Abstract
Silkworm (Bombyx mori L.) larvae were used as an ideal animal protein source for astronauts in the bioregenerative life support system (BLSS). Here, we compared the difference in bacterial communities of the silkworm larval gut between the BLSS rearing way (BRW) and the traditional rearing way (TRW) through culture-dependent approach, 16S rRNA gene analysis, and denaturing gradient gel electrophoresis (DGGE). The culture-dependent approach revealed that the numbers of gut bacteria of silkworm in the BRW significantly decreased compared with that of the TRW. The analysis of clone libraries showed that the gut microbiota in the BRW was significantly less diverse than that in the TRW. Acinetobacter and Bacteroides were dominant populations in the BRW, and Bacillus and Arcobacter dominated in the TRW. DGGE profiles confirmed the difference of silkworm gut bacterial community between two rearing ways. These results demonstrate that gut bacteria change from the BRW contributes to the decrease of silkworm physiological activity. This study increases our understanding of the change of silkworm gut microbiota in response to lettuce leaf feeding in the BRW. We could use the dominant populations to make probiotic products for nutrient absorption and disease prevention in the BLSS to improve gut microecology, as well as the yield and quality of animal protein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The insect gut is inhabited by a wide diversity of microorganisms as a result of its constituting intestinal microbial ecosystem. Studies have revealed that the gut microbiota is able to resist invasion of pathogenic microorganisms and to maintain normal ecological balance. Importantly, the intestinal microbiota in insect is involved in the digestion of insect it absorbs and may assist in insect immunity by forming a persistent infection within their hosts (McKillip et al. 1997; Dillon et al. 2005). The composition and structure of microbial are dynamic, which can be varied with changing nutrient availability, physiological environments, and the proximity to other organisms (Butler et al. 2003; Kiorboe et al. 2003; Militza et al. 2006). The modification of diets has been shown to be one of the most important factors leading to insect physiological activity and gut flora alterations.
Mulberry silkworm (Bombyx mori L.) is an important economical insect whose importance is reflected not only by its silk products used as textile raw materials, but also by its valuable nutritional composition. The idea of rearing mulberry silkworm larvae to provide animal protein for crew in the bioregenerative life support system (BLSS) required by long-term missions to the moon and Mars is widely accepted. This is because silkworm food has many positive characteristics such as high protein content, appropriate proportions of the amino acids, and unsaturated fatty acids that can satisfy human nutrition requirements. Furthermore, silkworm culture requires short time and small growing space with little odors and wastewater products (Yang et al. 2009). Due to limitation of space and resource, mulberry silkworm rearing method in BLSS was different from traditional rearing method which only uses mulberry leaves. In BLSS, mulberry silkworms of the first three instars (from the 1st day to 16th day) were fed with mulberry leaves and for those of the last two instars (from the 17th day to 25th day) were fed with lettuce leaves (Yu et al. 2008). In this rearing way, silkworms can grow to complete the life generation, but their growth rate and biological transformation rate were relatively slower compared with the traditional rearing way. Prior study has provided evidence that many silkworm intestinal bacteria produce digestive enzymes like amylase, caseinase, gelatinase, lipase, and urease (Kalpana et al. 1994). Thus, it is potentially possible that the change of gut flora due to lettuce leaf feeding may contribute to the decrease of physiological activity of the silkworms. However, how diet compositions in BLSS shape gut bacterial communities of mulberry silkworms is still unknown.
In this study, the gut bacterial communities of silkworm larvae reared with the traditional rearing way (TRW) and BLSS rearing way (BRW) were investigated using culture-dependent, clone library analysis of 16S rRNA gene and denaturing gradient gel electrophoresis (DGGE) approaches. The changes of intestinal flora ecology in silkworm larvae in response to lettuce leaf feeding were revealed. This study may promote the development of probiotic products of animal protein under BLSS.
Materials and methods
Silkworm strains and rearing methods
The silkworm eggs B. mori L. 872 × 871 were bought from Guangtong Silkworm seed Co. Ltd. (Shandong Province, China). The silkworm eggs were incubated under a 12-h light/12-h dark cycle in an artificial cultivation box at 25 °C and 80 % of relative humidity. When 20 % of eggs had little black dots on the surface, they were shaded with black gobo for about 2 days to ensure the larvae hatching out at one time. The silkworm larvae were reared with mulberry leaves from the first to third instar and then divided into two groups: the BLSS breeding group reared with stem lettuce leaves and the conventional breeding group still reared with mulberry leaves at the beginning of the fourth instar.
Sampling of silkworm gut, colony count, and DNA extraction
When the two groups of silkworms grew to the third day of the fifth instar, ten individuals of each group were selected and subjected to starvation overnight. Those silkworms were surface decontaminated by wiping with 70 % ethanol solution and scorched gently in a flame. The content of gut was taken and placed into sterile microcentrifuge tubes on ice under aseptic condition. According to the screening standard of bacteria, tenfold serial dilution was used for inoculation. On each nutrient agar plate, 0.1 mL of intestinal content of two groups with 105 dilution was spread and incubated at 37 °C for 5 days; all samples were repeated three times for counting colony. Pooled DNA samples of the fifth instar in the BLSS and the conventional breeding groups were composed of DNA extracted from the selected ten individuals, respectively. DNA was extracted with a Promega DNA Kit (Promega, USA), quantified with a BioPhotometer (Eppendorf), and stored at −20 °C until used.
PCR amplification and DGGE
Bacterial 16SrRNA gene was amplified by polymerase chain reaction (PCR). The primers specific for conserved sequences flanking the variable V6–V8 region of the 16S rDNA:799f (5-AACAGGATTA GATACCCTG-3) and 1492r (5-GGTTACCTTGTTACGACTT-3) were used. PCR amplification was performed in a total volume of 25 μL containing 5 μL of DNA extract, 1 μL of each primer (10 μL mL−1), 1 μL of deoxyribonucleotide triphosphate (dNTP) mixture (10 mmol mL−1), 3 μL of 10× Taq PCR buffer (containing Mg2+), and 0.5 μL of Taq DNA polymerase (TaKaRa, China). The touchdown PCRs were cycled in a thermal cycler with an initial denaturation at 94 °C for 5 min followed by 30 cycles of denaturation for 1 min at 94 °C, primer annealing for 1 min at 51 °C, primer extension for 1 min at 72 °C, followed by additional elongation at 72 °C for 10 min. A G+C clamp (5-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCG CGG GGG G-3) was added to the 5 end of the forward primer to stabilize the PCR product and to prevent strand dissociation during DGGE (Nakatsu et al. 2000). The primers were 968f (AA CGC GAA GAA CCT TAC) and 1378r (CGG TGT GTA CAA GGC CCG GGA ACG). The amplified total DNA products were used as the template. The PCR amplification was performed in a total volume of 100 μL containing 6 μL DNA template, 2 μL of each primer (10 μL mL−1), 8 μL dNTPs (10 mmol mL−1), 10 μL 10× Taq PCR reaction buffer (containing Mg2+), 1 μL Taq DNA polymerase (TaKaRa, China), and 71 μL ddH2O. The touchdown PCRs were cycled in a thermal cycler with an initial denaturation at 94 °C for 5 min, and after a single cycle of 94 °C melting for 5 min and 64 °C annealing for 1 min and 72 °C for 3 min, 19 cycles were performed in which the annealing temperature was decreased 1 °C every other cycle. Fifteen cycles were then performed using an extension of 55 °C, followed by a single cycle of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 10 min.
Sequence analysis of 16SrDNA and phylogenetic tree construction
Two groups of 16SrRNA genes were amplified with a primer pair of 799f and 1492r. The products were purified with PCR product purification kit and cloned by using the pMD-18T vector cloning system and introduced into competent Escherichia coli DH5a cells. Inserts of the expected size (approx. 700 bp) were amplified by PCR with the M13 forward and reverse primers. Three clones were randomly selected for sequencing in Sangon Biotech Co., Ltd (Shanghai, China). To determine the closest known relatives of the 16SrDNA sequences obtained, searches were performed with BLAST program at NCBI (http://www.ncbi.nlm.nih.gov/BLAST). The16SrRNA gene sequences obtained from the intestinal bacteria in mud crabs have been deposited to GenBank with the accession numbers from KF935549 to KF935648. Phylogenetic tree of the sequence analysis was constructed from a matrix of pairwise genetic distances by the neighbor-joining method (MEGA 5.0). The bootstrap analysis of 1,000 replicates was performed (Saitou and Nei 1987).
DGGE
DGGE was performed with a Bio-Rad DCode mutation detection system (Bio-Rad, USA). A forty-five microliter PCR product was deposited in each well of 8 % (m/v) polyacrylamide (acrylamide-bis-acrylamide, 37.5:1) gels containing a denaturant ranging from 30 to 55 % denaturant gradient. Electrophoresis was performed at a constant voltage of 30 V at 60 °C for 30 min, with an increase in the voltage to 200 V for another 5 h. After electrophoresis, the gels were stained in GelRed for 20 min and photographed under a UV transilluminator. DGGE band recovery and clustering were carried out as described previously.
Statistical analysis
Statistical analysis of microbial richness and diversity was performed based on 16SrDNA sequences. Microbial diversity indices were presented as the number and proportional abundance of clones. Briefly, Shannon’s index measures the proportional abundances of species in a community. Shannon’s index is calculated as follows:
(n i is the number of the clone and N is the number of all clones).
Coverage index is calculated as:
(n1 is the number of the out and N is the number of all clones).
Simpson’s index is calculated as:
(n i is the number of the clone and N is the number of all clones).
Results
Enumeration of the intestinal microflora
The results of bacterial counts on nutrient agar plates from the fifth instar silkworm larvae intestinal contents under the TRW and BRW are presented in Fig. 1. The gut bacterial concentration of the BRW samples ranged from 1.5 × 107 to 3.8 × 107 colony-forming units (CFU) mL−1 with an average of 3.2 × 107 CFU mL−1. In contrast, the average bacterial concentration of the TRW samples was 7.1 × 107 CFU mL−1, ranging from 5.6 × 107 to 9.9 × 107 CFU mL−1. Obviously, there was a significant difference in the gut bacterial number between TRW samples and BRW samples (p < 0.05), indicative of decreasing the numbers of intestinal bacteria of silkworm when fed with lettuce in the BRW.
Analysis of clone library sequences
The 16S rRNA gene libraries from the intestinal contents of the fifth instar silkworm larvae in the TRW and BRW samples were constructed to investigate the microbial diversity of silkworm gut, respectively. In total, 25 genotypes were found in both cases. The percentage of microorganism coverage in the BRW and TRW libraries was 70.21 and 78 % (Table 1), respectively, implying that each clone library covered the most of the gut microorganisms of silkworm larvae.
The clone analysis showed that the sequence population was less diverse in the BRW samples than that in the TRW samples. This was further confirmed by calculating the Shannon–Wiener (H) and Simpson indices (D) (1.968–2.374 and 83.6–90.8 %, respectively) (Table 1). These results showed that the bacterial diversity was higher in the guts of TRW silkworms than that of the BRW silkworms.
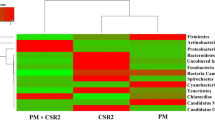
The sequences of a length of approximately 600 bp for clones representing individual genotypes were obtained from clone libraries of the TRW samples and BRW samples. The identified taxa for two clone libraries were mainly grouped into five different phyla: Firmicutes, Bacteroides, Proteobacteria, Tenericutes, and Actinomycetes. As shown in Fig. 2, Proteobacteria, Bacteroidetes, and Firmicutes were dominant in the intestinal bacterial community in both silkworm populations. However, there were some differences in the proportions of Proteobacteria, Bacteroidetes, and Firmicutes between mulberry silkworm of the BRW and TRW. The proportions of Proteobacteria and Firmicutes in the BRW samples were 30 and 34 %, respectively. In contrast, Proteobacteria and Firmicutes separately accounted for 31 and 43 % in the TRW samples. The proportion of Bacteroidetes in the BRW group was 55 %, which was higher by 10 % than that in the TRW group. Actinobacteria, as a unique phylum, only occurred in the TRW group with a lower percentage of 2 %. In libraries of the BRW group, however, Tenericutes was found to be a unique phylum with 2 % proportion. These data indicated that the diversity and dominant components in the gut bacteria of silkworms under two rearing ways were significantly different.
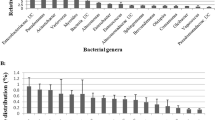
We next analyzed changes at genus level of silkworm gut microbes under the TRW and BRW ways. As shown in Fig. 3, 18 differently named bacterial genera were detected in the gut of silkworm under two rearing ways. Seven genera were common members in both samples: Bacteroides, Chryseobacterium, Clostridium, Enterococcus, Porphyromonas, Paenibacillus, and Devosia. In these seven genera, the most common bacterial genus was Bacteroides, with 22 % of clones in the TRW samples and 8.5 % of clones in the BRW samples. The next were Chryseobacterium (8 % of TRW clones and 10.6 % of BRW clones) and Clostridium (16 % of TRW clones and 2.1 % of BRW clones). The rest of the common genera comprised of <5 % of the total clones in both samples. Moreover, some unique genera were found in the gut microbiota of the two samples. The genera Acinetobacter (30 % of adequacy), Anaerofilum (8 %), Anaeroplasma (2 %), and Serratia (2 %) were exclusively found in the BRW samples, while the genera Peptostreptococcus (2.1 %), Janthinobacterium (12.8 %), Shigella (10.6 %), Arcobacter (17 %), Rothia (2.1 %), Streptococcus (4.3 %), and Bacillus (17 %) were only found in the TRW samples. Phylogenetic tree for the members of these genera within two types of samples were represented in Fig. 4. This analysis further confirmed that feeding with changed feedstuff at the beginning of the fourth instar resulted in great change in gut microbiota of silkworm in the BRW.
DGGE analysis of V6–V8 regions
The total genomic DNA was extracted from the gut of silkworm larvae. A 410 bp of V6–V8 region of the 16S rRNA gene was amplified by PCR from purified genomic DNA and DGGE profiles were performed in the three sample types, namely, 5M, gut content sample from the fifth instar larvae of silkworms in TRW; 5L, gut content sample from the fifth instar larvae of silkworms in BRW; and 4L, gut content sample from the fourth instar larvae of silkworms in BRW) (Fig. 5a). The number of bands in these three samples varied from 10 to 19, with the highest number occurring in the 5M sample. 4L sample was characterized by a limited number of bands (10). Each sample profile displayed a unique banding pattern. Cluster analysis revealed that gut microbial community structure in all three samples exhibited low similarity at a coefficient ranged from 45.7 % (between 4L and 5M) to 55.3 % (between 4L and 5L) (Fig. 5b). These results suggested that microbial community in the silkworm gut was altered due to lettuce leaf feeding.
Thirty-four representative bands from the DGGE gel were eluted and sequenced; 11 unique sequences were obtained eventually as shown in Table 2. Band 1, band 7, and band 10 appeared in all the silkworm instar gut samples under the two rearing ways and showed homology to Enterobacter sp., Pseudomonas fluorescens, and Pantoea sp., respectively. Band 2, band 4, and band 6, as unique bacteria in the silkworm gut of the TRW group, were affiliated to Pectobacterium carotovorum, Lysobacter sp., and Alcaligenes sp., respectively. Band 9 (Rhodococcus sp.) and band 11 (an uncultured bacterium) were exclusively found in the fourth and fifth instar gut of the BRW group. Band 3, Aeromonas hydrophila, occurred in gut content samples from 5M and 4L, while it disappeared in the samples from 5L. Band 5 and band 8, found only in samples 5L and 5M, were clustered in Moraxellaceae and Microbacterium.
Discussion
We compared the number of silkworm larval gut microbiota in response to two different rearing ways (the TRW and BRW) by classical culture techniques. Total cultivatable bacterial count of the entire digestive tract of silkworm fifth instar in the BRW was 3.2 × 107 CFU mL−1, which was significantly lower than that in the TRW (7.1 × 107 CFU mL−1). A prior study demonstrated that most of the cultivable bacteria from the silkworm larvae digestive tract were able to produce digestive enzymes to utilize plant polysaccharides such as cellulose, starch, xylan, and pectin (Anand et al. 2010). Thus, a remarkable decrease of cultivable bacteria in the BRW samples supports our hypothesis that the gut flora change due to lettuce leaf feeding leads to the decrease of silkworm physiological activity.
In order to show how the gut bacterial community structure of silkworm larvae changes under different rearing methods, we examined diet effect on the composition of gut bacteria of the silkworm fifth instar by detecting 16S rRNA gene libraries. Silkworms fed on mulberry leaves from the first instar to fifth instar had a significantly more diverse bacterial community than those fed on mulberry leaves from the first instar to third instar and on lettuce during the fourth instar and fifth instar. The traditional diversity indices including coverage, Simpson, and Shannon, which assess relative phylotype abundance (Eckburg et al. 2005), provide further evidence for these observations (Table 2).
16S rRNA gene libraries indicated that the gut microbiota of silkworms fed on lettuce leaves was relatively simple, consisting of 11 different phylotypes (Fig. 3). In contrast, there were14 different phylotypes in the gut microbiota of silkworms fed on mulberry leaves Among them, there are seven phylotypes in common bacteria: Clostridium, Bacteroides, Enterococcus, Chryseobacterium, Porphyromonas, Paenibacillus, and Devosia. Clostridium, a genus containing a wide range of anaerobic bacteria able to use cellulose, hemicellulose, and pectin (Weber et al. 2001), plays a key role in the lignocellulosic biomass degradation of the insect gut (Shi et al. 2011). Enterococcus is a predominant bacterium in the silkworm gut according to Xiang et al. (2007). Studies have shown that the gypsy moth larvae intestinal Enterococcus can decrease intestinal pH through metabolism effect, which protects the host from attack of some toxins (Broderick et al. 2004). In the silkworm larvae, Enterococcus is present at high frequency in the digestive tract, which can lower the gut pH and inhibit the suppression of Nosema bombycis germination, contributing to resistance to disease (Takizawa and Iizuka 1968; Lu and Wang 2002). Previous study pointed out that the Bacillus group (Bacillus pumilus, Bacillus atrophaeus, Bacillus licheniformis, and Bacillus amyloliquefaciens) in the gut of indigenous silkworm breeds was one of the functionally important beneficial microbes, which developed new probiotic formulation as pharmaceuticals for enhancing the development and growth of silkworm [(Subramanian et al. 2009; Ceuppens et al. 2013). In our study, Bacillus was exclusively found in the TRW samples. Moreover, genus Bacillus from the termite gut was able to use xylan, arabinogalactan, and carboxymethylcellulose as substrates (Schäfer et al. 1996). Therefore, it is reasonable that the function of Bacillus in the silkworm gut is associated with nutrient and energy digestion. Serratia can proliferate in the digestive tract of the insect host and invade the coelom, resulting in host infection and death (Apte-Deshpande et al. 2012). Serratia occurred as a unique microbe in the BRW group. Kalpana et al. reported that the percentage of Acinetobacter in the digestive tract of silkworm fed on mulberry leaves gradually decreased from the second to fourth instar and completely disappeared in the fifth star (Kalpana et al. 1994). Similarly, in our study, Acinetobacter was not found to occur in the digestive tract of silkworm fifth instar in the TRW group. However, Acinetobacter was a predominant bacterium in the gut of fifth instar silkworm under the BRW condition. This phenomenon indicates that Acinetobacter may play a positive role in digesting lettuce leaves in the silkworm gut. Anaerofilum and Anaeroplasma occurred only in the BRW group and were pathogenic bacteria which can reduce resistance to diseases of the insects fed with lettuce leaves (Ansari et al. 2003). Overall, our results demonstrate that gut bacterial community structure of silkworm larvae was changed dramatically after being fed by different food in the fourth instar. Moreover, the decrease of beneficial bacteria and appearance of pathogenic bacteria in the BRW samples confirmed that silkworm larvae had reduced digestive enzyme activity and physiological activity after feeding with lettuce leaves.
DGGE was used as a complementary method to further evaluate the gut bacterial community structure of silkworm larvae associated with two different rearing methods because DGGE profiles and clone library data depended on different primer sets. DGGE fingerprints showed a similar phenomenon to those observed by clone library. For instance, gut bacterial communities of silkworm fifth instar appeared markedly different between BRW (5M) and TRW (5L). In addition, DGGE profiles showed that gut flora of silkworms in the BRW changed with the instar. The fourth instar and fifth instar silkworm gut microbiota had a differentiation of 46.5 %. This is consistent with previous work where bacterial flora of the silkworm digestive tract exhibited a big change in population and diversity in different instars (Kalpana et al. 1994). Microbacterium oxydans and Rhodococcus sp. isolated from silkworm digestive tract in previous studies (Tian et al. 2007) were found to be the degraders of anthracene due to their metabolic versatility, genetic plasticity, and ability to survive in harsh environment (Salam et al. 2013). In this study, those two species (bands 8 and 9) were presented in sample 5L only. Thus, we conclude that the change of feedstuff made the silkworm gut not be in an environment suitable for growth in the BRW. A. hydrophila-degrading carbohydrates through glycolysis and pentose phosphate pathway (Seshadri et al. 2006) were detected in samples 4L and 5M (band 3). but not in sample 5L. This is similar to that reported by Kalpana et al. (1994), in which Aeromonas was found in the fifth instar, not in the fourth instar of silkworm fed on mulberry leaves. Therefore, the fact that a low number of Aeromonas was found on the fourth instar of silkworm gut in the BRW reveals that the gut microbiota did not completely change and had relevance with the gut of silkworm fed with mulberry leaves at the beginning of breeding silkworms with lettuce leaves. Overall, our results showed diversity decrease and imbalance of gut microbiota of silkworms in the BRW, indicating that alteration in the gut flora by feeding lettuce was attributed to digestive enzyme activity and physiological activity reduction of silkworms. However, the bacteria obtained by 16S rDNA sequencing did not agree with those obtained by sequencing of dominant bands detected in the DGGE. The reason is that the DGGE method only referred to about 200 bp fragment of 16S rDNA V6–V8 region, which could not cover the overall data information. Xiang et al. used clone libraries and 16S PCR-DGGE to study the bacteria community and the effects of diet on bacterial composition in larval midguts of the silkworm and only found one common bacterium Enterococcus (Xiang et al. 2007).
In summary, the appearance of profitless bacteria in the gut of silkworm under the BRW might break down the balance structure of healthy gut microbial community, resulting in reduced digestive enzyme activity and physiological activity. This hindered the growth of the silkworm. One way to solve this problem is to develop new probiotics using beneficial gut microbes of silkworm. The bacteria, dominated in the gut content from the silkworms well-developed in the BRW, may be a promising candidate as probiotics for improving animal protein yield and quality in BLSS. Detailed studies are needed to confirm the hypothesis that the perceived roles of these microbes are good for sericulture in BLSS.
References
Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T, Geoffrey CJ, Vendan SE (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10:1–20
Ansari MA, Tirry L, Moens M (2003) Entomopathogenic nematodes and their symbiotic bacteria for the biological control of Hoplia philanthus (Coleoptera: Scarabaeidae). Biol Control 28:111–117
Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN (2012) Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e404017. doi:10.1371/journal.pone.004040
Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70:293–300
Butler JL, Williams MA, Bottomley PJ, Myrold DD (2003) Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microbiol 69:6793–6800
Ceuppens S, Boon N, Uyttendaele M (2013) Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol Ecol 1:1–18
Dillon RJ, Vennard CT, Buckling A, Charnley AK (2005) Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–1298
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Kalpana S, Hatha AAM, Lakshmanaperumalsamy P (1994) Gut microflora of the larva of silkworm, Bombyx mori. Insect Sci Appl 15:499–502
Kiorboe T, Tang K, Grossart HP, Ploug H (2003) Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl Environ Microbiol 69:3036–3047
Lu X, Wang F (2002) Inhibition of cultured supernatant of Enterococci strains on germination of Nosema bombycis spores in vitro. Sci Sericulture 28:126–128 (In Chinese)
McKillip JL, Small CL, Brown JL, Brunner JF, Spence KD (1997) Sporogenous midgut bacteria of the leafroller, Pandemis pyrusana (Lepidoptera: Tortricidae). Environ Entomol 26:1475–1481
Militza CC, Nakatsu CH, Konopka A (2006) Effect of nutrient periodicity on microbial community dynamics. Appl Environ Microbiol 72:3175–3183
Nakatsu CH, Torsvikb V, Øvreås L (2000) Soil community analysis using DGGE of 16S rDNA polymerase chain reaction products. Soil Sci Soc AM J 64:1382–1388
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salam LB, Obayori OS, Olatoye NO (2013) Biodegradation of anthracene by a novel actinomycete, Microbacterium sp. isolated from tropical hydrocarbon-contaminated soil. World J Microbiol Biotechnol. doi:10.1007/s11274-013-1437-7
Schäfer A, Konrad R, Kuhnigk T, Kämpfer P, Hertel H, König H (1996) Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Bacteriol 5:471–478
Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF (2006) Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188:8272–8282
Shi W, Uzuner U, Huang L, Jesudhasan PR, Pillai SD, Yuan JS (2011) Comparative analysis of insect gut symbionts for composition–function relationships and biofuel application potential. Biofuels 5:529–544
Subramanian S, Gadhave KR, Mohanraj P, Thangamalar A (2009) Use of 16S rRNA probes for characterization of gut microflora of silkworm (Bombyx mori L.) breeds. Karnataka J Agric Sci 22:476–478
Takizawa Y, Iizuka T (1968) The aerobic bacterial flora in the gut of larvae of the silkworm Bombyx mori L. (I) The relation between media and the numbers of living cells. J Sericultural Sci Jpn 37:295–305
Tian Z, Hui F, Ke T, Kan Y, Wen Z (2007) Molecular analysis of the bacteria community composition in silkworm midgut. Sci Sericulture 33:592–595 (In Chinese)
Weber S, Stubner S, Conrad R (2001) Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl Environ Microbiol 67:1318–1327
Yang Y, Tang L, Tong L, Liu H (2009) Silkworms culture as a source of protein for humans in space. Adv Space Res 43:1236–1242
Xiang H, Li M, Zhao Y, Zhao L, Zhang Y, Huang Y (2007) Bacterial community in midguts of the silkworm larvae estimated by PCR\DGGE and 16SrDNA gene library analysis. Acta Entomol Sin 3:222–233
Yu X, Liu H, Tong L (2008) Feeding scenario of the silkworm (Bombyx mori L.) in the BLSS. Acta Astronaut 63:1086–1092
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012DFR30570) and National Natural Science Foundation of China (Grant No.31301706).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xue Liang and Yuming Fu contributed equally to this study.
Rights and permissions
About this article
Cite this article
Liang, X., Fu, Y., Tong, L. et al. Microbial shifts of the silkworm larval gut in response to lettuce leaf feeding. Appl Microbiol Biotechnol 98, 3769–3776 (2014). https://doi.org/10.1007/s00253-014-5532-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5532-y