suggested denitrification, sulfide and organic carbon source oxidation pathway under microaerobic conditions
During the first 4 h, 3.1 mM sulfide as well as 2.0 mM nitrate (nitrate to nitrite) was removed in the presence of organic carbon source. Meanwhile, only 1.8 mM sulfide and 0.6 mM nitrate were eliminated in the absence of substrate. Between 4 and 9 h, 2.6 mM sulfide was oxidized, and 3.6 mM nitrate (nitrate to nitrite) was reduced in S + N + C medium. Correspondingly, 3.8 mM sulfide and 0.9 mM nitrate (nitrate to nitrite) were removed in S + N medium. In the presence of organic carbon source, nitrite began to decline at 9 h, accompanied by continuous decline of nitrate. It only needed 24 h for the sulfide (8.2 mM) being completely removed and 36 h for full TN removal (10.3 mM) with sludge incubated in S + N + C medium. However, when the organic carbon source was absent, only 7.3 mM sulfide and 8.6 mM nitrate (nitrate to nitrite) were removed even in roughly 36 h.
Discussion
Microaerobic mediated synergistic relationships between SOB and NRB, accompanying the improvement of the efficiency of sulfide conversion to S0, are a promising strategy for the potential co-reduction of enriched sulfide and nitrate. For the operating conditions tested in the study, both high removal efficiencies of sulfide and nitrate and peak S0 formation efficiency (98.3 ± 2.6 %) were achieved in stage II. Compared with anoxic reactor, sulfide removal loading rate (8.72 ± 0.04 kg-S · m−3d−1) in microaerobic bioreactor was higher, and the performance of simultaneous sulfide and nitrate removal was also enhanced (Beristain-Cardoso et al. 2008; Chen et al. 2008, 2009; Reyes-Avila et al. 2004). Limited oxygen supply could not only improve the overall S0 conversion efficiency significantly but also substantially increased the sulfide and nitrate removal efficiencies. However, the efficiencies of S0 conversion and nitrate removal were decreased with the continuous increase of DO level. The variation of the performance of the microaerobic sulfide and nitrate removal bioreactor might be related to the evolution of the microbial functional communities with the changing DO concentration. This study comprehensively characterized the diversity, structure, and metabolic potential of the microbial communities and investigated the shift of functional species in response to elevated DO levels. PCR-DGGE data analysis showed that the overall diversity, structure, and composition of the microbial communities had a closed relationship to DO levels.
The NRB are generally found abundant in nitrate laden wastewater treatment processes. It had been commonly accepted that NRB were strict anaerobes. However, in recent years, a growing number of studies have demonstrated that some NRB could utilize both oxygen and nitrate as electron acceptors (Bergaust et al. 2012; Luo et al. 2014; Shi et al. 2013; Vackova et al. 2011; Zhao et al. 2013). Higher abundances of NRB functional species were detected in stage II than that in stage I, indicating that limited oxygen had promoted the activity of NRB in the bioreactor. The same behavior was also observed in other researches that the abundance of NRB in limited oxygen fed bioreactor was approximately three to fivefold higher than that in anoxic bioreactor (Xu et al. 2014b). Nevertheless, a higher level of DO (0.3–0.5 mg · L−1) was detrimental to both the growth rate and viability of NRB. It was suggested that oxygen appeared to be energetically preferable for facultative NRB. Oxygen could regulate the synthesis of nitrate reductase enzyme and inhibit the denitrification of facultative NRB, resulting in substrate electron flow to oxygen cytochromes (Wu et al. 1994). So, the lower nitrate reduction rate in higher DO concentration might be attributed to the competition between nitrate and oxygen for the same enzymes of transferred electron (Xu et al. 2014a). In early studies, it was also demonstrated that once the concentration of oxygen was lowered or removed, reduced cytochrome b might resume its transfer of electrons to cytochrome c, ultimately resulting in the reduction of nitrate (John 1977). DGGE data also showed the inhibition effect of high concentrations of DO (˃0.5 mg · L−1) on NRB, as lower numbers and significant lower abundances of NRB functional species were detected in stage VI than that in stage I, stage II, and stage III. The reduction of NRB species might be due to the suppression of the thriving fermentation microorganisms which could strongly compete for carbon resources with NRB. The low nitrate removal efficiency in stage VI might be mainly caused by the low abundance of NRB instead of the competition between the excessive oxygen and nitrate or the inhibition of the excessive oxygen to the synthesis and activity of nitrate reductase.
The SOB species were reported to be able to use oxygen as electron acceptor, and it was also demonstrated that the heterotrophic growth of SOB could increase the rate of S0 oxidation (Hassan et al. 2013). The abundances of SOB species significantly increased in stage II (0.1 mg · L−1 < DO < 0.3 mg · L−1) (Fig. 3), suggesting that the sulfide oxidation process might be stimulated at stage II. This inference was supported by the great increase in the S0 conversion efficiency during stage II. In addition, the rapid sulfide degradation caused by the thriving SOB mitigated the inhibition of sulfide for denitrifier, which partially contributed to the slight increase of the nitrate removal efficiency in stage II (Chen et al. 2009).
Interestingly, the abundance of carbon degradation species increased significantly in stage II when compared to stage I. And then the increasing DO concentration sustained a continued growth in the abundance of carbon degradation species. Stage IV had the highest abundance of carbon degradation species indicating the sludge potential capacity for removing a variety of types of organic carbon in bioreactor. It has been reported that the presence of organic carbon source could increase both the S0 oxidation rate and the nitrate removal rate (Chen et al. 2010a). So, the increased abundance of carbon degradation species might be due to the growing organic carbon requirement caused by the thriving of heterotrophic SOB and NRB. Additionally, although the abundance of SOB species had no obvious increase with the continuous increase of DO levels (DO ˃ 0.3 mg · L−1, Fig. 3), the decrease of S0 conversion efficiencies appeared (Figs. 1 and 2). It has been demonstrated that with excessive oxygen supply, sulfide was oxidized into S0 in the first place and then S0 continued to be oxidized into thiosulfate or sulfate (Jensen et al. 2011).
The bioreactor performance and functional species analysis of microbial community were joined with the specific removal rates of sulfide, nitrate, and carbon source in the batch tests to infer a possible explanation for the sulfide oxidation and denitrification metabolic pathway in continuous culture (Fig. 4c). It was observed that nitrate reduction proceeded in two steps: nitrate was first reduced to nitrite, and then nitrite was further reduced to nitrogen in the second step. The reduction rate of nitrate to nitrite (R3) with the oxidation of sulfide to sulfur is far lower than that with the oxidation of glucose (R5) (Fig. 4b). Meanwhile, the reduction rate of nitrite to N2 with the participation of sulfur oxidation (R4) is also significantly lower than that related to the oxidation of glucose (R6). In terms of the removal of sulfide, the batch culture results showed that the rate of sulfide oxidation to sulfur in the presence of oxygen (R1) is much higher than that caused by the reduction of nitrate to nitrite (R3) (Fig. 4a). In general, compared with the performance without organic carbon source medium, the specific consumption rates of sulfide with organic carbon source showed an increase of 1.72 times (highest), while denitrification rates with organic carbon source showed an increase of 2.98 times (highest). The behavior was also observed with the addition of acetate, in which both sulfide oxidation and denitrification rate were increased by roughly 55 % (Reyes-Avila et al. 2004). Those results demonstrated that mixotrophic denitrification rate was higher than the sulfur-based autotrophic denitrification rate. What is more, the production of sulfate (R4) was significantly depressed by the mixotrophic denitrification (R6) process (Sahinkaya and Dursun 2012). The key species analysis in the microaerobic bioreactor also confirmed the sulfide oxidation and denitrification metabolic pathway. As shown in Figs. 3 and 4, Tables 1 and 2, the functional species in stage II included Streptococcus sp., Clostridium sp., Facklamia sp., Methanogenic prokaryote, Pantoea vagans, Paracoccus sp., Thauera aromatica, and Azoarcus sp. It was likely that the metabolic pathway of the oxidation of sulfide to sulfur at stage II was based on the reactions R1 and R3, while nitrate was reduced by the reactions R3, R5, and R6.
Overall, the results obtained in this study demonstrated that the microbial community functional compositions and structures changed dramatically in different dissolved oxygen concentrations. The abundances of NRB species involved in nitrate reduction processes increased with DO levels gradually elevated from <0.1 to 0.1–0.3 mg · L−1. Nevertheless, when the DO levels were further increased, the abundances of NRB species were observed to decrease slightly, which became significant with DO concentration more than 0.5 mg · L−1. The abundances of SOB species participating in sulfur/sulfide oxidation processes significantly increased with DO concentration growing within the range from 0.1 to 0.3 mg · L−1 and remained stable with a further increase in DO concentration. Interestingly, the abundance of carbon degradation species continued to increase with the raising of DO levels. The variations of the abundances coincided with the performance of the microaerobic bioreactor.
Therefore, in the process of sulfide and nitrate removal, it is important to maintain an appropriate DO concentration to achieve the highest nitrate and sulfide removal efficiencies as well as the highest sulfur generation efficiency. Plenty of valuable information was provided by the PCR-DGGE analysis of functional microbial communities, which was meaningful for optimizing the performance of the microaerobic sulfide and nitrate removal reactor. Results obtained in the present research can be used as a design tool for removing enriched sulfide and nitrate efficiently in simultaneous flue gas desulfurization and denitrification process.
References
APHA (2005) Standard methods for the examination of water and wastewater, 21st edn. American Public Health Association, Washington, DC
Ben Hania W, Fraj B, Postec A, Fadhlaoui K, Hamdi M, Ollivier B, Fardeau ML (2012) Fusibacter tunisiensis sp. nov., isolated from an anaerobic reactor used to treat olive-mill wastewater. Int J Syst Evol Microbiol 62:1365–1368
Bergaust L, van Spanning RJ, Frostegard A, Bakken LR (2012) Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158:826–834
Beristain-Cardoso R, Texier A-C, Sierra-Álvarez R, Field JA, Razo-Flores E, Gómez J (2008) Simultaneous sulfide and acetate oxidation under denitrifying conditions using an inverse fluidized bed reactor. J Chem Technol Biotechnol 83:1197–1203
Bernat K, Wojnowska-Baryła I (2007) Carbon source in aerobic denitrification. Biochem Eng J 36:116–122
Chen C, Ren NQ, Wang AJ, Yu Z, Lee DJ (2008) Simultaneous biological removal of sulfur, nitrogen and carbon using EGSB reactor. Appl Microbiol Biotechnol 78:1057–1063
Chen C, Wang AJ, Ren NQ, Lee DJ, Lai JY (2009) High-rate denitrifying sulfide removal process in expanded granular sludge bed reactor. Bioresour Technol 100:2316–2319
Chen C, Ren NQ, Wang AJ, Liu L, Lee DJ (2010a) Enhanced performance of denitrifying sulfide removal process under micro-aerobic condition. J Hazard Mater 179:1147–1151
Chen C, Ren NQ, Wang AJ, Liu L, Lee DJ (2010b) Functional consortium for denitrifying sulfide removal process. Appl Microbiol Biotechnol 86:353–358
Chen C, Wang AJ, Ren NQ, Zhao QL, Liu LH, Adav SS, Lee DJ, Chang JS (2010c) Enhancing denitrifying sulfide removal with functional strains under micro-aerobic condition. Process Biochem 45:1007–1010
Costa C, Dijkema C, Friedrich M, Garcia-Encina P, Fernandez-Polanco F, Stams AJM (2000) Denitrification with methane as electron donor in oxygen-limited bioreactors. Appl Microbiol Biotechnol 53:754–762
de Graaff M, Klok JB, Bijmans MF, Muyzer G, Janssen AJ (2012) Application of a 2-step process for the biological treatment of sulfidic spent caustics. Water Res 46:723–730
Di Giacomo M, Paolino M, Silvestro D, Vigliotta G, Imperi F, Visca P, Alifano P, Parente D (2007) Microbial community structure and dynamics of dark fire-cured tobacco fermentation. Appl Environ Microbiol 73:825–837
Ding J, Zhong Q, Zhang S (2014) Simultaneous removal of NOx and SO2 with H2O2 over Fe based catalysts at low temperature. RSC Adv 4:5394–5398
Feng LJ, Xu J, Xu XY, Zhu L, Xu J, Ding W, Luan J (2012) Enhanced biological nitrogen removal via dissolved oxygen partitioning and step feeding in a simulated river bioreactor for contaminated source water remediation. Int Biodeterior Biodegrad 71:72–79
Hassan SH, Van Ginkel SW, Oh SE (2013) Effect of organics and alkalinity on the sulfur oxidizing bacteria (SOB) biosensor. Chemosphere 90:965–970
Huang Q, Fu PC, Yang C, Shimizu K (1998) Microaerobic lysine fermentations and metabolic flux analysis. Biochem Eng J 2:89–100
Huang C, Li ZL, Chen F, Liu Q, Zhao YK, Gao LF, Chen C, Zhou JZ, Wang AJ (2015a) Efficient regulation of elemental sulfur recovery through optimizing working height of upflow anaerobic sludge blanket reactor during denitrifying sulfide removal process. Bioresour Technol. doi:10.1016/j.biortech.2015.09.109
Huang C, Li ZL, Chen F, Liu Q, Zhao YK, Zhou JZ, Wang AJ (2015b) Microbial community structure and function in response to the shift of sulfide/nitrate loading ratio during the denitrifying sulfide removal process. Bioresour Technol 197:227–234
Hubert C, Voordouw G (2007) Oil field souring control by nitrate-reducing Sulfurospirillum spp. that outcompete sulfate-reducing bacteria for organic electron donors. Appl Environ Microbiol 73:2644–2652
Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2012) Identification and quantification of key microbial trophic groups of methanogenic glucose degradation in an anaerobic digester sludge. Bioresour Technol 123:599–607
Jensen HS, Lens PN, Nielsen JL, Bester K, Nielsen AH, Hvitved-Jacobsen T, Vollertsen J (2011) Growth kinetics of hydrogen sulfide oxidizing bacteria in corroded concrete from sewers. J Hazard Mater 189:685–691
John P (1977) Aerobic and anaerobic bacterial respiration monitored by electrodes. J Gen Microbiol 98:231–238
Knowles R (1982) Denitrification. Microbiol Rev 46(1):43–70
Krishnakumar B, Majumdar S, Manilal VB, Haridas A (2005) Treatment of sulphide containing wastewater with sulphur recovery in a novel reverse fluidized loop reactor (RFLR). Water Res 39:639–647
Lee DJ, Wong BT (2014) Denitrifying sulfide removal and nitrososulfide complex: Azoarcus sp. NSC3 and Pseudomonas sp. CRS1 mix. Bioresour Technol 166:616–619
Lee DJ, Wong BT, Adav SS (2014) Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl Microbiol Biotechnol 98:1301–1307
Luo J, Hao T, Wei L, Mackey HR, Lin Z, Chen GH (2014) Impact of influent COD/N ratio on disintegration of aerobic granular sludge. Water Res 62:127–135
Mickelson MN (1972) Glucose degradation, molar growth yields, and evidence for oxidative phosphorylation in Streptococcus agalactiae. J Bacterial 109:96–105
Monot F, Martin JR, Petitdemange H, Gay R (1982) Acetone and butanol production by Clostridium acetobutylicum in a synthetic medium. Appl Environ Microbiol 44:1318–1324
Mora M, Fernandez M, Gomez JM, Cantero D, Lafuente J, Gamisans X, Gabriel D (2015) Kinetic and stoichiometric characterization of anoxic sulfide oxidation by SO-NR mixed cultures from anoxic biotrickling filters. Appl Microbiol Biotechnol 99:77–87
Quentmeier A, Hellwig P, Bardischewsky F, Grelle G, Kraft R, Friedrich CG (2003) Sulfur oxidation in Paracoccus pantotrophus: interaction of the sulfur-binding protein SoxYZ with the dimanganese SoxB protein. Biochem Biophys Res Commun 312:1011–1018
Reyes-Avila J, Razo-Flores E, Gomez J (2004) Simultaneous biological removal of nitrogen, carbon and sulfur by denitrification. Water Res 38:3313–3321
Rhee SK, Lee GM, Yoon JH, Park HY, Bae HS, Lee ST (1997) Anaerobic and aerobic degradation of pyridine by a newly isolated denitrifying bacterium. Appl Environ Microbiol 63:2578–2585
Roosta A, Jahanmiri A, Mowla D, Niazi A (2011) Mathematical modeling of biological sulfide removal in a fed batch bioreactor. Biochem Eng J 58–59:50–56
Sahinkaya E, Dursun N (2012) Sulfur-oxidizing autotrophic and mixotrophic denitrification processes for drinking water treatment: elimination of excess sulfate production and alkalinity requirement. Chemosphere 89:144–149
Shi Z, Zhang Y, Zhou JT, Chen MX, Wang XJ (2013) Biological removal of nitrate and ammonium under aerobic atmosphere by Paracoccus versutus LYM. Bioresour Technol 148:144–148
Trüper HG, Schlegel HG (1964) Sulphur metabolism in Thiorhodaceae I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30:225–238
Vackova L, Srb M, Stloukal R, Wanner J (2011) Comparison of denitrification at low temperature using encapsulated Paracoccus denitrificans, Pseudomonas fluorescens and mixed culture. Bioresour Technol 102:4661–4666
Varas R, Guzmán-Fierro V, Giustinianovich E, Behar J, Fernández K, Roeckel M (2015) Startup and oxygen concentration effects in a continuous granular mixed flow completely autotrophic nitrogen removal over nitrite reactor. Bioresour Technol 190:345–351
Wang XW, Zhang Y, Zhou JT, Zhang TT, Chen MX (2015) Regeneration of elemental sulfur in a simultaneous sulfide and nitrate removal reactor under different dissolved oxygen conditions. Bioresour Technol 182:75–81
Wang XW, Zhang Y, Zhang TT, Zhou JT, Chen MX (2016) Waste activated sludge fermentation liquid as carbon source for biological treatment of sulfide and nitrate in microaerobic conditions. Chem Eng J 283:167–174
Wu Q, Knowles R, Niven DF (1994) O2 regulation of denitrification in Flexibacter canadensis. Can J Microbiol 40:916–921
Xu XJ, Chen C, Wang AJ, Guo HL, Yuan Y, Lee DJ, Ren NQ (2014a) Kinetics of nitrate and sulfate removal using a mixed microbial culture with or without limited-oxygen fed. Appl Microbiol Biotechnol 98:6115–6124
Xu XJ, Chen C, Wang AJ, Yu H, Zhou X, Guo HL, Yuan Y, Lee DJ, Zhou J, Ren NQ (2014b) Bioreactor performance and functional gene analysis of microbial community in a limited-oxygen fed bioreactor for co-reduction of sulfate and nitrate with high organic input. J Hazard Mater 278:250–257
Zhao YG, Huang J, Zhao H, Yang H (2013) Microbial community and N removal of aerobic granular sludge at high COD and N loading rates. Bioresour Technol 143:439–446
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No. 51078056), the Fundamental Research Funds for the Central Universities (No. DUT14LK37), and the Key Laboratory of Industrial Ecology and Environmental Engineering, China Ministry of Education (No. KLIEEE-08-09).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 123 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Zhang, Y., Zhang, T. et al. Effect of dissolved oxygen on elemental sulfur generation in sulfide and nitrate removal process: characterization, pathway, and microbial community analysis. Appl Microbiol Biotechnol 100, 2895–2905 (2016). https://doi.org/10.1007/s00253-015-7146-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7146-4