Abstract
The strain NSC3T, a novel, facultative, chemolithotrophic, denitrifying, alkaliphilic, sulfide-oxidizing bacterium isolated from a hot spring in Yang-Ming Mountain, Taiwan, was Gram negative, rod shaped, and motile by single polar flagella and grew facultatively by adopting a denitrifying metabolism. The 16S rRNA sequence analysis revealed that strain NSC3T belongs to beta subclass of the Proteobacteria and most closely related to Azoarcus evansii KB740T (95.44 %), Azoarcus toluvorans Td-21T (95.21 %), Azoarcus tolulyticus Tol-4T (95.08 %), and Azoarcus toluclasticus MF63T (94.94 %). The phylogenetic analyses based on 16S rRNA gene sequences indicated that the strain NSC3T formed a distinct lineage in the Betaproteobacteria and that it exhibited the highest level of sequence similarity with species of the genera Azoarcus (95.28–93.13 %). The major fatty acids of the type strain were C16:0 (26.9 %), C16:1w7c (28.9 %), C18:0 (9.6 %), and C18:1w7c/w6c (29.9 %). The DNA G+C content of genomic DNA was 63.7 mol%. On the basis of the 16S rRNA sequence similarity, phenotypic and genotypic characteristics, and chemotaxonomic data, the strain NSC3T could be differentiated from other species of the genus Azoarcus. Therefore, strain NSC3T (equal to BCRC 80111T and DSM 24109T) is proposed as a novel species in genus Azoarcus, for which the name Azoarcus taiwanensis sp. nov. is proposed. The strain NSC3T is deposited in Bioresource Collection and Research Center, Taiwan, under the reference number BCRC 80111T, and German Collection of Microorganisms and Cell Cultures, Germany (DSMZ), with DSM 24109T.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Eutrophication of rivers and deterioration of water sources, which are hazards to human health, have been reported due to discharge of nitrogenous compound containing waste into the environment. The World Health Organization established the limit for nitrates in drinking water at 50 mg NO3 −/L (11.3 mg NO3 −–N/L), while the US Environmental Protection Agency has set this limit at 10 mg NO3 − (2.3 mg NO3 −–N/L) (Directive 98/83/EC).
Oxidation of sulfide by chemolithoautotrophic denitrifying bacteria can lead to the formation of elemental sulfur or sulfate, depending on the physiological conditions. So far, genus Azoarcus is represented by validly published seven species. The genus Azoarcus primarily can be divided into two ecologically different groups of bacteria. The first group of bacteria is associated with grass roots which apparently does not survive well in root-free soil. Based on the nitrogen fixing potential, strains isolated from the rhizosphere microfora of Kallar grass were named as Azoarcus indigens and Azoarcus communis (Reinhold-Hurek et al. 1993). Second group could be soil-borne strains that can degrade aromatic hydrocarbons under denitrifying condition. These include aromatic compound degrading strain such as Azoarcus evansii (Anders et al. 1995), toluene-degrading denitrifiers as Azoarcus tolulyticus (Zhou et al. 1995), and resorcinol-degrading obligate denitrifying bacterium as Azoarcus anaerobius (Springer et al. 1998). The more detailed taxonomic examination and reclassification resulted to proposal of two additional species, Azoarcus toluvorans and Azoarcus toluclasticus (Song et al. 1999). In this study, a new member of genus Azoarcus, isolated from a hot spring in Yang-Ming Mountain, Taipei, Taiwan, is described and proposed as a novel member of the genus Azoarcus on the basis of the recommendations for the description of a novel species provided by Kampfer et al. (2003).
In this study, we demonstrate the sulfide-oxidizing ability under denitrifying condition by a new strain, Azoarcus sp. NSC3 isolated from Yang-Ming Mountain, Taipei County, Taiwan.
Materials and methods
Medium, isolation, and characterization
A sample of soil mixed with water was collected from hot sulfur spring in Yang-Ming Mountain, New Taipei City, Taiwan. To enrich culture in alkaliphilic condition, samples were mixed with nitrate, sulfide, and acetate containing medium under suitable stoichiometry (per liter): 0.755 g of KNO3, 1.5 g of Na2S·9H2O, 0.25 g of NH4Cl, 0.05 g of K2HPO4, 1.5 CU of NaHCO3, 0.44 g of CH3COONa·3H2O, and 1 mL of trace element solution (Pfennig and Lippert 1966). The pH value was 9.0. After several enrichment of the culture, it forms aggregate of elemental sulfur granules. The enrichment samples were diluted and spread onto the solidified agar plates containing sulfide and nitrate (NS medium) for isolating autotrophic bacteria. The cell morphology of the strain was observed by a phase-contrast microscope and transmission electron microscopy (Hitachi H-7100) by staining with 1 % (w/v) phosphotungstic acid. Cells of isolates grown before and after in NS medium were used in microscopic characterization. The widths and lengths given represent the averages of measurements of several cells.
Growth test
The strain growths were tested in temperature between 4 and 50 °C and pH between 6 and 11. NaCl tolerance of the strains was tested with supplement with 1 to 6 % (w/v) NaCl. Kinetic test and stoichiometry for the isolated were investigated for autotrophic growth. The tests described above were carried out at 37 °C under anoxic/anaerobic condition, flushed with argon for 20 min. For heterotrophic growth, mineral medium was supplemented with acetate instead of sulfide.
Cell characterization
Cell morphology and motility of exponentially grown cultures were observed by phase-contrast confocal laser scanning microscope (Leica TCS SP2 Confocal Spectral Microscope Imaging System, GmbH, Wetzlar, Germany). The presence of flagellum and surface morphology of the exponentially grown cultures were observed using transmission electron microscopy by staining with 1 % (w/v) phosphotungstic acid. Growth of strain NSC3T under denitrifying condition was tested using acetate as a carbon source or sulfide as electron donor. The strain NSC3T and its related type strains were characterized for its optimum growth at the temperature range of 0–50 °C, pH 4.5–11, requirement of NaCl, and its tolerance in minimal medium containing nitrate, acetate, and bicarbonate. Gram staining and oxidase and catalase activities were determined using standard methods described by Smibert and Krieg (1994), while catalase activity was also confirmed using hydrogen peroxide. The biochemical testing of strain NSC3T was carried out using API 20E and API 50 CH test strips, API 50 CHB/E medium, and the Biolog identification system. The carbon utilization capabilities of strain NSC3T was determined using Biolog EcoPlate™ (Biolog Inc., Hayward, CA) according to the manufacturer’s instructions and also confirmed using traditional cultivation methods using minimal medium supplemented with test substrates at 37 °C. The Biolog EcoPlate™ 96 (microwell plate) in triplicate was inoculated with test strains, incubated at 37 °C for 48–72 h, and absorbance was recorded using microplate reader. Antibiotic susceptibility of strain NSC3T was tested using disc diffusion technique using antibiotic discs (Oxoid, Cambridge, UK) containing penicillin G (10 U) and streptomycin (25 μg). The sensitivities to gentamicine (5 μg), streptomycin (10 μg), vancomycin (10 μg), rifampicin (5 μg), and amoxicillin (25 μg) were tested in minimal medium using traditional method. The fatty acids were extracted, saponified, and methylated according to the protocol of the Sherlock Microbial Identification System. The DNA G+C content was determined as described by Manaia et al. (2003).
Analytical methods
Thiosulfate concentrations were analyzed by cyanolysis (Kelly et al. 1969); sulfide concentrations were measured by methylene blue method according to the Standard Method (APHA 1998). Nitrate and nitrite were detected by high-performance liquid chromatography (HPLC) as described by Zecca et al. (1998). Samples before sulfur precipitation were filtered through glass fiber filter paper. The biomass protein was calculated according to the Standard Method (APHA 1998). The N2, CO2, and N2O gas contents in the headspace samples were analyzed by gas chromatography (GC-9A; Shimadzu, Tokyo, Japan) equipped with a thermal conductivity detector and column packed with molecular sieve 5A 80/100 mesh. Oven temperature was set at 70 °C, and helium was the carrier gas flowing at 30 ml min−1. Column, injector port, and detector temperatures were 70, 120, and 120 °C, respectively. Concentrations of ammonium ions were determined using Nessler’s method (APHA 1998). Sulfide concentrations were analyzed calorimetrically using the methylene blue scheme (Moest 1975). The VFA contents were analyzed by HPLC (LCP 4100 Pump, LCD 2083 Detector; ECOM, Prague) with an autosampler (HTA HT3000L; ECOM, Prague). The mobile phase was 25 mM phosphate buffer (pH 2.5) at a flowrate of 1 ml min−1. Biologically produced elemental sulfur was extracted into chloroform and analyzed using HPLC with wavelength of 263 nm and mobile phase of chloroform–methanol ratio of 1:9. To verify the existence of NO or NOS in the culture, a cell-permeable NO specific probe 4,5-diaminofluorescein diacetate (DAF-2DA) was used. Culture was withdrawn by syringe and centrifuged at 10,000 rpm to remove the supernatant. Ten micrometers of DAF-2DA was added and incubated for 60 min at 37 °C.
16S rDNA sequence determination and phylogenetic analysis
The 16S rRNA gene sequence was determined after PCR amplification using primers F27 (5′-CCA GAG TTT GAT CMT GGC TCA G-3′) and R1492 (5′-TAC CTT GTT ACG ACT T-3). The amplified PCR product was checked by electrophoresis and purified by GeneSpin™ (Protect Technology Co., Ltd., USA). The purified PCR-amplified 16S rDNA was sequenced using the ABI Prism model 3730 (version 3.2) DNA sequencer. This generated 1,406 bp gene sequence. The closest relatives were identified by comparing the 16S rRNA gene of strain with those of previously reported strains using the Ez Taxon (Chun et al. 2007) and NCBI BLAST program. The sequences were aligned by the multiple alignment ClustalW, and phylogenetic trees were generated by neighbor joining, maximum likelihood, minimum evolution, and unweighted pair group with arithmetic mean (UPGMA) by using the MEGA 4.0.2 software (Kumar et al. 2004) using bootstrap resampling methods with 1,000 replications (Tamura et al. 2007). The topology of phylogenic tree obtained by these methods were further confirmed using different out-groups.
The 16S rRNA sequence was deposited to GenBank under accession number GQ389714.
Results
Strain identification
The strain (NSC3T) was isolated from the soil sample from a hot sulfur spring in Yang-Ming Mountain, New Taipei City, Taiwan. Based on 16S rRNA gene sequence, the closest relatives of the strain NSC3T were A. evansii KB740T (95.44 %), A. toluvorans Td-21T (95.21 %), A. tolulyticus Tol-4T (95.08 %), A. toluclasticus MF63T (94.94 %), Azoarcus buckelii U120T (94.58 %), A. anaerobius LuFres1T (94.30 %), Thauera butanivorans Bu-B1211T (93.41 %), A. communis SWub3T (93.13 %), and Thauera phenylacetica B4PT (92.949 %). The phylogenetic tree based on 16S rRNA revealed that the strain formed a distinct robust lineage among type strains of Azoarcus spp. within beta-subclass of Proteobacteria (Fig. 1 and Figs. S1–S4 in Electronic supplementary material). According to Stackebrandt and Goebel (1994), if rRNA sequence similarities are 97 % or above, DNA–DNA relatedness studies of novel and closely related type strains are required for species differentiation. However, in the case of strain NSC3T, 16S rRNA sequence similarities below this level justify validity of16S rRNA-based description of a new species of the genus Azoarcus. The isolate has been deposited in Bioresource Collection and Research Center, Taiwan, under the reference number BCRC 80111T, and German Collection of Microorganisms and Cell Cultures, Germany (DSMZ), with DSM 24109T.
Strain characterization
The major fatty acids of the strain NSC3T were C16:0 (26.9 %), C16:1w7c (28.9 %), C18:0 (9.6 %), and C18:1w7c/w6c (29.9 %) (Table 1). The major fatty acid component C16:1w7c was 28.9 % in strain NSC3T, while it was 44.7 % in A. tolulyticus Tol-4T, 51.0 % in A. toluvorans Td21T, 51.8 % in A. toluclasticus MF63T, and 46.9 % in A. evansii KB 740T which distinguished strain NSC3T from these type strains. The higher content of C16:1 in A. toluvorans Td21T, A. toluclasticus MF63T, A. communis SWub3T, A. indigens VB32T, and A. evansii KB 740T differentiated strain NSC3T from these type strains.
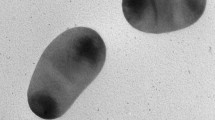
The biochemical, physiological, and morphological characteristics of strain NSC3T were summarized in Table 2. The denitrification and sulfide removal potential of strain is shown in Fig. S5. The strain grew well on minimal mineral media with NaCl (1–3 % w/v, optimum at 1 %), 1.5 g NaHCO3 and with optimum growth at 35–37 °C. No growth was observed at 4 °C and above 55 °C. The strain grew between pH 6.5 and 10.5 with optimal growth occurring at pH 9.0. The strain can grow chemolithotrophically with sulfide as electron donor and nitrate as electron acceptor. The strain NSC3T was positive to nitrate reductase, nitrite reductase, catalase, and oxidase; motile with single polar flagella (Fig. 2); Gram negative; and consistent with those of Azoarcus sp. The cells were sensitive to gentamicin (5 μg), streptomycin (10–25 μg), and vancomycin (10 μg) and resistant to penicillin G (10 U), rifampicin (5 μg), and amoxicillin (25 μg).
Strain description
The Azoarcus taiwanensis sp. nov. is a rod-shaped bacterium of 0.9–2.2 × 0.5–0.7 μm, Gram negative, appears as a single cell or in pairs, and motile by means of a single polar flagellum. The colonies are circular, smooth, convex, and transparent; are positive to catalase, oxidase, nitrate reductase, and sulfide oxidase; and grew at 15–40 °C, optimally at a temperature of 37 °C, pH 6.5–10.5, with optimum growth at pH 9.0. In Biolog microplate test, the strain assimilated d-alanine, l-alanine, l-alanylglycine, d-glucosamine, l-glutamine, glycine, l-methionine, l-ornithine, l-phenylalanine, l-proline, d-serine, l-serine, l-valine, β-d-allose, d-arabinose, l-arabinose, cellulose, 2-deoxy-d-ribose, d-fructose, l-fucose, d-glucose-6-phosphate, l-lyxose, maltotriose, d-melibiose, palatinose, d-raffinose, d-ribose, stachyose, sucrose, d-tagatose, acetic acid, butyric acid, capric acid, citric acid, formic acid, fumaric acid, l-lactic acid, dl-malic acid, l-malic acid, propionic acid, pyruvic acid, succinic acid, acetoacetic acid, γ-aminobutyric acid, δ-aminovaleric acid, bromosuccinic acid, 3-0-β-d-galactopyranosyl-d-arabinose, d-galacturonic acid, d-gluconic acid, d-glucuronic acid, d-glucosaminic acid, l-glutamic acid, glycyl-l-aspartic acid, glycyl-l-glutamic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, α-hydroxyglutaric acid-γ-lactone, itaconic acid, α-ketobutyric acid, 5-keto-d-gluconic acid, α-ketoglutaric acid, d-lactic acid methyl ester, 3-methylglucose, α-methyl-d-glucoside, α-methyl-d-mannoside, methyl pyruvate, β-methyl-d-xyloside, oxalomalic acid, l-pyroglutamic acid, sorbic acid, tricarballylic acid, amygdalin, sec-butylamine, dl-carnitine, chondroitin sulfate C, α-cyclodextrin, dextrin, gelatin, inosine, inulin, laminarin, dl-octopamine, tryptophan, Tween 20, Tween 80, urea, xylan, adonitol, 2-aminoethanol, dulcitol, I-erythritol, isopropanol, lactitol, maltitol, propanol, xylitol, anthrone, and 2,3-butanone. Under anaerobic denitrifying condition, the strain NSC3T utilized different reduced sulfur compounds such as sulfide, sulfite, tetrathionate, and thiosulfate and various aromatic compound included benzene, benzoic acid, o-cresol, p-cresol, m-cresol, 4-hydroxybenzoic acid, p-hydroxyphenylacetic acid, quinoline, pyridine, terephthalic acid, toluene (denitrifying), phenol, and m-xylene. The G + C content was 63.7 %. The cells are sensitive to gentamicin, streptomycin, amoxycilin, and vancomycin and resistant to penicillin G, rifampicin, and amoxicillin. The major fatty acids of the strain NSC3T were C16:0 (26.9 %), C16:1w7c (28.9 %), C18:0 (9.6 %), and C18:1w7c/w6c (29.9 %).
Discussion
Strain identity
In phenotypic characteristics, chemolithotrophic growth in presence of sulfide, high NaCl tolerance, and alkaliphilic nature could differentiate strain NSC3T from A. evansii KB740T, A. toluvorans Td-21T, A. tolulyticus Tol-4T, A. toluclasticus MF63T, A. buckelii U120T, A. anaerobius LuFres1T, and A. communis SWub3T. Based on GC content, strain NSC3T (63.7 %) could be distinguished from A. tolulyticus Tol-4T (66.9 %), A. toluvorans Td-21T (67.8 %), A. toluclasticus MF63T (67.3 %), A. indigens VB32T (66.6 %), A. evansii KB740T (67.5 %), and A. anaerobius LuFres1T (66.5 %). Strain NSC3Tcould be distinguished from A. tolulyticus Tol-4T, A. toluvorans Td-21T, and A. toluclasticus MF63T by positive to nitrate reductase and citrate assimilation, while it can be differentiated from A. communis SWub3T and A. indigens VB32T based on l-serine, sucrose, and toluene utilization potential. The antibiotic resistance to penicillin G (10 U), rifampicin (5 μg), and amoxicillin (25 μg) distinguished strain NSC3T from other strains of genus Azoarcus. The cells were sensitive to gentamicin (5 μg), streptomycin (10–25 μg), and vancomycin (10 μg). Resistance of reported type strains of Azoarcus spp. to antibiotics has not been extensively tested. The 16S rRNA sequence similarities and phylogenetic analysis by neighbor joining, maximum parsimony, maximum likelihood, and UPGMA suggested affiliation of strain NSC3T to genus Azoarcus with a robust clad with members of Azoarcus. The phylogenic tree topology and clustering of strain NSC3T were further confirmed by selecting different out-groups. The strain NSC3T was isolated from a hot spring, while strain A. tolulyticus Tol-4T was isolated from petroleum-contaminated soil, A. evansii KB740T from a creek sediment, A. indigens VB32T from stem bases/root of Kallar grass, A. communis SWub3T from root piece of Kallar grass, A. toluvorans Td-21T from petroleum-contaminated soil, and A. anaerobius LuFres1T from anoxic sewage sludge. Based on phenotypic features, biochemical tests, fatty acid composition comparisons, 16S rRNA gene sequence similarities, and phylogenetic analysis, the strain NSC3T represents a novel member of genus Azoarcus, for which the name A. taiwanensis sp. nov. is proposed.
Facultative autotrophic denitrifiers
The denitrifying sulfide removal (DSR) process was proposed to achieve simultaneous conversion of nitrate to N2, sulfide to elementary sulfur, and organic substances to CO2 via balanced growth of both autotrophic and heterotrophic denitrifiers (Chen et al. 2008a, b). Chen et al. (2008c) noted that when concentrations of sulfide were too high to inhibit activities of heterotrophic denitrifiers (Pseudomonas aeruginosa and Sulfurospirillum sp. in that study), nitrite would be accumulated so the activities of autotrophic denitrifiers (Sulfurovum sp. and Paracoccus denitrificans) would be inhibited, hence leading to complete breakdown of the DSR process. The facultative autotrophic bacteria (FAB) that can grow mixotrophically and heterotrophically with oxidization of reduced sulfur compounds may be used to lift the major limitations of balanced growth of autotrophic and heterotrophic bacteria in the DSR reactor (Lee et al. 2013). A group of FAB were proposed, including Thiobacillus delicates, Thiobacillus pantotropha, Pseudomonas stutzeri, and Paracocus denitrificans (Robertson and Kuenen 1983; Sorokin et al. 1996, 1999; Gupta 1997; Nemati et al. 2001; Sorokin 2003). The strain NSC3T represents a novel member of FAB that can have potential to be applied in DSR-related processes.
References
Anders HJ, Kaetzke A, Kampfer P, Ludwig W, Fuchs G (1995) Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int J Syst Bacteriol 45:327–333
APHA (1998) Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association. Washington DC, USA
Chen C, Ren NQ, Wang AJ, Yu ZG, Lee DJ (2008a) Microbial community of granules in EGSB reactor for simultaneous biological removal of sulfate, nitrate, and COD. Appl Microbiol Biotechnol 79:1071–1077
Chen C, Ren NQ, Wang AJ, Yu ZG, Lee DJ (2008b) Simultaneous biological removal of sulfur, nitrogen and carbon using EGSB reactor. Appl Microbiol Biotechnol 78:1057–1063
Chen C, Wang AJ, Ren NQ, Kan HJ, Lee DJ (2008c) Biological breakdown of denitrifying sulfide removal process in high-rate expanded granular bed reactor. Appl Microbiol Biotechnol 81:765–770
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Gupta AB (1997) Thiosphaera pantotropha: a sulphur bacterium capable of simultaneous heterotrophic nitrification and aerobic denitrification. Enzym Microb Technol 21:589–595
Kampfer P, Buczolits S, Albrecht A, Busse HJ, Stackebrandt E (2003) Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int J Syst Evol Microbiol 53:893–896
Kelly DP, Chambers LA, Thudinger PA (1969) Cyanolysis and spectrophotometric estimation of trithionate in mixtures with thiosulfate and tetrathionate. Anal Chem 41:898–901
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lee DJ, Pan XL, Wang AJ, Ho KL (2013) Facultative autotrophic denitrifiers in denitrifying sulfide removal granules. Bioresour Technol 132:356–360
Manaia CM, Nunes OC, Nogales B (2003) Caenibacterium thermophilum gen. nov., sp. nov., isolated from a thermophilic aerobic digester of municipal sludge. Int J Syst Evol Microbiol 53:1375–1382
Moest RR (1975) Hydrogen sulfide determination by the methylene blue method. Anal Chem 47:1204–1205
Nemati M, Jenneman GE, Voordouw G (2001) Mechanistic study of microbial control of hydrogen sulfide production in oil reservoirs. Biotechnol Bioeng 74:424–434
Pfennig N, Lippert KD (1966) Uber Das Vitamin B12-Bedurfnis Phototropher Schwefelbakterien. Arch Microbiol 55:245–256
Reinhold-Hurek B, Hurek T, Gillis M, Hoste B, Vancanneyt M, Kersters K, De Ley J (1993) Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth), and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int J Syst Bacteriol 43:574–584
Robertson LA, Kuenen JG (1983) Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulphur bacterium. J Gen Microbiol 129:2847–2855
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–655
Song B, Haggblom MM, Zhou JZ, Tiedje JM, Palleroni NJ (1999) Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int J Syst Bacteriol 49:1129–1140
Sorokin DY (2003) Oxidation of inorganic sulfur compounds by obligatively organotrophic bacteria. Microbiology 72:641–653
Sorokin DY, Lysenko AM, Mityushina LL (1996) Isolation and characterization of alkaliphilic chemoorganoheterotrophic bacteria oxidizing reduced inorganic sulfur compounds to tetrathionate. Microbiology 65:326–338
Sorokin DY, Teske A, Robertson LA, Kuenen JG (1999) Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group. FEMS Microbiol Ecol 30:113–123
Springer N, Ludwig W, Philipp B, Schink B (1998) Azoarcus anaerobius sp. nov., a resorcinol-degrading, strictly anaerobic, denitrifying bacterium. Int J Syst Bacteriol 48:953–956
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16s rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Zecca L, Rosati M, Renella R, Galimberti M, Ambrosini A, Fariello RG (1998) Nitrite and nitrate levels in cerebrospinal fluid of normal subjects. J Neural Transm 105: 627–633
Zhou J, Fries MR, Chee-Sanford JC, Tiedje JM (1995) Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol 45:500–506
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 222 kb)
Rights and permissions
About this article
Cite this article
Lee, DJ., Wong, BT. & Adav, S.S. Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl Microbiol Biotechnol 98, 1301–1307 (2014). https://doi.org/10.1007/s00253-013-4976-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4976-9