Abstract
Microorganisms play an important role in the growth and development of numerous insect species. The mulberry silkworm, Bombyx mori (Lepidoptera), harbors several bacteria in its midgut aiding the metabolic processes; however, the variability of bacterial spp. present in the midgut and their role(s) in the growth and development of the silkworm are poorly understood. The present work compares the diversity of midgut bacterial communities in silkworms of variable voltinism (Pure Mysore, PM: multivoltine; CSR2: bivoltine and PM × CSR2: crossbreed) through metagenomics. The predominance of Enterococcus (30.30%) followed by Bacillus (16.96%) was observed in PM, whereas Lactobacillus (56.56%) followed by Enterococcus (10.58%) was seen only in CSR2. Interestingly, crossbreed midgut harbored diverse bacterial communities (36.21% Lactobacillus, 25.94% Bacillus, 8.1% Enterococcus, and 18.37% uncultured bacteria). Metagenomic profiles indicate variability in the gut bacterial population in different kinds of silkworms influencing the physiological activities accordingly. The dominant bacteria, particularly lactobacilli, bacilli, and enterococci could be further explored for identifying the potential probiotic consortia based on a literature survey and potential involvement in nutrient absorption, disease/stress tolerance, and improved economic traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mulberry silkworm, Bombyx mori (B. mori), is an economically important insect domesticated for commercial production of silk. The quantity and quality of silk produced depend on breed/hybrid, agro-climatic conditions, and overall physiological function of the silkworm as well as mulberry leaf nutrient status. Even though several studies have demonstrated gut bacteria in silkworm, their precise role in silkworm growth, development, silk production, and disease/stress tolerance is not clearly understood [1,2,3,4]. However, it has been assumed that genetic machinery of the silkworm does not code for cellulase genes, and mulberry leaf cellulose is rather digested by the gut symbiotic bacteria, such as Enterobacter, Proteus vulgaris, Klebsiella pneumonia, and Citrobacter freundii [5,6,7,8]. Further, cofactors, particularly cobalamin forms, are neither synthesized by the silkworm nor obtained from the mulberry leaf, but play an essential role in propionate metabolism (propionate is a precursor for the biosynthesis of juvenile hormone); hence, they must have been obtained from the gut microbiota [9]. Several reports suggest key roles for gut bacteria in silkworm metabolism, growth, and development; yet, symbiotic relationships between the gut bacteria are far from fully understood.
The midgut of B. mori has lower microbial diversity than vertebrates due to shorter life span and controlled micro-environment for optimal growth and development. Moreover, the silkworm gut microbiota have to tolerate alkalinity as high as 11–12 pH [10] and continuous replacement of peritrophic matrix during molting, which adversely affects the growth and colonization of most microbial communities [11]. However, these adverse conditions may not entirely prevent microbial colonization, instead support the growth of alkaline-tolerant microorganisms, particularly, Firmicutes, Clostridium, Planctomycetes, and Microsporidians [12]. Low oxygen levels present in B. mori midgut allow the survival of facultative anaerobic microorganisms only [13].
Most of the silkworm microbiota studies focused either on culture-dependent methods with high variability or culture-independent molecular approaches (16S rRNA gene amplifications), possibly with a biased view on the composition of gut communities [14]. However, conventional culture-dependent techniques together with the modern metagenomic approach would provide a better picture of bacterial communities with reference to various silkworm races or breeds under specific environmental and geographical conditions for improved understanding of the microbes living in the gut of the silkworm [15]. Silkworm gut microbiome provides insights into the relationships between insect and gut bacterial communities for the development of beneficial microbial cultures as probiotics for commercial exploitation. Probiotic consortia have been exploited effectively in several organisms for the benefit of mankind and improved economic returns to the stakeholders [16].
Materials and Methods
Sample Collection
The silkworm breeds utilized in the study include Pure Mysore (PM; multivoltine), CSR2 (bivoltine), and crossbreed (PM × CSR2). Silkworms were reared on mulberry leaves from I to V instar under normal laboratory conditions (25–27 ± 1 °C temperature and 70 ± 5% relative humidity). Three healthy larvae from each batch were randomly selected on the second day of the V instar and subjected to overnight starvation to eliminate mulberry leaf material from the midgut. The selected larvae were sterilized by wiping with 70% ethyl alcohol and gentle flame exposure. They were then dissected under the sterilized condition and the midgut content was collected in microtube under aseptic condition and stored at − 20 °C for further DNA extraction.
Illumina Miseq Sequencing and Data Analysis
The total genomic DNA was extracted using DNeasy PowerSoil kit (Qiagen,) as per the manufactures instructions (quick-start protocol). Bacteria communities were barcoded and identified based on the ribosomal DNA (16S rRNA) sequencing. The sequencing libraries were prepared according to the Illumina 16S Metagenomic Sequencing Library protocols to amplify the V3 and V4 regions. The DNA quantity was measured by PicoGreen, and input gDNA (10 ng) was PCR amplified. The primer sequences used for the first amplification were as follows:
V3-F:5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′,V4-R:5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′.
The final purified product was then quantified using qPCR according to the qPCR quantification protocol guide (KAPA Library quantification kits for Illumina Sequencing platforms) and qualified using the Tape Station DNA screen tape (Agilent Technologies, Waldbronn, Germany). And then the paired-end (2 × 300 bp) sequencing was performed by the Macrogen Inc., Korea using the MiSeq™ platform (Illumina, San Diego, USA). The Illumina Miseq generates raw images with the MiSeq Control Software v2.2 for system control and base calling through integrated primary analysis software, Real Time Analysis.v1.18. The BCL (base calls) binary was converted into FASTQ utilizing Illumina package bcl2fastq v1.8.4. The adapter sequences and reads shorter than 36 bp were removed and clean data were produced using Scythe (v0.994) (https://github.com/vsbuffalo/scythe) and Sickle programs [17]. The reads were further categorized taxonomically by utilizing Kaiju web server (http://kaiju.binf.ku.dk/) for sensitive taxonomic classification of high-throughput sequencing reads from metagenomic or metatranscriptomic experiments. The classification was carried out in a greedy heuristic mode with SEG filter and parameters, such that minimum match length, minimum match score, and allowed mismatches were 11, 90, and 5, respectively [18]. The reads were assembled using Metavelvet [19] and checked for chimera using DECIPHER’s Find Chimeras web tool [20]. Sequences were clustered into operational taxonomic units (OTUs) defined at 97% similarity threshold. Taxonomical classification of OTUs was carried out by using Mothur’s version of the Ribosomal Database Project (RDP) at the genus level and NCBI BLAST at the species level. The trimmed Illumina Miseq metagenomic raw data were submitted and deposited in NCBI database with accession numbers: SAMN08848316, SAMN08912448, and SAMN09499250.
Statistical Methods
Statistical significance was tested using Fisher’s exact test for categorical data of individual bacterial genus present in two different voltinism-based silkworm breeds (PM, CSR2) and their crossbreed (PM × CSR2). Statistical analysis was carried out using GraphPad software (www.graphPad.com).
Results
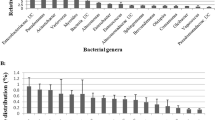
The analysis of rRNA gene sequences is the most common approach to determine microbial diversity. Silkworm gut microbiome was profiled through Illumina Miseq sequencing of 16S rRNA gene, which yielded a total of 4,09,608; 3,18,910; and 4,14,610 paired reads for PM, CSR2, and PM × CSR2, respectively. After trimming the adaptor sequences, the total number of clean reads for PM, CSR2, and PM × CSR2 was 4,09,210; 3,18,770; and 4,14,314, respectively (Table 1). At read level, the percentage of reads classified as bacteria of PM, CSR2, and PM × CSR2 was 99.75, 99.42, and 99.94, respectively. About 0.18, 0.41, and 0.04 percentage of reads was for eukaryota for PM, CSR2, and PM × CSR2, respectively. About < 0.1% of the total reads were unclassified in three silkworm samples, and their taxonomic abundance was classified at phylum level and Firmicutes was predominant, followed by Actinobacteria, Proteobacteria, etc. (Fig. 1).
Heatmap representing bacterial phyla found in Bombyx mori midgut. Light color represents the absence of bacterial phylum/group, and the darker colored tiles indicate presence of particular phylum. Silkworm breeds, PM, and CSR2 had different bacterial phyla; however, the crossbreed, PM × CSR2 exhibited combined phyla of PM and CSR2
The clean reads were assembled into contigs using Metavelvet, and 1105, 821, and 1156 contigs were found for PM, CSR2, and PM × CSR2, respectively. Detailed information on assembled contigs for three kinds of silkworms is shown in Table 1. The taxonomical classification of silkworm contigs shows that the predominant genera were Lactobacillus (40%), followed by Bacillus (15.3%), Enterococcus (15%), uncultured bacteria (13.7%), Staphylococcus (4%), Lysinibacillus (3.5%), Bifidobacterium (1.6%), Clostridium (1.2%), Enterobacter (0.8%), Klebsiella (0.6%), Micrococus (0.6%), Escheirichia coli (0.6%), and 3.2% other bacteria (Fig. 2).
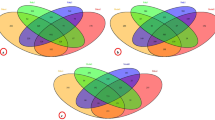
The midgut bacterial profile of individual mulberry silkworm breeds and crossbreed reveals the predominance of Lactobacillus (56.56%), followed by Enterococcus (10.58%), unculturable bacteria (10.21%), Staphylococcus (9.12%), Bacillus (8.3%), Lysinibacillus (2.55%), Micrococus (1.45%), and Escheirichia (1.09%) in popular bivoltine silkworm breed, CSR2. However, the multivoltine breed, PM, harbored Enterococcus (30.3%), followed by Lactobacillus (16.96%), Bacillus (15.15%), unculturable bacteria (14.54%), Klebsiella (9.69%), Lysinibacillus (9.09%), Bifidobacterium (3.03%), and Veillonella (1.21%). On the other hand, the crossbreed of multivoltine × bivoltine, PM × CSR2 harbored Lactobacillus (36.21%), followed by Bacillus (25.94%), uncultured bacteria (18.37%), Enterococcus (8.10%), Clostridium (3.78%), Bifidobacterium (2.7%), Enterobacter (2.7%), and 2.1% Klebsiella. The relative proportion of bacterial genera, such as Lactobacillus, Enterococcus, and Bacillus in CSR2 significantly varied from PM and PM × CSR2 (> 0.05). An analysis of the abundance of the frequency of individual bacterial spp. reveals that Lactobacillus plantarum, L. rhamnosus, L. paracasei, L. acidophilus, and Bacillus sp. were commonly shared between CSR2 and PM × CSR2 than PM, whereas Enterococcus faecium and E. faecalis were shared between PM and CSR2 than PM × CSR2. The shared bacterial communities of three predominant genera among the two silkworm breeds and crossbreed are represented in a Venn diagram (Fig. 3), which reveals that Lactobacillus is the most frequently shared bacteria followed by Enterococcus and Bacillus.
Discussion
Microorganisms are being supplemented as probiotics to humans, ruminants, poultry, and fisheries for beneficial effects. However, supplementation of probiotics to the silkworm is in its primitive stage, because the precise mechanism of beneficial effects of gut bacteria on silkworm physiology or interaction among the different bacterial strains present as microbiota is not well studied. To date, there are no correlative reports regarding the distribution and composition of the gut microbiome in silkworm breeds and their cross breed. Hence, the relationship between gut microbiota of popular silkworm breeds of different voltinism and crossbreed was explored through metagenomic approach for identifying potential probiotic bacterial species. The predominant bacterial genera observed in B. mori midgut in the present study were Enterococcus, Bacillus, and Lactobacillus. In contrast, Yuan et al. [21] identified several bacterial species in the intestinal tract of B. mori; the species predominantly belong to genera Arthrobacter, Lactobacillus, Escherichia, Pseudomonas, Bacillus, and Staphylococcus. However, the gut of the vertebrates and several insect species is composed of a larger proportion of uncultured bacteria [22]. Surprisingly, in the present study, results indicated that B. mori midgut contains relatively lower proportion (~ 15%) of unculturable bacteria; the bacterial diversity in midgut was also found to be comparatively lesser and comprises culturable bacterial spp. Enterococcus was predominant in multivoltine silkworm breed (PM) as compared with bivoltine (CSR2) and crossbreed (PM × CSR2) and plays an important role in the reduction of gut pH leading to the suppression of Nosema bombycis spore germination [23]. Lu et al. [24] identified several Enterococcus strains from healthy silkworm larvae and E. faecalis was identified as a major species and also the most abundant species in the bivoltine breed (CSR2) as observed in the current study; however, E. faecium outnumbered the other species in PM. A recent comparative study on the gut microbiota of a healthy silkworm and their changes when infected with B. mori cypovirus (BmCPV) revealed the presence of Enterococcus, Delftia, Pelomonas, Ralstonia, and Staphylococcus in healthy silkworms, whereas, BmCPV-infected silkworms had lesser bacterial diversity with an abundance of Enterococcus and Staphylococcus [25]. Further, the common bacterial genus observed in all the three silkworm breeds was Bacillus, which is thought to be an important producer of cellulases, proteases, and lipases as demonstrated by Subramanian et al. [14], Anand et al. [4], and Feng et al. [26]. Strains of Bacillus licheniformis could be considered as a possible candidate for a probiotic supplement to silkworms as they are well-known producers of extracellular enzymes [27].
The predominance of Lactobacillus in the gut of mulberry silkworm was not reported earlier [21]; however, in the present study, Lactobacillus was identified in three types of silkworm breeds and found predominantly in bivoltine (CSR2). Among the lactobacilli, the frequency was high in L. plantarum, followed by L. rhamnosus; however, PM had a higher proportion of L. rhamnosus as compared to other Lactobacillus species. There are a few reports on the possible role of probiotic Lactobacillus in the improvement of cocoon production in B. mori [28]. L. plantarum and L. rhamnosus were dominant bacteria in CSR2 as compared to PM, whereas the hybrid (PM × CSR2) also had similar abundances like CSR2 indicating that lactobacilli might be under direct natural selection. L. paracasei and L. acidophilus were higher in the crossbreed as compared with the parental breeds. Even though Lactobacillus requires acidic pH for optimal growth, its tolerance towards silkworm midgut alkaline environment is not fully understood as yet. Whether silkworm acquires these Lactobacillus spp. through the mulberry leaf or from rearing environment was also not yet studied [15]. However, Lactobacillus spp. are considered to be very important as they produce several antimicrobial substances (lactic acid, H2O2, bacteriocins, etc.) and coenzymes (folate and cobalamin) [29, 30]. Singh et al. [28] demonstrated that supplementation of L. plantarum helped to improve body weight, cocoon, shell, and pupation rate. Some of the lactobacilli species particularly L. plantarum, L. rhamnosus, L. paracasei, and L. acidophilus have been recognized as potential probiotics for humans and animals [31, 32], and also frequently occurred in the silkworm gut as noticed in the present investigation. The predominance of lactobacilli in silkworm gut would be considering them as possible probiotics for improved silkworm growth and development.
A recent metagenomic study on B. mori gut microbiota demonstrates that the composition of bacterial flora is closely related to the development stage, host plant, environment, and physiological status [33]. Similarly, Sun et al. [34] showed decreased levels of resistance, productivity, and cocoon quality when the silkworm was exposed to high temperature, humidity, and pathogens due to disturbed native gut microbiota. Moreover, the proportion of individual bacterial spp. varies with silkworm breeds as evidenced in the present study. Therefore, identification of specific probiotics and their application would help in the syngenical development of climate-resilient breeds with improved silk productivity and defense against pathogens.
Conclusion
The diversity and proportion of bacterial profile in silkworm breeds and their crossbreed were different, and the genus Lactobacillus was abundant in mulberry silkworm midgut, followed by Enterococcus and Bacillus. Most of the Lactobacillus and Bacillus spp. are well-known producers of coenzymes, antimicrobial substances, and extracellular enzymes. Possible bacterial species identified as probiotics in the present study include L. plantarum, L. rhamnosus, L. paracasei, L. acidophilus, and Bacillus; these species could be exploited further to supplement through a mulberry feed to improve the economic characteristics of the silkworm. The gut bacterial communities present in multivoltine and crossbreed might be considered as suitable probiotic consortia for productivity in bivoltine silkworm.
References
Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70(1):293–300
Xiang H, Wei GF, Jia SH, Huang J, Miao XX, Zhou ZH, Zhao LP, Huang YP (2007) Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can J Microbiol 52:1085–1092
Gao HJ, Lu GB, Cha CY, Liu JR, Mu ZM (2007) Isolation and screening of the enzyme-producing bacteria in the intestine of silkworm. Acta Sericol Sin 33(2):228–233
Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T, Geoffrey CJ, Vendan SE (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10:107
Ross P, Mayer R, Benziman M (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55(1):35–58
Lynd LR, Wyman CE, Gerngross TU (1999) Biocommodity engineering. Biotechnol Prog 17:777–793
Watanabe H, Tokuda G (2010) Cellulolytic systems in insects. Annu Rev Entomol 55:609–632
Pytelkova J, Hubert J, Lepsik M, Sobotnik J, Sendelyca R (2009) Digestive α- amylases of the flour math Ephestia kuehniella-adaptation to alkaline environment and plant inhibitors. FEBS J 276:3531–3546
Halarnkar PP, Blomquist GJ (1989) Comparative aspects of propionate metabolism. Comp Biochem Physiol 92B(2):227–231
Harrison JF (2001) Insect acid-base physiology. Annu Rev Entomol 46:221–250
Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37(5):699–735
Bignell DE, Roisin Y, Lo N (2010) Biology of termites: a modern synthesis. Springer, Berlin, pp 375–412
Johnson KS, Barbehenn RV (2000) Oxygen levels in the gut lumens of herbivorous insects. J Insect Physiol 46:897–903
Subramanian S, Ramesh KG, Mohanraj P, Thangamalar A (2009) Use of 16S rRNA probes for characterization of gut microflora of silkworm (Bombyx mori L.) breeds. Karnataka J Agric Sci 22:476–478
Thirupathaiah Y, Bhuvaneswari E, Vaijayanthi PV, Kumar V, Sivaprasad V (2018) Diversity and functional role of gut microbiota of silkworm Bombyx mori L. J Sericologia 58(1):1–11
Kerry RJ, Patra JK, Gouda S, Park Y, Shin HS, Das G (2018) Benefaction of probiotics for human health: a review. J Food Drug Anal 26:927–939
Joshi NA, Fass JN (2011) Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. (software) Version 1.33
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257
Namiki T, Hachiya T, Tanaka H, Sakakibara Y (2012) MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40(20):e155
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78(3):717–725
Yuan ZH, Lan XQ, Yang T, Xiao J, Zhou ZY (2006) Investigation and analysis of the bacteria community in silkworm intestine. Acta Microbiol Sin 46(2):285–291
Bahrndorff S, Alemu T, Alemneh T, Nielsen JL (2016) The microbiome of animals: implications for conservation biology. Int J Genomics 2016:1–7
Lu XM, Wang FW (2002) Inhibition of cultured supernatant of Enterococcus strains on germination of Nosema Bombycis spores in vitro. Acta Sericol Sin 28:126–128
Lu X, Hashimoto Y, Shimizu S (1994) Taxonomical studies on enterococci isolated from the intestine of the silkworm, Bombyx mori. J Sericul Sci Japan 63(6):481–487
Sun Z, Lu Y, Zhang H, Kumar D, Liu B, Gong Y (2016) Effects of BmCPV infection on silkworm Bombyx mori intestinal bacteria. PLoS One 11(1):1–17
Feng W, Wang XQ, Zhou W, Liu GY, Wan YJ (2011) Isolation and characterization of lipase-producing bacteria in the intestine of the silkworm, Bombyx mori, reared on different forage. J Insect Sci 11:1–10
Mala M, Vijila K (2018) Beneficial effects of Bacillus licheniformis and Bacillus niabensis on growth and economic characteristics of silkworm, Bombyx mori L. IJCS 6(2):1750–1754
Singh KK, Chauhan RM, Pande AB, Gokhale SB, Hedge NG (2005) Effect of use of Lactobacillus plantarum as a probiotics to improve cocoon production of mulberry silkworm, Bombyx mori L. J Basic Appl Sci 1:1–8
Carina AM, Torres MJ, Sabaté DC, Ibarguren C, Apella MC (2011) Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol Res 166(1):1–13
LeBlanc JG, Laino JE, del Valle MJ, Vannini V, Van Sinderen D, Taranto MP, De Valdez GF, De Giori GS, Sesma F (2011) B-group vitamin production by lactic acid bacteria--current knowledge and potential applications. J Appl Microbiol 111(6):1297–1309
Bauer C, Lopis M, Antolín M, Monedero V, Mata M, Zuniga M, Francisco G, Gaspar Perez G (2013) Lactobacillus paracasei and Lactobacillus plantarum strains downregulate proinflammatory genes in an ex vivo system of cultured human colonic mucosa. Genes Nutr 8(2):165–180
Thirupathaiah Y, Hemalatha RR, Vasundara D (2017) Vaginal lactobacilli profile in pregnant women with normal and abnormal vaginal flora. Ind J Med Res 146:534–540
Chen B, Du K, Sun C, Vimalanathan A, Liang X, Li Y, Wang B, Lu X, Li L, Shao Y (2018) Gut bacterial and fungal communities of the domesticated silkworm (B. mori) and wild mulberry-feeding relatives. ISME J 12(9):2252–2262
Sun Z, Kumar D, Cao G, Zhu L, Liu B, Zhu M, Liang Z, Kuang S, Chen F, Yongjie F, Hu X, Xue R, Gong C (2017) Effects of transient high temperature treatment on the intestinal flora of the silkworm Bombyx mori. Sci Rep 7:3349
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yeruva, T., Vankadara, S., Ramasamy, S. et al. Identification of Potential Probiotics in the Midgut of Mulberry Silkworm, Bombyx mori Through Metagenomic Approach. Probiotics & Antimicro. Prot. 12, 635–640 (2020). https://doi.org/10.1007/s12602-019-09580-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-019-09580-3