Abstract
Improving the production of ethanol from xylose is an important goal in metabolic engineering of Saccharomyces cerevisiae. Furthermore, S. cerevisiae must produce ethanol in the presence of weak acids (formate and acetate) generated during pre-treatment of lignocellulosic biomass. In this study, weak acid-containing xylose fermentation was significantly improved using cells that were acclimated to the weak acids during pre-cultivation. Transcriptome analyses showed that levels of transcripts for transcriptional/translational machinery-related genes (RTC3 and ANB1) were enhanced by formate and acetate acclimation. Recombinant yeast strains overexpressing RTC3 and ANB1 demonstrated improved ethanol production from xylose in the presence of the weak acids, along with improved tolerance to the acids. Novel metabolic engineering strategy based on the combination of short-term acclimation and system-wide analysis was developed, which can develop stress-tolerant strains in a short period of time, although conventional evolutionary engineering approach has required long periods of time to isolate inhibitor-adapted strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ethanol production from plant biomass has received considerable attention because bioethanol reduces demand for petroleum-based fuels (Cardona and Sanchez 2007). In particular, lignocellulosic biomass, such as wood and agricultural residues, represents a sustainable energy resource that does not compete with food and animal feed production. The hydrolysis of lignocellulosic biomass (consisting of cellulose, hemicellulose, and lignin) liberates sugars, primarily glucose and xylose, which are subsequently fermented by microorganisms to produce ethanol (Hasunuma and Kondo 2012). The yeast Saccharomyces cerevisiae is a promising candidate for industrial bioethanol production due to its robustness and capacity for high-level ethanol production. However, a major drawback is that S. cerevisiae cannot utilize xylose, which is the most common pentose sugar in hemicellulose, comprising a sizable fraction of lignocellulosic hydrolysate.
Advances in genetic engineering approaches over the past few decades have enabled researchers to devise S. cerevisiae strains capable of utilizing xylose through the expression of heterologous genes for xylose reductase (XR) and xylitol dehydrogenase (XDH) from Scheffersomyces stipitis, together with its endogenous gene for xylulokinase (XK) (Chu and Hung 2007; Eliasson et al. 2000). XR first reduces xylose to xylitol, which is then oxidized to xylulose by the action of XDH. Xylulose is subsequently phosphorylated by XK to xylulose 5-phosphate, which is metabolized through the pentose phosphate pathway (PPP) and glycolysis to ethanol under oxygen-limited conditions. As another challenge in the fermentation of xylose, a xylose isomerase (XI) gene derived from the anaerobic fungi, Piromyces and Orpinomyces, has also been introduced into S. cerevisiae (Kuyper et al. 2003; Lee et al. 2014; Madhavan et al. 2009). XI converts xylose to xylulose in one step, although the rate of xylose consumption is lower in the XI-expressing strain. In xylose-fermenting S. cerevisiae strains, fermentation inhibitors such as formic and acetic acids, which are generated during pre-treatment of lignocellulosic biomass, have a more significant impact on xylose utilization than glucose utilization (Bellissimi et al. 2009; Hasunuma et al. 2011a; Sanda et al. 2011).
The structure of lignocellulosic biomass is recalcitrant to hydrolysis, which necessitates chemical and physicochemical pre-treatment in order to swell the material before saccharification and fermentation (de Costa Sousa et al. 2009; Chandel et al. 2012). However, the harsh conditions used in pre-treatment release fermentation inhibitors, including weak organic acids, furan derivatives, and phenolics (Almeida et al. 2007). In particular, high concentrations of acetic acid (1~10 g/L) are released into the lignocellulosic hydrolysate during the solubilization and hydrolysis of hemicellulose. Although formic acid is typically present at lower concentrations than acetic acid, it is more toxic to S. cerevisiae than acetic acid (Hasunuma et al. 2011b; Martin et al. 2007). Weak acids such as formic and acetic acids negatively affect microbial growth and metabolism, thereby decreasing ethanol titer. Especially, ethanol production from xylose is severely inhibited in the presence of weak acids (Hasunuma et al. 2011a).
To overcome the effects of fermentation inhibitors on ethanol production, more robust yeast strains have been developed through evolutionary engineering, mutagenesis, genome shuffling, meiotic recombination, and metabolic engineering (Demeke et al. 2013a, b; Hasunuma and Kondo 2012; Xiao and Zhao 2014). Yeast strains subjected to high concentrations of inhibitors for an extended period can gain substantial tolerance as a result of changes in their genetic background. Long-term adaptation is one way to mimic natural selection resulting from environmental pressures. Due to the complexity of the underlying molecular mechanisms associated with inhibitor tolerance, evolutionary engineering strategies have been employed as an alternate means of improving the fermentation capabilities of yeast strains in the presence of inhibitory compounds.
An additional strategy for overcoming the effects of fermentation inhibitors is to relieve the stress associated with the inhibitor through pre-cultivation. Cells living in highly variable environments adapt to sudden environmental changes that can stress cellular systems (Berry and Gasch 2008; Bertilsson et al. 2009). For instance, cells grown at 36 °C are less affected by heat shock at 52 °C than cells grown at 23 °C (Walton and Pringle 1980). Yeast cells pre-cultivated on the hydrolysate from steam-pre-treated spruce exhibit enhanced ethanol production during simultaneous saccharification and fermentation (Alkasrawi et al. 2006). Recently, pre-cultivation of a yeast strain in the presence of 67 mM acetate enabled its aerobic growth and anaerobic ethanol production from glucose in the presence of 100 mM acetate (Sànchez i Nogué et al. 2013). Thus, stress responses observable at the phenotypic level can be initiated through pre-exposure to a given stress, which seems to prepare cells to withstand future stress exposure. However, the physiological mechanisms underlying stress responses in yeast have yet to be fully elucidated.
In the present study, we found that pre-incubating a xylose-fermenting S. cerevisiae strain in the presence of weak acids improves xylose fermentation in weak acid–containing hydrolysates. Transcriptome analysis was used to isolate the genes responsible for this short-term acclimation. A significant increase in the expression of the genes RTC3 (involved in RNA metabolism) and ANB1 (involved in mRNA translation) was observed with coordinated addition of formic and acetic acids. Elucidation of the mechanisms underlying yeast acclimation to weak acids allowed for enhancing inhibitor tolerance through metabolic engineering. This is the first report of enhanced xylose fermentation through inverse metabolic engineering based on the short-term stress response.
Materials and methods
Strains and media
Yeast strains were routinely cultivated at 30 °C in synthetic medium (SD medium; 6.7 g/L yeast nitrogen base without amino acids [Difco Laboratories, Detroit, MI], 20 g/L glucose) supplemented with appropriate amino acids and nucleotides and in YPD medium (20 g/L peptone, 10 g/L yeast extract, 20 g/L glucose). Escherichia coli strain NovaBlue (Novagen, Inc., Madison, WI) was used as the host for recombinant DNA manipulation and was grown in Luria–Bertani medium (10 g/L peptone, 5 g/L yeast extract, 5 g/L sodium chloride) containing 100 mg/L ampicillin. The growth media and culture conditions for these strains were described previously (Hasunuma et al. 2011a).
Batch fermentation under oxygen-limited conditions
After cultivation under aerobic conditions for 48 h at 30 °C in YPD medium containing a specified concentration of fermentation inhibitor (30 mM acetic acid or 20 mM formic acid), yeast cells were collected by centrifugation at 1000×g for 5 min at 4 °C, washed twice with distilled water, and then inoculated into fermentation medium (10 g/L yeast extract, 20 g/L peptone) containing 50 g/L xylose or 50 g/L glucose and a specified concentration of fermentation inhibitor (acetic or formic acid). The initial cell concentration was adjusted to 50 g wet cells/L. The initial pH of the fermentation was 5.9, 4.5, and 4.3 under no acid, 30 mM acetic acid, and 20 mM formic acid conditions, respectively. All fermentations were performed at 30 °C under oxygen-limited conditions with an agitation speed of 500 rpm in 100-mL closed bottles equipped with a bubbling CO2 outlet and a stir bar, as described previously (Fujitomi et al. 2012). Wet cell weight was determined by weighing cell pellets that were harvested by centrifugation from 48-h YPD cultures. The concentrations of ethanol, glycerol, xylitol, and xylose in the fermentation medium were determined as described previously (Hasunuma et al. 2011a).
Analysis of intracellular metabolites
Samples for intracellular metabolite analysis were prepared as described previously (Hasunuma et al. 2011a). Briefly, 5 mL of broth was withdrawn and injected (<1 s) into a tube containing 7 mL of pure methanol pre-cooled to −40 °C. The mixture was quickly vortexed and placed back in the cryostat (−40 °C). Extracellular medium was removed by centrifugation at 5000×g at −20 °C for 5 min. After decanting, 7.5 μL of 1 mM 1,4-piperazinediethanesulfonic acid was added to the samples as an internal standard. Ethanol (75 % [v/v]) pre-heated in a water bath to 95 °C was quickly poured over the cell pellet; the mixture was immediately vortexed, and the sample was placed in the water bath for 3 min. After centrifugation at 14,000×g at 4 °C for 5 min, intracellular metabolites were obtained by evaporation of the supernatant under vacuum using a FreeZone 2.5 Plus freeze dry system (Labconco, Kansas City, MO). Dried metabolites were dissolved in 15 μL of Milli-Q water before capillary electrophoresis–mass spectrometry (CE-MS) analysis. All CE-MS experiments were performed using an Agilent CE capillary electrophoresis system, an Agilent G6220AA LC/MSD TOF system, and an Agilent 1200 series isocratic HPLC pump (Agilent Technologies, Palo Alto, CA), as described previously (Hasunuma et al. 2011a). Agilent Chem-Station software for CE and MassHunter software for the Agilent TOF-MS were used for system control and data acquisition. CE separation was performed in a fused silica capillary (1 m × 50 μm i.d.) filled with 30 mM ammonium formate (pH 10) as the electrolyte. Prior to the first use, each capillary was washed by running the electrolyte for 20 min with application of 50 mbar of pressure. Before injection in each analysis, the capillary was equilibrated first for 2 min with 1 M formic acid and then for 4 min with pre-conditioning buffer (15 mM ammonium acetate, 75 mM sodium phosphate, pH 7.5), 4 min with water, and 6 min with electrolyte, which was replenished after every run using a buffer replenishment system configured with the Agilent CE instrument. Sample was injected at a pressure of 50 mbar for 10 s (approximately 10 nL). The CE polarity was such that the electrolyte vial (inlet) was at the anode, and the electrospray ionization (ESI) probe (outlet) was at the cathode. The voltage applied to the CE capillary was set at 30 kV, with 0.3 min of ramp time. Electrophoresis was performed for 50 min. The capillary temperature was maintained at 20 °C, and the sample tray was cooled to below 6 °C. An HPLC pump equipped with a 1:100 splitter was used to deliver 8 μL/min of 50 % (v/v) methanol/water containing reference masses of the standards purine ([M-H]−, m/z 119.03632) and hexakis (1H,1H,3H-tetrafluoroproxy) phosphazine ([M-H]−, m/z 966.000725) to the CE interface, where the solvent was used as a sheath fluid around the outside of the CE capillary to provide a stable electrical connection between the tip of the capillary and the grounded electrospray needle. ESI-MS was conducted in the negative ion mode, with the capillary voltage set at 3.5 kV. The nebulizing gas (heated dry nitrogen [heater temperature, 300 °C]) was switched off during the pre-conditioning step, and a pressure of 10 psi was applied 1 min after sample injection using the CE system’s timetable.
Global gene expression analysis
Total RNA was obtained after 4 h of fermentation in the presence of 20 mM formic acid or 30 mM acetic acid using a Total RNA Isolation Mini Kit (Agilent Technologies) according to the manufacturer’s protocol. RNA concentration and quality were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. cDNA was reverse-transcribed and labeled with cyanine 3-CTP using a Low-Input Quick Amp Labeling Kit (Agilent Technologies) for hybridization in S. cerevisiae 4 × 44 microarrays. Hybridization was performed at 65 °C for 17 h. The arrays were scanned using an Agilent Single-Color DNA Microarray Scanner (Agilent Technologies). Gene expression levels were normalized per chip. GeneSpring GX ver. 11.5.1 software (Agilent Technologies) was used to analyze fold change in expression data. Every biological sample for the microarray analysis was analyzed in duplicate.
Plasmid construction and yeast transformation
The RTC3 gene was amplified using genomic DNA of S. cerevisiae strain YPH499 deposited as ATCC® 204679™ as a template and the primer set RTC3-01 (5′-CTCGCTAGCGTCGACATGTCTACTGTAACCAAATACTTTTACAAG-3′)/RTC3-02 (5′-CTCAGATCTGGATCCTCAATTGTAGGCTTTGGTTCCGGCGTTA-3′). The SalI and BamHI sites are underlined in the preceding sequences. PCR-amplified RTC3 was digested with SalI and BamHI and then ligated into the SalI-BamHI site of pGK425 (Ishii et al. 2009) to yield plasmid pGK425-RTC3. Similarly, the ANB1 gene was amplified from the same genomic DNA using the primer set ANB1-01 (5′-CTCGCTAGCCTCGAGATGTCTGACGAAGAACACACCTTTGAAAAT-3′)/ANB1-02 (5′-CTCCCCGGGGGATCCCTAATCAGATCTTGGAGCTTCCTTGAAGGA-3′). The XhoI and BamHI sites are underlined in the preceding sequences. PCR-amplified ANB1 was digested with XhoI and BamHI and then ligated into the XhoI-BamHI site of pGK425 to yield plasmid pGK425-ANB1. YPH499XU was used as the parental strain (Ishii et al. 2012). Plasmids pGK425, pGK425-RTC3, and pGK425-ANB1 were transformed into S. cerevisiae strain YPH499XU to yield YPH499XU/pGK425, YPH499XU/pGK425-RTC3, and YPH499XU/pGK425-ANB1, respectively. The resulting transformants were selected on SD medium supplemented with appropriate amino acids and nucleotides.
Cell viability
Cell viability was assessed microscopically (Zeiss, Jena, Germany) using a disposable hemocytometer chamber (C-Chip, Seoul, Korea), as previously described (Ismail et al. 2014). Trypan blue was used for vital staining.
Results
Ethanol production from xylose in acid-acclimated yeast cells
A xylose-fermenting recombinant yeast strain, YPH499XU, was constructed by integrating into the genome of strain YPH499 the Scheffersomyces stipitis genes Xyl1 and Xyl2, encoding xylose reductase (XR) and xylitol dehydrogenase (XDH), respectively, and the S. cerevisiae gene Xks1, encoding xylulokinase (XK) (Ishii et al. 2012). Expression of Xyl1, Xyl2, and Xks1 was controlled by the S. cerevisiae TDH3 promoter (Ishii et al. 2012). Before xylose fermentation, YPH499XU was aerobically pre-cultivated in YPD medium in either the absence or presence of an inhibitory compound (20 mM formic acid or 30 mM acetic acid) for 72 h (Fig. 1). By addition of inhibitory compound, cell growth was retarded during pre-cultivation (Fig. S1 in the Supplementary Material). Yeast cells pre-cultivated in the presence and absence of inhibitory compounds are hereafter referred to as acclimated cells and control cells, respectively.
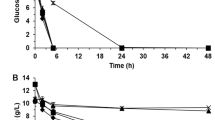
Xylose fermentation was performed in YP medium containing 50 g/L of xylose as the sole carbon source, with or without addition of 20 mM formic acid or 30 mM acetic acid, under oxygen-limited conditions (Fig. 2). Control cells, pre-cultivated in the absence of an inhibitory compound, completely consumed the xylose, producing 17.05 g/L of ethanol after 72 h of fermentation, whereas xylose consumption was reduced by the addition of 20 mM formic acid or 30 mM acetic acid (Fig. 2a–c). Ethanol production was 7.46 and 12.76 g/L in the presence of 20 mM formic acid and 30 mM acetic acid, respectively. Glycerol production and xylitol production were similar in the presence and absence of the acids. As shown in Fig. 2d, e, cells acclimated to formate demonstrated 2.3-fold higher ethanol production than control cells in the presence of 20 mM formate. In the absence of formic acid, there was no difference in ethanol production between formate-acclimated cells and control cells. Acetate-acclimated cells demonstrated higher volumetric ethanol productivity than control cells without acid addition (Fig. 2f and g). In the presence of 30 mM acetic acid, 19.64 g/L of ethanol was produced by the acetate-acclimated cells after 48 h of xylose fermentation, which was 1.5-fold more ethanol that was produced by control cells (12.76 g/L; Table 1). These results indicate that short-term acclimation improves xylose fermentation in the presence of formic or acetic acids.
Ethanol production from xylose by non-acclimated control cells in the absence (a) or presence of 20 mM formic acid (b) or 30 mM acetic acid (c). Ethanol production from xylose by formate-acclimated cells in the absence (d) or presence (e) of 20 mM formic acid. Ethanol production from xylose by acetate-acclimated cells in the absence (f) or presence (g) of 30 mM acetic acid. Symbols: filled circles, xylose; open circles, ethanol; triangles, xylitol; squares, glycerol. Values represent the average (±SD) of three independent experiments
Analysis of metabolites and gene expression
The levels of intracellular metabolites involved in the PPP were determined after 2 and 24 h of fermentation. In the absence of formate or acetate, non-acclimated control cells consumed intracellular erythrose-4-phosphate (E4P), ribulose-5-phosphate (Ru5P), and sedoheptulose-7-phosphate (S7P) during the fermentation, whereas the consumption of these metabolites was reduced by the addition of 20 mM formic acid or 30 mM acetic acid (Fig. 3). In particular, the levels of E4P and S7P, abundant PPP metabolites, were almost constant during 24 h of fermentation in the presence of the inhibitors. In contrast, as shown in Table 2, formate-acclimated cells exhibited a 47, 73, 54, and 30 % decrease in the levels of E4P, R5P, Ru5P, and S7P, respectively, in the presence of 20 mM formic acid. In acetate-acclimated cells, the levels of these metabolites accumulated after 24 h of fermentation were less than one tenth of their 2-h concentrations.
Intracellular content of pentose phosphate pathway metabolites in control cells after 2 h (black bars) and 24 h (white bars) of fermentation in the absence or presence of 30 mM acetate or 20 mM formate. Values represent the average (±SEM) of three independent experiments. Abbreviations: E4P erythrose-4-phosphate, R5P ribose-5-phosphate, Ru5P ribulose-5-phosphate, S7P sedoheptulose-7-phosphate, DCW dry cell weight
Microarray analyses were performed to investigate the differences in global gene expression between acid-acclimated cells and control cells. RNA was obtained after 4 h of fermentation in the presence of 20 mM formic acid or 30 mM acetic acid. The number of genes for which the level of transcription was increased by more than twofold by formate and acetate acclimation was 213 and 223, respectively. A total of 44 genes were upregulated in both formate- and acetate-acclimated cells (Fig. S2 in the Supplementary Material). The ten most upregulated and downregulated genes in formate- and acetate-acclimated cells are listed in Tables 3 and 4, respectively. For instance, the transcript levels of genes involved in transcriptional and translational machinery, such as RTC3 and ANB1, were higher in acclimated cells. Transcription of RTC3 (YHR087W), which encodes a protein of unknown function involved in RNA metabolism (Gomar-Alba et al. 2012), was enhanced 3.67- and 5.38-fold by formic acid and acetic acid by acclimation to formic and acetic acid, respectively. Transcription of ANB1 (YJR047C), which encodes translation elongation factor eIF-5A (Saini et al. 2009), was increased more than threefold in acclimated cells.
Construction of recombinant strains based on global gene expression profiles
In an effort to improve the resistance of yeast to the fermentation inhibitors, genes upregulated in acclimated cells were integrated into the genome of the xylose-fermenting strain along with the constitutive PGK1 promoter. Thus, RTC3- and ANB1-overexpressing strains (YPH499XU/pGK425-RTC3 and YPH499XU/pGK425-ANB1) were constructed. Transformation of YPH499XU was performed with an empty vector, pGK425, to obtain a vector control strain, YPH499XU/pGK425. After pre-cultivation in YPD medium without addition of formic or acetic acids, fermentation of 50 g/L of xylose was performed in either the absence or presence of 20 mM formic acid or 30 mM acetic acid (Fig. 4). Xylose fermentation by the control strain, YPH499XU/pGK425, was inhibited by the addition of 20 mM formic acid or 30 mM acetic acid (Fig. 4a). In the presence of 20 mM formic acid, xylose consumed by the vector control strain was 36.37 g/L after 72 h of fermentation. In contrast, YPH499XU/pGK425-RTC3 completely consumed 50 g/L of xylose, producing 20.08 g/L of ethanol under the same conditions (Fig. 4b). Furthermore, the volumetric ethanol productivity of YPH499XU/pGK425-RTC3 was 61 % higher than that of the vector control (Table 5). In the presence of 30 mM acetic acid, RTC3-overexpressing strains showed 47 % higher volumetric ethanol productivity than the control strain. YPH499XU/pGK425-RTC3 showed higher xylitol production than the control strain (Table 5), whereas there was almost no difference in glycerol production between the two strains (Fig. S3 in the Supplementary Material). In the absence of formic and acetic acids, YPH499XU/pGK425-RTC3 showed a higher rate of xylose consumption and volumetric ethanol productivity than YPH499XU/pGK425 in the absence of formic and acetic acids (Fig. 4a), although there was no difference in final ethanol production between YPH499XU/pGK425-RTC3 and YPH499XU/pGK425. YPH499XU/pGK425-ANB1, which overexpresses ANB1, demonstrated higher ethanol production from xylose than YPH499XU/pGK425 in the presence of 20 mM formic or 30 mM acetic acid, although without addition of the weak acids, xylose consumption and ethanol production by YPH499XU/pGK425-ANB1 were similar to those of YPH499XU/pGK425.
Ethanol (open symbols) production from xylose (filled symbols) in the absence (a) or presence of 20 mM formic acid (b) or 30 mM acetic acid (c) by YPH499XU/pGK425 (squares), YPH499XU/pGK425-RTC3 (circles), and YPH499XU/pGK425-ANB1 (triangles). Values represent the average (±SD) of three independent experiments
As shown in Fig. 5, in the presence of 20 mM formic acid or 30 mM acetic acid, the viability of control strain cells declined during the 72-h fermentation. However, the rate of decline in cell viability was lower in the RTC3- and ANB1-overexpressing strains. These data indicate that overexpression of RTC3 and ANB1 enhances the tolerance of yeast to both formic and acetic acids.
Fermentation of 50 g/L glucose was performed in the presence of 45 mM formic acid or 100 mM acetic acid. In the control strain, the fermentation was inhibited by the acid addition (Fig. S4 in the Supplementary Material). The inhibition of both glucose consumption and ethanol production was not alleviated by the overexpression of RTC3 and ANB1.
Discussion
To develop acid-tolerant yeast strains, novel metabolic engineering strategy based on the combination of short-term acclimation and system-wide metabolic analysis was developed. In this study, global gene expression analysis demonstrated that levels of transcripts for transcriptional and translational machinery–related genes, such as RTC3 and ANB1, were enhanced by short-time acclimation. Overexpression of genes upregulated by the short-time acclimation improved ethanol production from xylose in the presence of weak acids as well as yeast tolerance to weak acids.
Several reports describe increased tolerance of S .cerevisiae strains to fermentation inhibitors through long-term adaptation and directed evolution (Dragosits and Mattanovich 2013; Koppram et al. 2012; Wallace-Salinas and Gorwa-Grauslund 2013). For instance, the adaptation of xylose-fermenting S. cerevisiae strains to sugarcane bagasse by cultivation over 353 h in media containing increasing concentrations of fermentation inhibitors led to a significant improvement in ethanol production from bagasse hydrolysate (Martín et al. 2007). An evolutionarily engineered yeast strain obtained by sequential cultivation in wheat straw hydrolysate with increasing concentrations of inhibitors exhibited 20 % higher final ethanol yield and 65 % higher xylose consumption than its parental strain in the fermentation of steam-pretreated wheat straw (Tomás-Pejó et al. 2010). The evolutionary engineering approach has required long periods of time to isolate inhibitor-adapted strains. However, metabolic engineering based on the systems biology analysis of short-term acclimation improved yeast tolerance in a short period of time.
According to a previous report (Sanda et al. 2011), the concentrations of formic acid and acetic acid contained in lignocellulosic hydrolysate obtained by hydrothermal pre-treatment of rice straw are 20.06 and 27.11 mM, respectively. Therefore, the concentrations used in this study are of practical relevance for lignocellulose-based bioethanol production processes. As shown in Fig. 2, short-term acclimation of yeast cells to 20 mM formic acid increased ethanol production and volumetric productivity in xylose fermentation in the presence of 20 mM formate by 2.3- and 1.3-fold, respectively. Acclimation to 30 mM acetic acid increased ethanol production and volumetric productivity in the presence of 30 mM acetate by 1.5- and 2.6-fold, respectively. Interestingly, acetate acclimation improved the rates of both xylose consumption and ethanol production, even in the absence of 30 mM acetate (Fig. 2).
As shown in Fig. 3, in the absence of added acids, the levels of non-oxidative PPP intermediates such as E4P, R5P, Ru5P, and S7P decreased with time during xylose fermentation. However, the rates of decrease in the levels of these metabolites were lower in the presence of 20 mM formic acid or 30 mM acetic acid. This result indicates that the flux in non-oxidative PPP intermediates in the xylose-fermenting strain slows in the presence of weak acid, which is consistent with the results of a previous study (Hasunuma et al. 2011a). In contrast, the content of non-oxidative PPP intermediates declined significantly in formate- and acetate-acclimated cells during fermentation (Table 2). In particular, the level of S7P, the most abundant intermediate, declined 18-fold in cells subjected to short-term acetate acclimation. These results suggest that short-term acclimation to formic and acetic acids during pre-cultivation relieves the bottleneck in the metabolic pathway during fermentation.
Short-term exposure of the laboratory strain S. cerevisiae S288c to environmental stresses such as NaCl, H2O2, and extreme temperature was shown to trigger the expression of a large number of genes (Berry and Gasch 2008). Pre-cultivation of a yeast strain in the presence of 67 mM acetate improved glucose utilization in the presence of 100 mM acetate under anaerobic condition (Sànchez i Nogué et al. 2013), although xylose utilization is more severely affected by the addition of acetate than glucose utilization (Hasunuma et al. 2011a). However, the mechanism behind the increased tolerance acquired by pre-exposure to fermentation inhibitor–induced stress has yet to be elucidated. As shown in Table 3, transcriptome analysis of microarray data demonstrated significant increases in the expression of wide variety of genes. The transcription of 213 and 223 genes increased more than twofold in cells acclimated to formate and acetate, respectively. RTC3 encodes a protein of unknown function involved in RNA metabolism (Gomar-Alba et al. 2012). The expression of one well-studied hypoxia response gene, ANB1 (Saini et al. 2009), was also high in acclimated cells. The metabolic pathway genes GOR1 and GPH1 encode glyoxylate reductase and glycogen phosphorylase, respectively (Rintala et al. 2007; Stanford et al. 2004; Sunnarborg et al. 2001). The enzyme Gph1p catalyzes the phosphorolysis of α-1,4-linked glucan in glycogen, releasing glucose-1-phosphate. Expression of GPH1 is regulated by stress-response elements and the Hog1p-MAP kinase pathway (Sunnarborg et al. 2001). DSE2 encodes a daughter cell–specific secreted protein similar to glucanases, which degrades the cell wall from the daughter side, causing the daughter cell to separate from the mother cell (Colman-Lerner et al. 2001). CIS3 and TIP1 encode cell wall mannoproteins (Castillo et al. 2003; Kondo and Inouye 1991; Kowalski et al. 1995). GRX2 encodes Grx2p (a member of the two-cysteine subfamily of glutaredoxins), which catalyzes glutathione-disulfide oxidoreductions. Grx2p, which plays a role in resistance to oxidative stress, appears to also play a role in defense against reactive oxygen species by means of its dethiolase activity (Luikenhuis et al. 1998).
The expression of 559 and 481 genes decreased more than twofold in formate- and acetate-acclimated cells, respectively. As shown in Table 4, genes highly downregulated in acclimated cells included PHO89, AQY1, FET3, ADY2, PUT4, and PHO84, which are involved in the transportation of metal ions, phosphate, acetate, and proline. Although the mechanism of fermentation inhibition by weak acids remains unclear due to their molecular heterogeneity and lack of highly accurate quantitative analyses, weak acids reportedly induce intracellular acidification (with negative consequences for the activity of metabolic enzymes) and cause dissipation of the plasma membrane potential (Pereira et al. 2014).
As shown in Fig. 4, overexpression of RTC3 and ANB1 enhanced ethanol production from xylose in the recombinant strain in the presence of formic and acetic acids. Figure 5 shows the improved tolerance to formic and acetic acids in RTC3- and ANB1-expressing strains. This is the first report of overexpression of RTC3 and ANB1 in xylose-fermenting S. cerevisiae strains. The systematic name of RTC3 is YHR087W. The expression of this gene is induced in response to osmotic stress caused by high sugar concentrations (Gomar-Alba et al. 2012). In a laboratory strain, disruption of RTC3 was shown to result in growth delay, lower viability, and reduced glucose consumption in the presence of 25 and 30 % glucose (Jiménez-Martí et al. 2011). Overexpression of this gene results not only in enhanced viability in the presence of high glucose concentrations but also in better fermentative activity in wine yeast strains under vinification conditions (Jiménez-Martí et al. 2009). Gomar-Alba et al. (2012) reported that the expression of YHR087W is controlled by the transcription factors Msn2/4p under heat shock and by the transcription factors Hot1p and Sko1p during osmotic stress. Although the role of YHR087Wp in yeast cells remains unclear, genetic interaction between YHR087W and several genes encoding proteins involved in transcription and transcription control, such as Nut1p, Set3p, Ntl3p, Cna1p, and Bcy1p, has been reported (Costanzo et al. 2010). Tandem affinity purification experiments indicated interaction between YHR087Wp and proteins involved in translation initiation (Gomar-Alba et al. 2012). In addition, deletion of YHR087W was shown to cause both growth defects in the presence of translation inhibitors and a reduction in translation recovery after exposure of yeast cells to high-glucose stress (Gomar-Alba et al. 2012). These results indicate that YHR087Wp is involved in RNA translation as well as transcription under stress conditions. ANB1, the transcription of which is tightly regulated by the presence of oxygen, encodes the translation elongation factor eIF-5A (Saini et al. 2009). This study raises the possibility that modification of the transcriptional and translational machinery might improve xylose fermentation by S. cerevisiae in the presence of inhibitory acids.
Disruption of PHO13, which encodes p-nitrophenylphosphatase, was shown to improve both the titer and volumetric productivity of ethanol from xylose in the presence of fermentation inhibitors, including formate, acetate, and furfural, although the physiological role of PHO13 remains unclear (Fujitomi et al. 2012). Overexpression of HAA1, which encodes an acetic acid–responsive transcriptional activator, was shown to improve cell growth and ethanol titer from xylose under aerobic and oxygen-limited conditions, respectively, in the presence of acetic acid (Sakihama et al. 2014). The expression of Haa1p regulons such as SAP30 (which encodes a histone deacetylase complex subunit) and HRK1 (which encodes a protein kinase implicated in activation of the plasma membrane H+-ATPase) was shown to protect against acetic acid–associated stress (Mira et al. 2010a). However, the levels of PHO13 and HAA1 transcripts were not significantly changed in our short-term acclimation experiments. Pereira et al. (2014) performed genome-wide screening of genes required for tolerance to key inhibitors of lignocellulosic fermentation using EUROSCARF haploid mutant correction; however, the 10 most upregulated genes in acclimated cells in the present study (Table 3) were not included in their list of candidate genes. Mira et al. (2010b) screened the disruptome of the EUROSCARF mutant to identify genes responsible for acetate tolerance. With the exception of GPH1, the genes listed in Table 3 were not isolated in their disruptome screening. The short-term acclimation strategy utilized in the present study thus identified additional genes responsible for tolerance to weak acids in yeast.
Various systems biology approaches, such as disruptome screening, transcriptomics, proteomics, and metabolomics, have been applied to gain insights into the molecular and genetic traits involved in the adaptation to fermentation inhibitors and to aid in generating inhibitor-tolerant yeast strains (Gorsich et al. 2006; Hasunuma et al. 2011a, b; Heer et al. 2009; Lin et al. 2009). Nielsen and co-workers reviewed a systems-level approach for metabolic engineering of yeast strains, including rational metabolic engineering and inverse metabolic engineering (Kim et al. 2012). Systems-wide analyses of yeast cell biology can assist in the design of strategies to functionally improve strains. In inverse metabolic engineering, initial selection of cellular systems with a desired phenotype is followed by systems-wide comparative analysis and construction of novel recombinant strains based on the results of the analysis. Hong and Nielsen (2012) identified a specific mutation in adaptively evolved yeast strains that improves galactose utilization. In the present study, metabolome and transcriptome analyses were used to examine the response of S. cerevisiae in order to uncover the mechanism underlying yeast short-term acclimation to fermentation inhibitors. The results of genome-wide analyses made it possible to identify phenotype-specific genes expressed under selective conditions; these genes could serve as targets for subsequent metabolic engineering studies aimed at obtaining more robust yeast strains. Through the targeted modification of intracellular metabolism, super-yeast strains exhibiting significantly enhanced production of lignocellulosic ethanol would be developed.
References
Alkasrawi M, Rudolf A, Lidén G, Zacchi G (2006) Influence of strain and cultivation on the performance of simultaneous saccharification and fermentation of steam pretreated spruce. Enzym Microb Technol 38:279–286
Almeida JRM, Modig T, Petersson A, Hahn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349
Bellissimi E, van Dijken JP, Pronk JT, van Maris AJA (2009) Effects of acetic acid on the kinetics of xylose fermentation by an engineered, xylose isomerase-based Saccharomyces cerevisiae strain. FEMS Yeast Res 9:358–364
Berry DB, Gasch AP (2008) Stress-activated genome expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19:4580–4587
Bertilsson M, Olofsson K, Lidén G (2009) Prefermentation improves xylose utilization in simultaneous saccharification and co-fermentation of pretreated spruce. Biotechnol Biofuels 2:8
Cardona CA, Sanchez OJ (2007) Fuel ethanol production: process design trends and integration opportunities. Bioresour Technol 98:2415–2457
Castillo L, Martinez AI, Garcerá A, Elorza MV, Valentín E, Sentandreu R (2003) Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: the repetitive sequence is needed for binding to the cell wall β-1,3-glucan. Yeast 20:973–983
Chandel AK, Chandrasekhar G, Silva MB, Silvério da Silva S (2012) The realm of cellulases in biorefinery development. Crit Rev Biotechnol 32:187–202
Chu BCH, Hung L (2007) Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv 25:425–441
Colman-Lerner A, Chin TE, Brent R (2001) Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739–750
Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pál C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C (2010) The genetic landscape of a cell. Science 327:425–431
da Costa SL, Chudawat SPS, Balan V, Dale BE (2009) ‘Cradle-to-grave’ assessment of existing lignocellulose pretreatment technologies. Curr Opin Biotechnol 20:339–347
Demeke MK, Dietz H, Li Y, Foulquié-Moreno MR, Mutturi S, Deprez S, Abt TD, Bonini BM, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein JM (2013a) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89
Demeke MK, Dumortier F, Li Y, Broeckx T, Foulquié-Moreno MR, Thevelein JM (2013b) Combining inhibitor tolerance and D-xylose fermentation in industrial Saccharomyces cerevisieae for efficient lignocellulose-based bioethanol production. Biotechnol Biofuels 6:120
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution – principles and applications for biotechnology. Microb Cell Fact 12:64
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Fujitomi K, Sanda T, Hasunuma T, Kondo A (2012) Deletion of the PHO13 gene in Saccharomyces cerevisiae improves ethanol production from lignocellulosic hydrolysate in the presence of acetic and formic acids, and furfural. Bioresour Technol 111:161–166
Gomar-Alba M, Jiménez-Martí E, del Olmo M (2012) The Saccharomyces cerevisiae Hot1p regulated gene YHR087W (HGI1) has a role in translation upon high glucose concentration stress. BMC Mol Biol 13:19
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349
Hasunuma T, Kondo A (2012) Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol Adv 30:1207–1218
Hasunuma T, Sanda T, Yamada R, Yoshimura K, Ishii J, Kondo A (2011a) Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microb Cell Fact 10:2
Hasunuma T, Sung KM, Sanda T, Yoshimura K, Matsuda F, Kondo A (2011b) Efficient fermentation of xylose to ethanol at high formic acid concentrations by metabolically engineered Saccharomyces cerevisiae. Appl Microbiol Biotechnol 90:997–1004
Heer D, Heine D, Sauer U (2009) Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxidoreductase. Appl Environ Microbiol 75:7631–7638
Hong KK, Nielsen J (2012) Recovery of phenotypes obtained by adaptive evolution through inverse metabolic engineering. Appl Environ Microbiol 78:7579–7586
Ishii J, Izawa K, Matsumura S, Wakamura K, Tanino T, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J Biochem 145:701–708
Ishii J, Yoshimura K, Hasunuma T, Kondo A (2012) Reduction of furan derivatives by overexpressing NADH-dependent Adh1 improves ethanol fermentation using xylose as sole carbon source with Saccharomyces cerevisiae harboring XR-XDH pathway. Appl Microbiol Biotechnol 97:2597–2607
Ismail KS, Sakamoto T, Hasunuma T, Zhao XQ, Kondo A (2014) Zinc, magnesium, and calcium ion supplementation confers tolerance to acetic acid stress in industrial Saccharomyces cerevisiae utilizing xylose. Biotechnol J 9:1519–1525
Jiménez-Martí E, Zuzuarregui A, Riduara I, Lozano N, del Olmo M (2009) Genetic manipulation of HSP26 and YHR087W stress genes may improve fermentative behavior in wine yeasts under vinification conditions. Int J Food Microbiol 130:122–130
Jiménez-Martí E, Zuzuarregui A, Gomar-Alba M, Gutiérrez D, Gil C, del Olmo M (2011) Molecular response of Saccharomyces cerevisiae wine and laboratory strains to high sugar stress conditions. Int J Food Microbiol 145:211–220
Kim IK, Roldão A, Siewers V, Nielsen J (2012) A systems-level approach for metabolic engineering of yeast cell factories. FEMS Yeast Res 12:228–248
Kondo K, Inouye M (1991) TIP1, a cold shock-inducible gene of Saccharomyces cerevisiae. J Biol Chem 266:17537–17544
Koppram R, Albers E, Olsson L (2012) Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels 5:32
Kowalski LRZ, Kondo K, Inouye M (1995) Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol Microbiol 15:341–353
Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MS, De-Laat WT, Den-Ridder JJ, Op den Camp HJ, van Dijken JP, Pronk JT (2003) High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res 4:69–78
Lee SM, Jellison T, Alper HS (2014) Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7:122
Lin FM, Qiao B, Yuan YJ (2009) Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, lignocellulosic inhibitory compound. Appl Environ Microbiol 75:3765–3776
Luikenhuis S, Perrone G, Dawes IW, Grant CM (1998) The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol Biol Cell 9:1081–1091
Madhavan A, Tamalampudi S, Ushida K, Kanai D, Katahira S, Srivastava A, Fukuda H, Bisaria VS, Kondo A (2009) Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol 82:1067–1078
Martin C, Alriksson B, Sjöde A, Nilvebrant N-O, Jönsson LJ (2007) Dilute sulfuric acid pretreatment of agricultural and agro-industrial residues for ethanol production. Appl Biochem Biotechnol 137:339–352
Martín C, Marcet M, Almazán O, Jönsson LJ (2007) Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresour Technol 98:1767–1773
Mira NP, Becker JD, Sá-Correia I (2010a) Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 14:587–601
Mira NP, Palma M, Guerreiro JF, Sá-Correia I (2010b) Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79
Pereira FB, Teixeira MC, Mira NP, Sá-Correia I, Domingues L (2014) Genome-wide screening of Saccharomyces cerevisiae genes required to foster tolerance towards industrial wheat straw hydrolysates. J Ind Microbiol Biotechnol 41:1753–1761
Rintala E, Pitkänen JP, Vehkomäki ML, Penttilä M, Ruohonen L (2007) The ORF YNL274c (GOR1) codes for glyoxylate reductase in Saccharomyces cerevisiae. Yeast 24:129–136
Saini P, Eyler DE, Green R, Dever TE (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459:118–121
Sakihama Y, Hasunuma T, Kondo A (2014) Improved ethanol production from xylose in the presence of acetic acid by the overexpression of the HAA1 gene in Saccharomyces cerevisiae. J Biosci Bioeng 119:297–302
Sànchez i Nogué V, Narayanan V, Gorwa-Grauslund MF (2013) Short-term adaptation improves the fermentation performance of Saccharomyces cerevisiae in the presence of acetic acid at low pH. Appl Microbiol Biotechnol 97:7517–7525
Sanda T, Hasunuma T, Matsuda F, Kondo A (2011) Repeated-batch fermentation of lignocellulosic hydrolysate to ethanol using a hybrid Saccharomyces cerevisiae strain metabolically engineered for tolerance to acetic and formic acids. Bioresour Technol 102:7917–7924
Stanford DR, Whitney ML, Hurto RL, Eisaman DM, Shen W-C, Hopper AK (2004) Division of labor among the yeast Sol proteins implicated in tRNA nuclear export and carbohydrate metabolism. Genetics 168:117–127
Sunnarborg SW, Miller SP, Unnikrishnan I, LaPorte DC (2001) Expression of the yeast glycogen phosphoylase gene is regulated by stress-response elements and by the HOG MAP kinase pathway. Yeast 18:1505–1514
Tomás-Pejó E, Ballesteros M, Oliva JM, Olsson L (2010) Adaptation of the xylose fermenting yeast Saccharomyces cerevisiae F12 for improving ethanol production in different fed-batch SSF processes. J Ind Microbiol Biotechnol 37:1211–1220
Wallace-Salinas V, Gorwa-Grauslund MF (2013) Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151
Walton EF, Pringle JR (1980) Effect of growth temperature upon heat sensitivity in Saccharomyces cerevisiae. Arch Microbiol 124:285–287
Xiao H, Zhao H (2014) Genome-wide RNAi screen reveals the E3 SUMO-protein ligase gene SIZ1 as a novel determinant of furfural tolerance in Saccharomyces cerevisiae. Biotechnol Biofuels 7:78
Acknowledgments
The authors thank Ms. Yoshimi Hori for technical assistance. This work was supported by Special Coordination Funds for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interests.
Ethical approval
This study does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 491 kb)
Rights and permissions
About this article
Cite this article
Hasunuma, T., Sakamoto, T. & Kondo, A. Inverse metabolic engineering based on transient acclimation of yeast improves acid-containing xylose fermentation and tolerance to formic and acetic acids. Appl Microbiol Biotechnol 100, 1027–1038 (2016). https://doi.org/10.1007/s00253-015-7094-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7094-z