Abstract
Development of rapid and sensitive methods to detect pathogens is important to food and water safety. This study aimed to detect and discriminate important food- and waterborne bacteria (i.e., Escherichia coli O157:H7, Staphylococcus epidermidis, Listeria monocytogenes, and Enterococcus faecelis) by surface-enhanced Raman spectroscopy (SERS) coupled with intracellular nanosilver as SERS substrates. An in vivo molecular probing using intracellular nanosilver for the preparation of bacterial samples was established and assessed. Satisfactory SERS performance and characteristic SERS spectra were obtained from different bacterial samples. Distinctive differences were observed in SERS spectral data, specifically in the Raman shift region of 500–1,800 cm−1, and between bacterial samples at the species and strain levels. The detection limit of SERS coupled with in vivo molecular probing using silver nanosubstrates could reach the level of single cells. Experiments with a mixture of E. coli O157:H7 and S. epidermidis for SERS measurement demonstrate that SERS could be used for classification of mixed bacterial samples. Transmission electron microscopy was used to characterize changes of morphology and cellular composition of bacterial cells after treatment of intracellular nanosilver. The results indicate that SERS coupled with intracellular silver nanosubstrates is a promising method for detection and characterization of food- and waterborne pathogenic and non-pathogenic bacterial samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the primary safety concerns in the food industry is the presence of pathogenic microorganisms in foods and water, such as Escherichia coli O157:H7, Listeria monocytogenes, and other food- and waterborne pathogens. Rapid methods with high sensitivity and specificity for bacterial detection are of critical importance to food safety and human health. Rapid and early detection can prevent and control pathogen-related diseases and potential bioterrorism attacks (Jarvis and Goodacre 2008).
To date, many detection methods have been developed and applied in detection and identification of microorganisms, including biochemical, immunological, DNA hybridization, and conductance methods (Noble and Weisberg 2005). However, these methods are time-consuming, labor-intensive, and many of them require at least 24 to 48 h to obtain testing results (Maquelin et al. 2002). For example, although real-time PCR methods have been reported for the detection of foodborne pathogens (Wang et al. 2009), these methods require highly purified template nucleic acids to prevent the contamination from non-template nucleic acids. Therefore, there is a high demand for rapid, sensitive, and accurate methods that can be used to detect and identify pathogens in food and water.
In recent years, advances in the development of vibrational spectroscopy (infrared and Raman spectroscopy) provide the basis for non-destructive detection of microorganisms (Kneipp et al. 2002). In particular, surface-enhanced Raman spectroscopy (SERS) is a sensitive analytical tool that can be used in testing chemical and biochemical samples when coupled with SERS-active substrates. SERS is a specialized type of Raman spectroscopy that is based upon significantly enhanced Raman signals acquired from samples deposited on nanosubstrates with roughened surface. SERS coupled with statistical analyses has been increasingly used in detection and discrimination of bacterial cells, spores, and viruses (Jarvis et al. 2004; Jarvis and Goodacre 2004; Yan and Vo-Dinh 2007; He et al. 2008; Fan et al. 2010). Gold and silver are two most commonly used materials for fabricating nanostructured metallic substrates for SERS measurement. Compared with gold, silver produces stronger SERS enhancement (Freeman et al. 1996; Otto et al. 1992).
The objectives of this study were to detect food- and waterborne bacteria by SERS coupled with intracellular nanosilver as nanosubstrates for in vivo molecular probing and to study the detection limit and specificity of the new SERS method in detecting pure and mixed bacterial cultures.
Materials and methods
Preparation of bacterial samples and internal deposition of silver nanoparticles
Bacterial strains were obtained from the culture collection in the Food Microbiology Laboratory at the University of Missouri, Columbia, Missouri, USA. They included E. coli ATCC 15597, E. coli K-12 ATCC 23716, E. coli O157:H7 H2439 (from CDC, Odwalla apple cider isolate), Staphylococcus epidermidis ATCC 12228, Enterococcus faecalis ATCC 7080, and L. monocytogenes Scott A. Bacteria were grown aerobically at 37°C for 12 to 16 h in tryptone soy broth (TSB, Difco, MD, USA). The bacterial samples were harvested and washed twice with MilliQ water at room temperature through centrifugation at 130,000×g in a swinging bucket rotor (Beckman SW41, Beckman Coulter, Inc., Brea, CA). The concentrations of the bacterial samples were 8–9 log CFU ml−1. These samples were further diluted ten times in deionized water for use. Thereafter, an aliquot of bacteria in water was applied onto a gold-coated microscope slide (Thermo Fisher Scientific, Waltham, MA, USA) for SERS spectral acquisition. For the internally deposited methods, bacterial cells were washed and treated with silver nitrate and sodium borohydride sequentially by suspending washed bacterial cells (∼1 × 108–109 CFU ml−1) in 1 ml 0.05 N silver nitrate solution and then suspending in 1 ml 0.1 N sodium borohydride solution for 1 min. One more washing step was then conducted for the sample preparation with intracellular nanosilver deposition (Zeiri et al. 2002).
The reproducibility study was conducted using two bacterial strains (E. coli O157:H7 H2439 and L. monocytogenes Scott A). Preparation and treatment of silver nanosubstrates were performed three times for each bacterial sample. During SERS measurement, two or three drops of sample solutions were placed on the gold plate and multiple SERS spectra were obtained from different positions on one drop of mixture.
Instrumentation
A Renishaw RM1000 Raman spectrometer system (Gloucestershire, UK) equipped with a Leica DMLB microscope (Wetzlar, Germany) was used in this study. This system is equipped with a 785-nm near-infrared diode laser source. During the measurement, light from the high power (maximum at 300 mW) diode laser was directed and focused onto the sample on a microscope stage through a 50× objective. Raman scattering signals were detected by a 578 × 385 pixel CCD array detector. The size of each pixel was 22 × 22 μm. Spectral data were collected by WiRE 1.3 software (Gloucestershire, UK). Spectra of each bacterial sample were collected using a 50× objective with a detection range from 500 to 1,800 cm−1 in the extended mode. The measurement was conducted with a 10-s exposure time and ~20 mW laser power.
Transmission electron microscope
Transmission electron microscope (TEM) was used to characterize the silver nanoparticles and their interactions with bacterial cells. E. coli O157:H7 cultures were incubated overnight at 37°C followed by the treatments with the silver nanoparticles as described above. Both the control and treated E. coli O157:H7 cells were stored in primary fixative (2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 mol l−1, Na-cacodylate buffer, pH 7.35) for further processing. The fixed samples were microwaved (MW) under vacuum conditions in a Pelco Biowave (Ted Pella, Inc., Redding, CA, USA) at 120 W. The samples were rinsed several times with 0.1 mol l−1 cacodylate buffer and embedded in histogel, followed by secondary MW fixation with a buffered (0.1 mol l−1 cacodylate, 0.01 mol l−1 2-mercaptoethanol, 0.13 mol l−1 sucrose) 1% osmium tetroxide. The samples were then quickly rinsed three times with 0.1 mol l−1 cacodylate, 0.01 mol l−1 2-mercaptoethanol, 0.13 mol l−1 sucrose sequentially, and then rinsed three times (5 min each) with ultrapure water. This was followed by dehydration with ethanol solutions (20%, 50%, 70%, 90%, and 100%) and last with a 100% acetone solution. The bacterial samples were infiltrated with Spurr's resin and polymerized at 60°C for 24 h. The sample blocks were processed into 85 nm thin sections with Leica Ultracut UCT ultramicrotomes (Leica Microsystems GmbH, Wetzlar, Germany). The sections were placed onto 200 mesh thin bar grids and post-stained for 20 min with 5% uranyl acetate and 10 min with Sato's triple lead stain. The stained samples were observed in a JEOL 1400 (JEOL, LTD., Tokyo, Japan) operating at 80 kV.
Data analysis
Data analysis was conducted using the Delight software (DSquared Development Inc., LaGrande, OR, USA). Pre-processing algorithms were employed to analyze the data, such as smoothing and second-derivative transformation. Smoothing eliminates high-frequency instrumental noises by averaging neighboring data points. Second-derivative transformation separates some overlapping bands and removes baseline offsets. Multivariate statistical analysis (principal component analysis (PCA)) was used to analyze the spectral data. PCA reduces a multidimensional data set to its most dominant features, removes random variation (noise), and retains the principal components (PCs) that capture the variation between samples treatments (Al-Holy et al. 2006; He et al. 2008).
Results
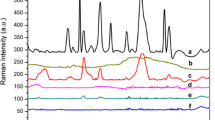
A comparison between the SERS spectra and normal Raman spectra of Gram-negative E. coli O157:H7 is shown in Fig. 1a, b. Normal Raman spectra (Fig. 1b) exhibit characteristic peaks at 554, 724, 782, 1,002, 1,099, 1,233, 1,338, 1,452, and 1,665 cm−1. Figure 1c, d show the comparison between SERS and normal Raman spectra of Gram-positive L. monocytogenes. Peaks at 543, 734, 793, 1,003, 1,129, 1,235, 1,325, 1,390, 1,459, and 1,568 cm−1 were clearly enhanced in the SERS spectra. Figure 2 shows average SERS spectra acquired from three bacterial species (L. monocytogenes, S. epidermidis, and E. faecalis). Typical peaks in the range between 500 and 1,700 cm−1 were clearly observed in the spectra of the three bacterial species, demonstrating vibrational information from cell structural components.
Comparison between average (n = 7) SERS (a) and normal Raman (b) spectra of E. coli O157:H7 cells and between average (n = 7) SERS (c) and normal Raman (d) spectra of L. monocytogenes. The intensity of the normal Raman spectra (b, d) has been multiplied by a factor of 10,000 to allow direct comparison with SERS spectra. Measurements were conducted from 500 to 1,800 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1, baseline adjustment by subtracting a second order polynomial function and normalization
Average SERS spectra (n = 7) of L. monocytogenes (a), S. epidermidis (b), and E. faecalis (c). Measurements were conducted from 500 to 1,800 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1 and baseline adjustment by subtracting a second order polynomial function
Average SERS spectra acquired from three E. coli strains (E. coli O157:H7, E. coli K-12, and E. coli ATCC 15597) are shown in Fig. 3a–c. Typical peaks at 548, 713, 928, 1,003, 1,100, 1,130, 1,239, 1,322, 1,392, 1,450, and 1,583 cm−1 are observed in three E. coli strains. Figure 4 illustrates the second derivative transformation of bacterial spectra in the region between 1,200 and 1,600 cm−1.
Average SERS spectra (n = 7) of E. coli O157:H7 (a), E. coli K-12 (b), and E. coli ATCC 15597 (c). Measurements were conducted from 500 to 1,800 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1 and baseline adjustment by subtracting a second order polynomial function
Representative second derivative transformation for SERS spectra of the control, S. epidermidis, E. coli O157:H7 and their 1:1 ratio mixture. Measurements were conducted from 1,200 to 1,600 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1 and second derivative transformation
PCA was used to differentiate the spectra acquired from bacterial samples. In this study, a mean-centered PCA was carried out on the second derivative transformed SERS spectra over the range of 500–1,800 cm−1. Figure 5 shows PCA clustering results from the SERS spectra of 1 ml S. epidermidis (∼1 × 108–109 CFU ml−1), 1 ml E. coli O157:H7 (∼1 × 108–109 CFU ml−1), and 1 ml of their mixture. Segregations between pure and mixed cultures of S. epidermidis and E. coli O157:H7 were clearly observed, and the data cluster of the mixed culture was in between the clusters of two pure bacterial samples. The first five PCs explain 80% of total data variances.
The reproducibility study using different batches of silver nanoparticles was conducted. Figure 6 shows the normalized SERS spectra of E. coli O157:H7 (Fig. 6a) and L. monocytogenes (Fig. 6b) acquired from two separate experiments for each species. The bacterial samples were treated with two batches of silver nanosubstrates individually. Detection of single bacterial cells by SERS was achieved. It is relatively easier to obtain two-cell chains which commonly exist in E. coli O157:H7 cell cultures. SERS spectra acquired from E. coli O157:H7 two-cell chains are shown in Fig. 7a, b and a SERS spectrum of multiple cells is shown in Fig. 7c. The corresponding white light images in the laser focal region are also shown in this figure.
Reproducibility focusing on two batches of silver nanostructures for detection of two bacterial strains: average SERS spectra (n = 7) of E. coli O157:H7 from two batches of silver nanostructures (a) and average SERS spectra (n = 7) of L. monocytogenes from two batches of silver nanostructures (b). Measurements were conducted from 500 to 1,800 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1 and baseline adjustment by subtracting a second order polynomial function
Limit of detection by SERS for E. coli O157:H7 cells: the SERS spectra of two-cell chains (a, b), the SERS spectra of multiple cells (c). Measurements were conducted from 500 to 1,800 cm−1 with a 10-s exposure time and ∼20 mW laser power. Spectra were presented with smoothing at 4 cm−1 and baseline adjustment by subtracting a second order polynomial function
TEM coupled with thin sections allows direct characterization of bacterial cells before and after treatment with silver nanoparticles, which provides critical information about the interaction between bacterial cells and internally deposited silver nanoparticles. Figure 8a, b shows the control (E. coli O157:H7 cell without treatment), while Fig. 8c, d shows E. coli O157:H7 cell after treatment with silver nanoparticles.
Discussions
Peaks in SERS spectra (Fig. 1) can be assigned to different functional groups as shown in Table 1, such as 1,002 cm−1 to phenylalanine (the symmetric ring breathing mode), 1,099 cm−1 to CC skeletal and COC stretching from glycosidic link, 1,233 cm−1 to CH2 deformation, 1,338 cm−1 to adenine, 1,452 cm−1 to C–H deformation, and 1,665 cm−1 to amide I. Likewise, similar peaks at 548, 713, 928, 1,003, 1,130, 1,239, 1,322, 1,392, 1,450, and 1,590 cm−1 were readily observed in SERS spectrum (Fig. 1a), which were also linked to different functional groups: 1,003, 1,239, and 1,590 cm−1 from proteins or amino acids, 713 and 1,322 cm−1 from nucleic acids, 1,239 and 1,450 cm−1 from lipids, and 928 cm−1 from carbohydrates (Schwartzberg et al. 2004; Otto et al. 1986; Podstawka et al. 2004; Kattumuri et al. 2006). These enhanced peaks could be attributed to structural components including proteins, nucleic acids, lipids, and carbohydrates. Although the normal Raman spectra of E. coli O157 and L. monocytogenes show high similarities, their SERS spectra differ greatly in peak pattern and their corresponding peak intensities. The SERS spectrum of the control (gold-coated microscope slides without bacterial cells) exhibited a flat line and showed no significant peaks.
The bacterial cell density for normal Raman measurement (Fig. 1b) was 100 times higher than that for SERS measurement (Fig. 1a). Moreover, the laser power for normal Raman measurement was ten times stronger than that for SERS measurement. However, the intensity of the peak at ∼1,002 cm−1 in the SERS spectra is ten times stronger than that in the normal Raman spectra. The enhancement factor of internally deposited silver nanoparticles was calculated to be around 1 × 104. Not all the peaks in the normal Raman spectra were enhanced with the same magnitude. Some molecular groups were preferentially enhanced. According to the electromagnetic and charge-transfer enhancement mechanisms (Moskovits 1985; Picorel et al. 1992), the SERS enhancement is dependent upon several factors, including the distance and the orientation towards the substrates. In this study, silver nanoparticles were used as the substrate to enhance Raman signals. Other studies reported that silver particles could block some important sites in the respiratory chain in bacterial cell membrane and the SERS spectra of silver-treated bacteria could be assigned to flavins, which are present in bacterial cell wall and participate in the respiratory processes (Zeiri et al. 2004; Sengupta et al. 2006).
In Fig. 2, a peak at 543 cm−1 could be assigned to S–S stretch, 734 cm−1 to adenine, 1,003 cm−1 to phenylalanine (the symmetric ring breathing mode), 1,235 cm−1 to CH2 deformation, 1,450 cm−1 to C–H deformation, and 1,583 cm−1 to tyrosine (Table 1). Interestingly, a peak at 928 cm−1 was clearly present in the spectra of S. epidermidis (Fig. 2b) and E. faecalis (Fig. 2c), but not in that of L. monocytogenes (Fig. 2a); while a band around 656 cm−1 was only present in the spectra of E. faecalis. The peak at 1,101 cm−1 is distinctive in the spectra of S. epidermidis and E. faecalis, but not in that of L. monocytogenes. Differences between the spectra of three bacterial species come from the differences in quantity and distribution of cellular components such as proteins, phospholipids, nucleic acids, and carbohydrates.
High similarity in SERS spectral patterns was observed among three E. coli strains (Fig. 3). However, a peak at 895 cm−1 is present only in the spectrum of E. coli ATCC 15597 (Fig. 3c) and not in the other two strains, while the intensity of the band around 1,100 cm−1 in the spectrum of E. coli ATCC 15597 (Fig. 3c) is much higher than that of the other two strains. Variations in the peak intensity are observed as well: take the example of peak intensity at 1,322 cm−1, E. coli ATCC 15597 (Fig. 3c) exhibits the highest intensity among these three bacterial strains, while the E. coli O157:H7 (Fig. 3a) has the lowest. These discrepancies could be explained by the differences in the distribution and quantity of cellular components among E. coli strains.
In Fig. 4, differences between E. coli O157:H7, S. epidermidis, and the mixed culture can be seen in the bands at 1,334 cm−1 (carbohydrate), 1,450–1,470 cm−1 (lipids), and at 1,570 cm−1 which is attributed to ring stretching vibrations of nucleobases such as guanine and adenine.
PCA is a multivariate statistical analysis widely used to capture and explain SERS spectral data variances. Data are usually shown as clustered samples depending upon differences in the spectral patterns that result from variations in the samples. In Fig. 5, the data variations among bacterial species resulted from differences in major cellular components such as nucleic acids, proteins, and phospholipids. These results demonstrate that mixed bacterial samples could also be differentiated by SERS in conjunction with the use of intracellular silver nanosubstrates.
For most studies using SERS to detect bacteria, a major challenge is inconsistent performance of substrates mainly due to heterogeneous structures of nanosubstrates and the natural growth cycle of bacteria (Premasiri et al. 2004). In this study, internally deposited silver nanoparticles were applied to treat bacterial cells. Although it is difficult to achieve even distribution of nanoparticles inside bacterial cells, reproducible SERS signals could be obtained since the overall information of bacterial cells were acquired during SERS measurement. The consistency of the internal nanosubstrate deposition within different batches was achieved by precisely controlling the exact amount of chemical agents added, the operating time at each step, and the well-mixed bacterial suspensions. Consistent and reproducible spectral data were obtained from duplicate experiments for two bacterial species (Fig. 6).
In Fig. 7, the spectral patterns for two-cell chains and multiple cells are comparable, and the intensity of multiple cells is significantly higher than the two-cell chains. The laser spot diameter was around 5 μm and the number of bacterial cells covered by the laser spot varies depending on the concentrations of bacteria cells. In the study of limit detection, the laser spot only covered selected two-cell chains as shown in Fig. 7. The SERS spectra of two-cell chains are quite similar to each other and to the average SERS spectra (Fig. 1a), indicating that the limit of detection of SERS coupled with internally deposited silver nanosubstrates could reach the single cell level.
Formation and deposition of silver nanoparticles within E. coli O157:H7 cells were observed by comparing Fig. 8a, c. Various sizes of silver nanoparticles accumulated within the cells. Silver nanoparticles deposited closer to the cell membranes were bigger than those accumulated deep inside the cells. TEM images show that the sizes of the internally deposited silver nanoparticles were mostly less than 50 nm.
Our results demonstrate that SERS method coupled with intracellular silver nanoparticles was was able to detect and differentiate bacteria quickly and accurately, which is in agreement with previous studies using silver nanoparticles in SERS to differentiate between different bacterial samples. The Efrima group classifed bacteria using SERS and identified several spectral regions important for bacterial identification (Efrima and Bronk 1998; Zeiri et al. 2002) in which silver nanoparticles were coated on the bacterial cells and accumulated inside the bacteria cells. Treated bacterial samples were then used for SERS measurement and intensely enhanced Raman signals were acquired. Jarvis and Goodacre (2004) employed a different method by preparing aggregated silver colloid substrate for discrimination of bacteria by SERS. The limit of detection of this method could reach the strain level, which was confirmed by statistical analysis (Jarvis and Goodacre 2004).
In summary, there is great potential to use SERS for detection of food- and waterborne pathogens. Coupled with intracellular silver nanoparticles, the improved SERS method was capable of rapidly detecting and differentiating bacteria with the limit of detection down to single cells. Unique and distinctive vibrational spectral information was obtained at both species and strain levels, which provides a sensitive and efficient tool for food and water safety control.
References
Al-Holy MA, Lin M, Cavinato AG, Rasco BA (2006) The use of Fourier transform infrared spectroscopy to differentiate Escherichia coli O157:H7 from other bacteria inoculated into apple juice. Food Microbiol 23:162–168
Efrima S, Bronk BV (1998) Silver colloids impregnating or coating bacteria. J Phys Chem B 102(31):5947–5950. doi:10.1021/jp9813903
Fan C, Hu ZQ, Riley LK, Purdy GA, Mustapha A, Lin M (2010) Detecting food- and waterborne viruses by surface-enhanced Raman spectroscopy. J Food Sci 75(5):M302–M307. doi:10.1111/j.1750-3841.2010.01619.x
Freeman RG, Hommer MB, Grabar KC, Jackson MA, Natan MJ (1996) Ag-clad Au nanoparticles: novel aggregation, optical, and surface-enhanced Raman scattering properties. J Phys Chem 100(2):718–724. doi:10.1021/jp951379s
He L, Liu Y, Lin M, Mustapha A, Wang Y (2008) Detecting single Bacillus spores by surface enhanced Raman spectroscopy. Sens & Instrument Food Qual 2:247–253
Jarvis RM, Brooker A, Goodacre R (2004) Surface-enhanced Raman spectroscopy for bacterial discrimination utilizing a scanning electron microscope with a Raman spectroscopy interface. Anal Chem 76(17):5198–5202. doi:10.1021/ac049663f
Jarvis RM, Goodacre R (2004) Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal Chem 76(1):40–47
Jarvis RM, Goodacre R (2008) Characterisation and identification of bacteria using SERS. Chem Soc Rev 37(5):931–936
Kattumuri V, Chandrasekhar M, Guha S, Raghuraman K, Katti KV, Ghosh K, Patel RJ (2006) Agarose-stabilized gold nanoparticles for surface-enhanced Raman spectroscopic detection of DNA nucleosides. Appl Phys Lett 88(15):153114
Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS (2002) Surface-enhanced Raman scattering and biophysics. J Phys Condens Matter 14:597
Maquelin K, Kirschner C, Choo-Smith L-P, van den Braak N, Endtz HP, Naumann D, Puppels GJ (2002) Identification of medically relevant microorgansims by vibrational spectroscopy. J Microbiol Meth 51:255–271
Moskovits M (1985) Surface-enhanced spectroscopy. Rev Mod Phys 57(3):783
Noble RT, Weisberg SB (2005) A review of technologies for rapid detection of bacteria in recreational waters. J Water Health 03(4):381–392
Otto A, Mrozek I, Grabhorn H, Akemann W (1992) Surface-enhanced Raman scattering. J Phys Condens Matter 4(5):1143
Otto C, van den Tweel TJJ, de Mul FFM, Greve J (1986) Surface-enhanced Raman spectroscopy of DNA bases. J Raman Spectrosc 17:289–298
Picorel R, Bakhtiari M, Lu T, Cotton TM, Seibert M (1992) Surface-enhanced resonance Raman scattering spectroscopy as a surface topography probe in plant photosynthetic membranes. Photochem Photobiol 56(2):263–270. doi:10.1111/j.1751-1097.1992.tb02156.x
Podstawka E, Ozaki Y, Proniewicz LM (2004) Part I: surface-enhanced Raman spectroscopy investigation of amino acids and their homodipeptides adsorbed on colloidal silver. Appl Spectrosc 58:570–580
Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LD (2004) Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J Phys Chem B 109(1):312–320. doi:10.1021/jp040442n
Schwartzberg AM, Grant CD, Wolcott A, Talley CE, Huser TR, Bogomolni R, Zhang JZ (2004) Unique gold nanoparticle aggregates as a highly active surface-enhanced Raman scattering substrate. J Phys Chem B 108(50):19191–19197. doi:10.1021/jp048430p
Sengupta A, Mujacic M, Davis EJ (2006) Detection of bacteria by surface-enhanced Raman spectroscopy. Anal Bioanal Chem 386(5):1379–1386
Wang L, Li Y, Mustapha A (2009) Detection of viable Escherichia coli O157:H7 by ethidium monoazide real-time PCR. J Appl Microbiol 107(5):1719–1728
Yan F, Vo-Dinh T (2007) Surface-enhanced Raman scattering detection of chemical and biological agents using a portable Raman integrated tunable sensor. Sens Actuators B Chem 121(1):61–66
Zeiri L, Bronk BV, Shabtai Y, Czégé J, Efrima S (2002) Silver metal induced surface enhanced Raman of bacteria. Colloids Surf, A Physicochem Eng Asp 208(1–3):357–362
Zeiri L, Bronk BV, Shabtai Y, Eichler J, Efrima S (2004) Surface-enhanced Raman spectroscopy as a tool for probing specific biochemical components in bacteria. Appl Spectrosc 58(1):33–40
Acknowledgements
This study was supported by the US EPA Science to Achieve Results (STAR) project 83384001.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, C., Hu, Z., Mustapha, A. et al. Rapid detection of food- and waterborne bacteria using surface-enhanced Raman spectroscopy coupled with silver nanosubstrates. Appl Microbiol Biotechnol 92, 1053–1061 (2011). https://doi.org/10.1007/s00253-011-3634-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3634-3