Abstract
Riemerella anatipestifer infection causes high mortality for ducks which results in major economic losses in the duck industry. In this study, we identified a mutant strain RA-M1 by Tn4351 transposon mutagenesis, in which the M949_1603 gene encoding glycosyl transferase was inactivated. PCR analysis revealed that M949_1603 gene is specifically existed in R. anatipestifer serotype 1 strains. RA-M1 presented no reactivity to the anti-lipopolysaccharide (LPS) MAb in an indirect ELISA. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting demonstrated that RA-M1 LPS had a deficiency in ladder-like binding pattern to rabbit antiserum against R. anatipestifer serotype 1 strain CH3, indicating that the O-antigen structure of RA-M1 was changed. RA-M1 showed significant attenuated virulence in ducks and higher sensitivity to normal duck serum, compared with its parent strain CH3. Furthermore, cross-protection of RA-M1 for R. anatipestifer serotypes 1, 2, and 10 strains was evaluated. Ducks that received two immunizations with inactivated RA-M1 vaccine were 100 % protected from challenge with R. anatipestifer serotype 1 strain WJ4, serotype 2 strain Yb2, and serotype 10 strain HXb2. No changes were observed in the liver, heart, or spleen samples from the protected ducks during autopsy and histological examination. Furthermore, vaccination generated high antibody titers of 1:12,800 against serotypes 1, 2, and 10 strains and enhanced production of interleukin 2 (IL-2) and IL-4 in ducks. These results suggested that M949_1603 gene is associated with serotype 1 O-antigen biosynthesis, and mutant RA-M1 could be used as a novel cross-protection vaccine candidate to protect ducks against R. anatipestifer infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Riemerella anatipestifer is a gram-negative, non-motile, non-spore-forming, rod-shaped bacterium that causes duck infection, accounts for major economic losses to the duck industry worldwide through poor feed conversion and high mortality rates. Infected ducks show clinical signs containing fibrinous pericarditis, perihepatitis, and meningitis (Sandhu and Rimler 1997; Segers et al. 1993). R. anatipestifer primarily affects young ducks under 8 weeks of age, less frequently, geese (Pierce and Vorhies 1973) and turkeys (Helfer and Helmboldt 1977). Once the bacteria infect a duck flock, it can become endemic and eradication can be difficult with repeated infectious episodes possible. Up to date, 21 serotypes of R. anatipestifer have been identified with no significant cross-protection reported (Loh et al. 1992; Pathanasophon et al. 1995; Pathanasophon et al. 2002). R. anatipestifer serotypes 1, 2, and 10 strains are responsible for most of the major outbreaks in China (Hu et al. 2010).
Lipopolysaccharide (LPS) is an essential component in the outer membrane and provides the structural integrity of the outer membrane in most gram-negative bacteria (Nikaido and Vaara 1985). LPS is a major virulence factor of most gram-negative bacteria which consists of lipid A, O-antigen, and core oligosaccharide, of which O-antigen is highly immunogenic and varies among different subgroups. Genes for synthesis of LPS have been characterized in several species of bacteria (Brown et al. 2012; Aquilini et al. 2014; Perepelov et al. 2014). In R. anatipestifer, however, only the AS87_04050 gene was characterized to be responsible for LPS synthesis (Wang et al. 2014). In this study, M949_1603 gene deletion R. anatipestifer mutant RA-M1 was characterized as a LPS-deficient strain, and the cross-protection among R. anatipestifer serotypes 1, 2, and 10 strains was also evaluated. Our data suggest that RA-M1 could be used as a novel cross-protection vaccine candidate to protect ducks against R. anatipestifer infection.
Materials and methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. R. anatipestifer CH3 is the wild-type strain used in this study, and the mutant strain RA-M1 was derived from the random tranposon mutant library for the defective in the reactivity with anti-LPS MAb in an indirect ELISA. Strains CH3 and RA-M1 were deposited in China General Microbiological Culture Collection Center with the numbers of CGMCC 10683 and CGMCC 10147, respectively). The library containing 2520 random Tn4351 transposon mutants was constructed previously in our laboratory by biparental mating with BW19851 (pEP4351) and R. anatipestifer strain CH3 (Hu et al. 2012). Biparental mating was performed as described previously (Bagdasarian et al. 1981) with modifications. R. anatipestifer strains were grown on tryptic soy agar (TSA, Difco, USA) or tryptic soy broth (TSB, Difco) at 37 °C in 5 % CO2. Antibiotics were used at the given concentrations when needed: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), erythromycin (0.5 μg/ml), kanamycin (50 μg/ml), and cefoxitin (5 μg/ml).
Development of anti-LPS MAb
R. anatipestifer strain CH3 was cultured to logarithmic phase and inactivated with formalin as described (Liu et al. 2013). BALB/c mice were immunized with the inactivated bacterial cells at 1 × 108 colony-forming units (CFUs) for three times. Hybridoma technique was performed, and single clones were screened using an indirect ELISA, which plates were coated with purified CH3 LPS (10 μg/well). Positive clone 8A9 was subjected to subclone for three times and used for MAb production.
Characterization of R. anatipestifer mutant strain RA-M1
R. anatipestifer mutant strain RA-M1 was obtained by screening the library with anti-LPS MAb 8A9. Southern blot analysis was used to identify the insertion of Tn4351 in the mutants as described (Hu et al. 2010). The plasmid pEP4351 and genomic DNA of wild-type strain CH3 were also subjected to hybridization analysis, which were used as the positive and negative control, respectively. The site of transposon insertion in the mutant strain was determined with a genome walking kit (TaKaRa) using four random primers (AP1, AP2, AP3, AP4) provided in the kit and three specific primers (SP1, SP2, and SP3), according to the manufacturer’s instructions. Polymerase chain reaction (PCR) amplified gene was inserted to T-vector pMD19 (TaKaRa). DNA sequencing was performed by Shanghai HuaGene Biotech Co., Ltd. The identified gene sequence was searched for homologous sequences and putative functions on the BLAST from National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST/).
Real-time RT-PCR was performed to measure expressions of Tn4351-disrupted upstream and downstream genes at transcriptional level. Gene-specific primers were designed using primer3 online software v.0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and described in Table S1. The expression of M949_1603 gene, its upstream gene M949_1602, and downstream gene M949_1604 were measured using primer pairs M949_1603 F/M949_1603 R, M949_1602 F/M949_1602 R, and M949_1604 F/M949_1604 R, respectively (Table S1), and the expression of 16S rRNA was used as an internal control. Reactions were performed in triplicate and run on the Mastercycler ep realplex4 apparatus (Eppendorf, Germany).
To detect the distribution of M949_1603 gene among R. anatipestifer serotype 1, 2, and 10 strains, serotype 1 strains CH1, WJ4, JY4, and CQ5; serotype 2 strains Yb2, NJ3, Th4, GD5, and YXb1; and serotype 10 strains HXb2 and YXb11 were cultured for PCR amplification using primers M949_1603 ORF-F/M949_1603 ORF-R. CH3 and the mutant strain RA-M1 were used as positive control and negative control, respectively.
The growth curves of wild-type strain CH3 and mutant strain RA-M1 were determined as described previously (Hu et al. 2002). Briefly, equal amount of each bacterial culture was transferred into fresh TSB medium at a ratio of 1:100 (v/v) for further growing at 37 °C for 10 h with shaking. The bacterial growth was measured by monitoring the optical density at 600 nm (OD600) at 1-h intervals using a spectrophotometer (Bio-Rad, USA). The statistical significance of the data was determined by one-way ANOVA in Graphpad Prism 6 software (GraphPad Software, Inc., CA, USA). A p value of <0.05 was considered to be statistically significant.
Serum killing assays
Normal duck serum was collected from 10 healthy 17-day-old Cherry Valley ducks without anti-R. anatipestifer antibody, pooled, and filter-sterilized (0.22 mm). The bactericidal assay was performed in a 96-well plate as described previously with modifications (McQuillen et al. 1994). Duck serum was diluted to 5, 12.5, 25, 50, and 100 % in pH 7.2 PBS. The bacterial suspension containing 106 CFU in 10 μl PBS were added into 190 μl of the diluted serum, 100 % heat-inactivated serum, or PBS alone and then incubated at 37 °C for 30 min. The bacteria in the wells were 10-fold serial diluted and plated onto TSB plates. Colonies were counted after 28-h incubation.
Adhesion and invasion capacities to Vero cells
Bacterial adherence and invasion assays were performed on Vero cells (ATCC CCL-81) as described (Hu et al. 2011). Briefly, Vero cells were seeded to the wells in 24-well tissue culture plates in Dulbecco’s modified Eagle medium (DMEM) (Biowest, France) containing 10 % fetal bovine serum 1 day before infection. The cells of each well were infected with CH3 and RA-M1 at 50 multiplicity of infection. The plates were incubated at 37 °C for 1.5 h and washed with PBS; 0.1 % trypsin (100 μl/well) was then added and incubated at 37 °C for 10 min. The cell suspensions were 10-fold diluted and plated onto TSA plates for bacterial counting. For the invasion assay, the extracellular bacteria were killed by incubation with DMEM medium supplemented with 100 μg/ml gentamicin for an additional 1 h, following the incubation with bacteria and three washes with PBS. All of the above assays were performed in triplicate and replicated three times.
Determination of bacterial median lethal dose (LD50)
Bacterial LD50 was measured as described (Hu et al. 2010). Briefly, for each strain, 17-day-old Cherry Valley ducks were divided evenly into five groups (eight ducks/group). The ducks were injected intramuscularly with bacteria at 106, 107, 108, 109, or 1010 CFU, respectively. Moribund ducks were killed humanely and counted as dead. Dead ducks were subjected to R. anatipestifer identification. The mortality of the ducks was recorded daily for a period of 7-days post infection. LD50 was calculated using the Reed–Muench method (Reed and Muench 1938).
Extraction, purification, and quantification of LPS
LPSs of wild-type strain CH3 and mutant RA-M1 were extracted using phenol-water method as described (Gu et al. 1998). The protein and nucleic acid contents in the LPS were determined less than 2 % (Smith et al. 1985; Warburg and Christian 1942). Purified LPS was identified using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining and Western blotting. Coomassie blue staining was used to exclude the contamination of protein in purified LPS. Western blotting was conducted to verify the binding activity of LPS with anti-LPS MAb and anti-CH3 rabbit serum. Briefly, purified LPS (100 μg/ml) was suspended in digestion buffer (1 M Tris [pH 6.8], 2 % SDS, 4 % 2-mercaptoethanol, 10 % glycerol, and 0.004 % bromophenol blue), and boiled at 100 °C for 10 min. Then, 10 μl of the sample was added to the well and subjected to SDS-PAGE on a Mini Protein II gel system (Bio-Rad, Richmond, CA, USA) with 15 % polyacrylamide in the separating gel. After electrophoresis (100 V, 2 h), purified LPSs of CH3 and RA-M1 were separated on a 15 % Bis-Tris gel (Invitrogen Life Technologies) using electrode buffer, transferred to nitrocellulose membranes (Millipore), and blocked with PBS-5 % skim milk at 4 °C overnight. Membranes were then incubated at 37 °C with anti-LPS MAb (1 μg/ml in PBS-0.05 % Tween 20) for 2 h. Goat anti-rabbit IgG (LI-COR) was used as the secondary antibody. The binding was shown using Odyssey infrared imaging system (Gene Company Limited).
Preparation of inactivated RA-M1 and trivalent R. anatipestifer vaccines
To investigate whether mutant strain RA-M1 could be used as a vaccine candidate, inactivated RA-M1 and trivalent R. anatipestifer vaccines were prepared for immunization. Briefly, RA-M1 bacteria were cultured in TSB at 37 °C for 16 h with shaking. Bacterial CFU was measured by detecting the optical density at 600 nm (OD600) (an OD600 of 1 equals 2.5 × 109 CFU of bacteria according to our calculations) and adjusting to 4 × 109 CFU/0.3 ml. The bacteria were then inactivated with 0.4 % (v/v) formalin at 37 °C for 16 h. Inactivated RA-M1 vaccine was made by blending 3 volumes of inactivated RA-M1 and 7 volumes of Montanide ISA 70 VG adjuvant (Seppic Special Chemical Corporation, Shanghai, China) according to the manufacturer’s protocol. Serotype 1, 2, and 10 trivalent R. anatipestifer vaccine was prepared as described previously (Liu et al. 2013).
Immunization, sample collection, and challenge experiment
Twelve treatment groups, eight ducks each, were given two immunizations (subcutaneous injection on the neck) with inactivated RA-M1 vaccine (groups 1–4), trivalent R. anatipestifer vaccine (groups 5–8), or saline in adjuvant (groups 9–12). Blood samples from groups 1, 5, and 9 were collected at day 7, 14, 28, and 42 post second immunization for antibody and cytokine determination.
To test the protection of vaccination, immunized ducks in groups 2, 6, and 10 were challenged with R. anatipestifer serotype 1 strain WJ4; ducks in groups 3, 7, and 11 were challenged with serotype 2 strain Yb2, and ducks in groups 4, 8, and 12 were challenged with serotype 10 strain HXb2 at 10 LD50 at day 14 post second immunization, which challenge doses were 3.25 × 109, 1.07 × 106, and 820 CFU/ml, respectively, as described previously (Liu et al. 2013). Ducks were monitored daily for clinical symptoms and death until 7 days post challenge. Furthermore, liver, heart, and spleen were collected for histological examinations from died ducks instantly or healthy ducks at day 7 post challenge.
Determination of serum antibody titers
The antisera were collected on day 7, 14, 28, and 42 post second vaccination to examine the antibody titers against R. anatipestifer strains WJ4, Yb2, and HXb2 with an indirect ELISA, as described previously (Liu et al. 2013). Briefly, 96-well ELISA plates were coated with 5 × 106 CFU/well of each strain in 50 μl bicarbonate buffer (pH 9.6) and heat-dried overnight at 50 °C. The duck sera were diluted in 2-fold steps from 1:100 to 1:25,600 for the experiment. The resulting OD450 was obtained with a plate reader (Synergy 2; BioTek). The highest dilution of the sera with an OD450 value over 2.1 times of the negative control wells was valued as the ELISA titers. All the samples were performed in triplicate. Data are presented as the means of three independent experiments.
Determination of cytokine production
Production of interleukin 2 (IL-2), IL-4, and interferon γ (IFN-γ) in the serum was measured at day 7 post second immunization using ELISA kits (Hermes Criterion Biotechnology, Elixir Canada Medicine Company, Ltd., Vancouver, BC, Canada) according to the manufacturer’s instructions. The serum samples were diluted in 5-fold steps in PBS-0.5 % Tween 20 (PBST) for the detections. The OD490 value was read with a plate reader (BioTek) within 15 min. PBST was used as the negative control. All experiments on standard dilutions, controls, and experimental samples were carried out in triplicate on the same plate, and each reaction plate contained the standard curve for the cytokine in the same preparation.
Statistical analysis
Statistical analyses were carried out using the GraphPad Software (La Jolla, CA, USA). One-way analysis of variance (ANOVA) was used for analyses of growth curves, serum bactericidal efficiency, adhesion capacity and invasion, serum antibody levels and cytokine production data, and two-way ANOVA was performed for analysis of the qRT-PCR results. The mean values are shown in the figures. Statistical significance was established at p < 0.05.
Results
Identification of the mutant strain RA-M1
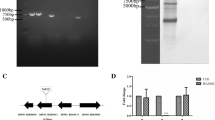
The mutant strain RA-M1 defected in reaction with anti-LPS MAb 8A9 was screened from the transposon insertion mutants using an indirect ELISA. Southern blot confirmed only one insertion of transposon Tn4351 in RA-M1 genome (Fig. 1a). The transposon-inserted gene was identified to be M949_1603 by genome walking as described previously (Hu et al. 2010) which is 873 nucleotides in length and codes for glycosyl transferase family 2 consisting of 290 amino acids. The insertion site was located at 197 bp of M949_1603 gene in wild-type strain CH3.
a Southern blot analysis of the transposon Tn4351 insertion. Lane 1, 10 μg of pEP4351 digested with XbaI (positive control). Lane 2, 10 μg of chromosomal DNA from wild-type strain CH3 digested with XbaI (negative control). Lane 3, 10 μg of chromosomal DNA from mutant strain RA-M1 digested with XbaI. Each digested sample was resolved on a 0.7 % agarose gel, and Southern blot was performed using a TnDIG labeled probe. b Real-time PCR analysis of the gene expression. The mRNA level of upstream M949_1602 gene and the downstream M949_1604 gene was measured. The changes of mRNAs were expressed as fold expression and calculated using the comparative CT(2−∆∆CT) method. Data were normalized to the housekeeping gene 16S rRNA and expressed as fold changes. The expression of M949_1603 in mutant strain RA-M1 was disrupted. Error bars represent standard deviations from three replicates. c Determination of the bacterial growth curves. Strains CH3 and RA-M1 were grown in TSB at 37 °C with shaking respectively, and the OD600 values were measured at 1-h intervals. This experiment was repeated three times, and the data were presented as the means. Error bars represent standard deviations. No significant difference was observed
As shown in Fig. 1b, the mRNA level of upstream gene M949_1602, which encodes hypothetical protein and the downstream gene M949_1604, which encodes glycosyl transferase family 1 showed no significant changes compared with wild-type strain CH3 by qRT-PCR. The transcription of M949_1603 gene was disrupted in the mutant strain RA-M1.
The growth curves of the wild-type strain CH3 and the mutant strain RA-M1 showed no significant difference during culture in TSB (Fig. 1c).
M949_1603 gene is involved in CH3 LPS synthesis and exists in R. anatipestifer serotype 1 strains
Western blotting confirmed that the binding activity of RA-M1 to rabbit anti-CH3 antiserum was changed. As shown in Fig. 2a, a deficiency in ladder-like binding pattern of CH3 LPS, which represents repeating units present in the O chain of LPS, was found in RA-M1. Furthermore, PCR amplification showed that the M949_1603 gene was lacked in R. anatipestifer serotype 2 and 10 strains detected (Fig. 2b), indicating that M949_1603 gene is type-specific for R. anatipestifer serotype 1 strains.
a Western blotting analysis of the LPS binding patterns. Each lane was sampled with 1 μg purified LPS and detected using rabbit serum against CH3. Lane 1: CH3 LPS and lane 2: RA-M1 LPS. b PCR analysis of the distribution of M949_1603 gene in Riemerella anatipestifer serotypes 1, 2, and 10 strains. Lane M: DM2000 DNA Marker (CWBIO, Beijing, China). Lanes 1–4: A 873-bp fragment was amplified from R. anatipestifer serotype 1 strains CH1, WJ4, JY4, and CQ5 using primer pairs M949_1603 ORF-F/M949_1603 ORF-R. Lanes 5–9: No M949_1603 gene was amplified from R. anatipestifer serotype 2 strains Yb2, NJ3, Th4, GD5, and YXb1. Lanes 10–11: No M949_1603 gene was amplified from R. anatipestifer serotype 10 strains HXb2 and YXb11. Lane 12: positive control of CH3; lane 13: negative control of RA-M1
Inactivation of M949_1603 gene reduced bacterial virulence and resistance to normal duck serum
The bacterial virulence of mutant strain RA-M1 was measured using Cherry Valley ducks. The results showed that the LD50 of RA-M1 was higher than 1010 CFU, more than 50 times attenuated virulence than the wild-type strain CH3, of which LD50 was 2 × 108 CFU.
The serum bactericidal assay was carried out to compare the ability of the mutant strain RA-M1 to resist the complement-mediated killing with its wild-type strain CH3. As shown in Fig. 3a, a 12.5 % serum was effective in killing the mutant strain RA-M1, which displayed higher sensitive to serum killing than the wild-type strain CH3, which was killed with 100 % serum.
a Bacterial serum resistance assay. Bacteria were incubated with the normal duck sera at different dilution at 37 °C and were enumerated at 30-min incubation. A significant reduced resistance of RA-M1 to the normal duck sera than that of CH3 was shown. ***p < 0.01. b–c Bacterial adherence and invasion assays. Strains CH3 and RA-M1 were tested on Vero cells. b Adherence assay. c Invasion assay. The data represent the number of bacteria bound to or invaded into Vero cells in each well of 24-well plate. The error bars represent mean ± standard deviations from three independent experiments. The capacities of both adherence and invasion for RA-M1 mutant were significantly increased in comparison with its wild-type strain CH3
Deletion of M949_1603 gene increased the bacterial adherence and invasion abilities
To determine whether deletion of M949_1603 gene affects the bacterial adherence and invasion activity, CH3 and RA-M1 were tested on Vero cells. When infected at 50 multiplicity of infection, the RA-M1 bacterial numbers accounted for adherence is 3,633,333 ± 700,000 CFU/well, 10 times higher than that of wild-type strain CH3 (358,889 ± 48,889 CFU/well) (p < 0.001) (Fig. 3b). After an additional 1 h of incubation with gentamicin (100 mg/ml), the RA-M1 bacterial counts for invasion were 1,457,778 ± 251,111 CFU/well, which were significantly increased in comparison with 111,778 ± 8111 CFU/well of wild-type strain CH3 (p < 0.001) (Fig. 3c).
Cross-protection for R. anatipestifer serotypes 1, 2, and 10 strains of inactivated RA-M1 vaccination
At day 14 post second vaccination, RA-M1 vaccinated ducks were challenged with virulent R. anatipestifer serotype 1 strain WJ4, serotype 2 strain Yb2, and serotype 10 strain HXb2 at 10 LD50, respectively. The results showed that RA-M1 vaccinated ducks were totally protected, which matched the efficiency level of the trivalent R. anatipestifer vaccine (Table 1). One week after challenge, ducks in immunized group remained healthy while ducks in control group appeared with clinical symptoms including apathy, loss of appetite, serositis, fibrinous pericarditis, perihepatitis, and necrosis foci in the spleen, as shown in Fig. 4.
Gross lesion observation. Row A samples collected from non-vaccinated ducks without challenge, normal control. Row B samples collected from non-vaccinated ducks with challenge of Yb2. Row C samples collected from vaccinated ducks without challenge. Row D samples collected from vaccinated ducks at 7 days post challenge of Yb2. Typical gross lesions of R. anatipestifer infection containing serositis, perihepatitis, fibrinous pericarditis, and marble-like lines in the spleen were shown in row B, while no pathological lesions in rows A, C, and D
Determination of serum antibody levels
The antibodies against strains WJ4, Yb2, and HXb2 were tested at day 7, 14, 28, and 42 post second immunization. The sera isolated from normal ducks before immunization were used as negative controls. As shown in Fig. 5, the antibodies against the three strains were all significantly enhanced post second immunization and maintained at high levels up to day 42.
Determination of antibody ELISA titers. a Antibody titers to strain WJ4. b Antibody titers to strain Yb2. c Antibody titers to strain HXb2. Data were converted and expressed as log titers. The logarithmic mean antibody titers (mean ± SD) are from one representative experiment with eight animals (n = 8). **p < 0.01; *p < 0.05
Cytokine expressions
Cytokine production in the sera of vaccinated ducks was detected by an ELISA kit at day 7 post second immunization. As shown in Fig. 6, the vaccinated ducks produced significantly larger amounts of IL-2 and IL-4 but lower amounts of IFN-γ, compared with the control group (p < 0.05).
Discussion
In the present study, M949_1603 gene deletion R. anatipestifer mutant RA-M1 was characterized and the cross-protection for serotype 1, 2, and 10 strains was evaluated. The mutant does not bind to anti-LPS MAb in an indirect ELISA. Western blotting indicated an obvious missing of ladder-like pattern in RA-M1 LPS, compared to those in CH3 LPS. Deletion of M949_1603 gene in RA-M1 leads to significant attenuated virulence in duck model and increased sensitive to normal duck serum. Sequence analysis revealed that the transposon was inserted into the M949_1603 gene at 197 bp in strain CH3 (Wang et al. 2015).
The M949_1603 gene encodes a glycosyl transferase, belonging to glycosyl transferase family 2. The specificity of glycosyl transferases contributes to O-antigen specificity in gram-negative bacteria. For example, in Escherichia coli O114, the second step of the repeating-unit assembly is catalyzed by α β-1,3-glycosyl transferase (β3-GalT) of transferase family 2 (Zhou et al. 2013). The M949_1603 gene exists in R. anatipestifer serotype 1 strains only, indicating that its encoding product is a type-specific glycosyl transferase. The ladder-like bands of bacterial LPS reflect the number of repeating units present in the O chain of LPS (Fomsgaard et al. 1990). The defect in ladder-like pattern of mutant LPS indicated that the M949_1603 gene is associated with the biosynthesis of intact O-antigen in R. anatipestifer.
O-antigen plays a key role in protecting bacteria from complement-mediated killing (Grozdanov et al. 2002). In this study, inactivation of the M949_1603 gene resulted in significantly increased sensitivity to normal duck serum killing, which is similar to results of other O-antigen deletion mutants (Wang et al. 2014). Innate immune response is believed to be the first step for host to eliminate invasive bacteria, and O chain is required for resistance to complement-mediated bactericidal activities. The attenuated virulence of RA-M1 (more than 50 times attenuated in comparison with its wild-type strain CH3, based on LD50 evaluation) may be caused by the defect in O-antigen.
In an effort to explore the role of O-antigen in the interaction between R. anatipestifer and epithelial cells, we have investigated the adherence and invasion capability of mutant strain RA-M1 on Vero cells as described previously (Hu et al. 2011). The results confirmed that about 10-fold increased numbers of adhesion and invasion of mutant strain RA-M1 than that of wild-type strain CH3. That is to say, O-antigen mutant strain RA-M1 changed the phenotype of CH3 and take different way to adherent and invade cells as reported for Brucella abortus (Pei and Ficht 2004).
Animal experiments demonstrated that inactivated RA-M1 vaccine was effective in protecting the ducks from challenge with serotype 1, 2, and 10 strains, which protection index was similar with the trivalent vaccine. Therefore, RA-M1 vaccine may act as a substitute for the trivalent R. anatipestifer vaccine for its simple manufacture process in the future study. Conserved LPS epitope(s) can exist among different strains, species, and genera of non-enteric gram-negative human pathogens (Campagnari et al. 1990). One explanation is that the deletion of the M949_1603 gene resulted in the exposure of the highly conversed epitope on the RA-M1 LPS by eliminating the latent steric effect, which may be concealed in the parent strain CH3. However, based on these data alone, it is not clear which moiety of LPS would serve as a conserved antigen among different R. anatipestifer serotype strains; further analysis of LPS structure is needed. At least, our current data revealed that deficiency of the type-specific O chain contributes to forming a highly conserved epitope among different serotype strains. Inactivated RA-M1 vaccine with cross-protection for R. anatipestifer strains WJ4 (serotype 1), Yb2 (serotype 2), and HXb2 (serotype 10) was successfully developed. Animal experiments demonstrated that the protective rate of the RA-M1 vaccine reached as high as 100 % after the second immunization. In addition, RA-M1 vaccine induced high levels of antibodies in ducks against R. anatipestifer serotype 1, 2, 10 strains, and high levels of cytokines IL-2 and IL-4 as the trivalent R. anatipestifer-inactivated vaccine did post second immunization. Cytokines play an important role in the activation of adaptive immune response toward a Th1- or Th2-type response, indicating that inactivated RA-M1 vaccine can induce both humoral and cellular immunity. The production of IFN-γ, however, was decreased post immunization with both inactivated RA-M1 and trivalent vaccine. IFN-γ mainly promotes cellular immunity, decreased IFN-γ level means that induced immune response of inactivated vaccine focus mainly on humoral immunity to develop long-term memory B cells. IFN-γ was consistently a subdominant effector response with many immunizations (Orr et al. 2015).
In summary, we have identified the mutant strain RA-M1, in which the O-antigen was defected due to the knockout of M949_1603 gene. Our study demonstrated that inactivated RA-M1 vaccine protected ducks from challenge with R. anatipestifer serotype 1, 2, and 10 strains. Therefore, RA-M1 can be further investigated as a vaccine candidate against R. anatipestifer for clinical applications. This study has important implications for the effective control of R. anatipestifer infections in China.
References
Aquilini E, Merino S, Regue M, Tomas JM (2014) Genomic and proteomic studies on Plesiomanas shigelloides lipopolysaccharide core biosynthesis. J Bacteriol 196(3):556–567
Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN (1981) Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16(1-3):237–247
Brown DB, Forsberg LS, Kannenberg EL, Carlson RW (2012) Characterization of galacturonosyl transferase genes rgtA, rgtB, rgtC, rgtD and rgtE responsible for lipopolysaccharide synthesis in nitrogen-fixing endosymbiont Rhizobium leguminosarum: lipopolysaccharide core and lipid galacturonosyl residues confer membrane stability. J.Biochem 287(2):935–949
Campagnari AA, Spinola SM, Lesse AJ, Kwaik YA, Mandrell RE, Apicella MA (1990) Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb Pathog 8(5):353–362
Fomsgaard A, Freudenberg M, Galanos C (1990) Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol 28(12):2627–2631
Grozdanov L, Zahringer U, Blum-Oehler G, Brade L, Henne A, Knirel Ya, Schombel U, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Rietschel ET, Dobrindt U (2002) A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J Bacteriol 184(21):5912–5925
Gu X-X, Chen J, Barenkamp SJ, Robbins JB, Tsai C-M, Lim DJ, Battey J (1998) Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarrhalis. Infect Immun 66(5):1891–1897
Helfer DH, Helmboldt CF (1977) Pasteurella anatipestifer infection in turkeys. Avian Dis 21(4):712–715
Hu Q, Chen H, Liu X, Zhan M, Zhang Z, Deen S, Zhang Y (2002) Determination of growth curve of Riemerella anatipestifer. Chin Anim Husb Vet Med 34:8–9
Hu Q, Han X, Zhou X, Ding S, Ding C, Yu S (2010) Characterization of biofilm formation by Riemerella anatipestifer. Vet Microbiol 144(3):429–436
Hu Q, Han X, Zhou X, Ding C, Zhu Y, Yu S (2011) OmpA is a virulence factor of Riemerella anatipestifer. Vet Microbiol 150(3):278–283
Hu Q, Zhu Y, Tu J, Yin Y, Wang X, Han X, Ding C, Zhang B, Yu S (2012) Identification of the genes involved in Riemerella anatipestifer biofilm formation by random transposon mutagenesis. PLoS ONE 7(6) doi:10.1371/journal.pone.0039805
Liu H, Wang X, Ding C, Han X, Cheng A, Wang S, Yu S (2013) Development and evaluation of a trivalent Riemerella anatipestifer-inactivated vaccine. Clin Vaccine Immunol 20(5):691–697
Loh H, Teo T, Tan HC (1992) Serotypes of Pasteurella anatipestifer isolates from ducks in Singapore: a proposal of new serotypes. Avian Pathol 21(3):453–459
McQuillen DP, Gulati S, Rice PA (1994) Complement-mediated bacterial killing assays. Methods Enzymol 236:137–147
Nikaido H, Vaara M (1985) Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49(1):1–32
Orr MT, Plessner Windish H, Beebe EA, Argilla D, Huang PW, Reese VA, Reed SG, Coler RN (2015) IFN-gamma and TNF are not essential parameters of CD4 T cell responses for vaccine control of tuberculosis. J Infect Dis. doi:10.1093/infdis/jiv055
Pathanasophon P, Sawada T, Tanticharoenyos T (1995) New serotypes of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 24(1):195–199
Pathanasophon P, Phuektes P, Tanticharoenyos T, Narongsak W, Sawada T (2002) A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 31(3):267–270
Pei J, Ficht TA (2004) Brucella abortus rough mutants are cytopathic for macrophages in culture. Infect Immun 72(1):440–450
Perepelov AV, Wang Q, Levina EA, Ovchinnikova OG, Qian Y, Shashkov AS, Wang L, Knirel YA (2014) Structure and gene cluster of the O-antigen of Escherichia coli O36. Carbohydr Res 390:46–49
Pierce RL, Vorhies MW (1973) Pasteurella anatipestifer infection in geese. Avian Dis 17(4):868–870
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27(3):493–497
Sandhu TS, Rimler R (1997) Riemerella anatipestifer infection. Diseases of Poultry 161–166
Segers P, Mannheim W, Vancanneyt M, De Brandt K, Hinz K-H, Kersters K, Vandamme P (1993) Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int J Syst Bacteriol 43(4):768–776
Smith P, Krohn RI, Hermanson G, Mallia A, Gartner F, Provenzano M, Fujimoto E, Goeke N, Olson B, Klenk D (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150(1):76–85
Wang X, Ding C, Wang S, Han X, Hou W, Yue J, Zou J, Yu S (2014) The AS87_04050 gene is involved in bacterial lipopolysaccharide biosynthesis and pathogenicity of Riemerella anatipestifer. PLoS ONE 9(10), e109962. doi:10.1371/journal.pone.0109962
Wang X, Ding C, Han X, Wang S, Yue J, Hou W, Cao S, Zou J, Yu S (2015) Complete genome sequence of Riemerella anatipestifer serotype 1 strain CH3. Genome Announc. doi:10.1128/genomeA.01594-14
Warburg O, Christian W (1942) Isolation and crystallization of enolase. Biochem Z 310:384–421
Zhou D, Utkina N, Li D, Dong C, Druzhinina T, Veselovsky V, Liu B (2013) Biochemical characterization of a new beta-1,3-galactosyltransferase WbuP from Escherichia coli O114 that catalyzes the second step in O-antigen repeating-unit. Carbohydr Res 381:43–50. doi:10.1016/j.carres.2013.08.021
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31272591).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(PDF 10 kb)
Rights and permissions
About this article
Cite this article
Zou, J., Wang, X., Ding, C. et al. Characterization and cross-protection evaluation of M949_1603 gene deletion Riemerella anatipestifer mutant RA-M1. Appl Microbiol Biotechnol 99, 10107–10116 (2015). https://doi.org/10.1007/s00253-015-6848-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6848-y