Abstract
Bio-catalytic calcification (BCC) reactors utilise microbial urea hydrolysis by autochthonous bacteria for the precipitation-removal of calcium, as calcite, from industrial wastewater. Due to the limited knowledge available concerning natural ureolytic microbial calcium carbonate (CaCO3) precipitation, the microbial ecology of BCC reactors has remained a black box to date. This paper characterises BCC reactor evolution from initialisation to optimisation over a 6-week period. Three key parameters were studied: (1) microbial evolution, (2) the (bio)chemical CaCO3 precipitation pathway, and (3) crystal nucleation site development. Six weeks were required to establish optimal reactor performance, which coincided with an increase in urease activity from an initial 7 mg urea l−1 reactor h−1 to about 100 mg urea l−1 reactor h−1. Urease activity in the optimal period was directly proportional to Ca2+ removal, but urease gene diversity was seemingly limited to a single gene. Denaturing gradient gel electrophoresis of 16S rRNA genes revealed the dynamic evolution of the microbial community structure of the calcareous sludge, which was eventually dominated by a few species including Porphyromonas sp., Arcobacter sp. and Bacteroides sp. Epi-fluorescence and scanning electron microscopy showed that the calcareous sludge was colonised with living bacteria, as well as the calcified remains of organisms. It appears that the precipitation event is localised in a micro-environment, due to colonisation of crystal nucleation sites (calcareous sludge) by the precipitating organisms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biological mineral precipitation represents an interesting alternative means for the treatment of industrial wastewaters as well as groundwater contaminated with excessive amounts of "unusual" pollutants such as heavy metals, radionuclides, phosphate, and salts (Lloyd and Lovley 2001). For example, both Fujita et al. (2001) and Warren et al. (2001) confirmed the potential of ureolytic co-precipitation for the in situ treatment of contaminants such as 90Sr2+, UO2 2+ and Co2+ in groundwater, while Lloyd and Lovley (2001) reported the applicability of Ralstonia eutropha for the removal/recovery of mercury and other divalent heavy metals (as metal-carbonates) from wastewater. The European paper recycling industry requires up to 1.4 billion m3 water annually, while producing effluents containing 10–40 mM Ca2+. Bio-catalytic calcification (BCC) reactors have been developed as an alternative to chemical precipitation technologies, specifically for the removal of soluble calcium from industrial wastewater in the form of calcite (Hammes et al. 2003a, 2003b). These reactors are based on ureolytic microbial calcium carbonate (CaCO3) precipitation (MCP), but, as with the natural process, precious little is known about the microbial ecology governing the precipitation event.
CaCO3 precipitation requires sufficient calcium ions (Ca2+) and carbonate ions (\( {\rm{CO}}_3^{2 - } \)) so that the ion activity product (IAP) exceeds the solubility constant (K s) (Eq. 1), as well as preferably the presence of crystal nucleation sites (Stumm and Morgan 1981). The concentration of carbonate ions is furthermore related to the concentration of dissolved inorganic carbon (DIC) and the pH of a given aquatic system (Eq. 2).
Micro-organisms can influence these four precipitation parameters. Intracellular calcium metabolism as well as the binding of cations to electron negative cell membranes and extracellular polymeric substances could all contribute to elevated localised concentrations of Ca2+ (Schultze-Lam et al. 1996; McConnaughey and Whelan 1997), with the latter two factors rendering bacterial cell walls as ideal crystal nucleation sites (Schultze-Lam et al. 1996). More importantly, micro-organisms can change the DIC and pH of a given environment through metabolic processes, thus catalysing precipitation (Douglas and Beveridge 1998). One such process is urea hydrolysis. In this reaction, urea is cleaved intracellularly to yield carbonic acid and ammonia. These products spontaneously equilibrate at physiological pH, yielding bicarbonate, ammonium and hydroxide ions, thus giving rise to a pH increase as well as a simultaneous increase in DIC (Eqs. 3, 4, 5) (Mobley and Hausinger 1989; Burne and Chen 2001). The urease enzyme (urea amidohydrolase; EC 3.5.1.5) is commonly found in various aerobic and anaerobic microbial species (Mobley and Hausinger 1989; Burne and Chen 2001; Reed 2001).
The aim of this work was firstly to characterise the biological catalyst of a BCC reactor in terms of the population and urease enzyme development over time, and secondly to underpin the exact mechanism of precipitation, and the role therein of micro-organisms.
Materials and methods
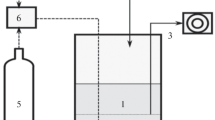
Reactor set-up, operation and water analysis
The wastewater used during experimentation was anaerobic effluent from a full-scale up-flow anaerobic sludge bed (UASB) reactor at a paper-recycling industrial site (VPK, Oudegem, Belgium). Water was collected weekly and stored anaerobically in sealed plastic containers at 5°C until use. Two replicate sequential batch reactors (hereafter referred to as BCC reactors) consisting of 3 l Erlenmeyer flasks with 2.5 l effective reactor volume were operated in parallel. Both reactors were inoculated with 0.25 l activated sludge, taken from the aerobic wastewater treatment system at the site, and this sludge volume was maintained in the reactors throughout the experimental period. For the first week, the reactors were fed 1 g l−1 urea (mixed liquor initial concentration), and operated with a hydraulic residence time (HRT) of 24 h. For the second week, the urea concentration was decreased to 0.5 g l−1, while maintaining an HRT of 24 h. Thereafter, the HRT was decreased to 8 h, allowing for three reactor cycles per day. A standard reactor cycle consisted of 15 min feeding, during which 2.25 l influent (wastewater) was fed to both reactors with a double-headed pump (LMI, Milton Roy, Ivyland, Pa.), and concentrated urea (75 g l−1) (Riedel-de Haën, Seelze, Germany) was fed with a separate double-headed pump (Watson Marlow 313S; Watson Marlow Bredel, Falmouth, UK) to the chosen fed-batch concentration. The reactors were then mixed for 6 h using magnetic stirring (100 rpm; Labinco, Breda, The Netherlands), followed by a 1.5 h settling period, and subsequent water discharge, using a double-headed effluent pump (LMI, Milton Roy). The entire cycle was controlled with three separate electronic time switches. A sludge residence time (SRT) of approximately 14 days was used. The influent, as well as the effluents from both reactors were analysed regularly for pH, temperature, Ca2+ (total and soluble), Mg2+ (total and soluble), total phosphate, alkalinity, total chemical oxygen demand (COD), and Kjeldahl and ammonium-nitrogen (Greenberg et al. 1992). The evolution of urease activity in the reactors was followed by measuring the amount of total ammonium-nitrogen (TAN) exactly 2 h after feeding. Mixed liquor suspended solids samples were taken, filtered (0.45 µm Whatman filters), and the total ammonium-nitrogen was determined spectrophotometrically (425 nm) according to the Nessler method (Greenberg et al. 1992). Effective reactor urease activity was expressed as mg urea l−1 reactor h−1.
Single reactor cycle analysis
During a period of optimal calcium removal, the evolution of pH, Ca2+, and TAN was followed within a single cycle in the reactors. Mixed liquor samples were taken approximately every 30–60 min, filtered immediately (0.45 µm Whatman filters), and analysed for TAN as described above. Simultaneously, some of the filtered sample was diluted (milliQ water), acidified (1 N HNO3) and analysed for Ca2+ with flame atomic adsorption spectrometry (Perkin Elmer Flame, Überlingen, Germany). pH evolution was detected and recorded automatically every 5 min using an R305 instrument (Consort, Turnhout, Belgium) equipped with two separate electrodes. This was repeated with the same set-up and sludge, but without the addition of any urea to the reactors, to serve as a negative control for the reaction.
DNA extraction of activated sludge samples
Sludge samples (10 ml) were taken approximately every 7 days from both reactors and stored at −80°C until use. An extraction protocol was adapted from Griffiths et al. (2000) and Kowalchuk et al. (1998). Briefly, in a 2 ml Eppendorf tube, 0.5 g RNase-free 0.1-mm diameter Zirconia/silica beads, 0.5 ml calcareous sludge, 0.5 ml CTAB buffer [hexadecyltrimethylammonium bromide 5% w/v, 0.35 M NaCl, 120 mM potassium phosphate buffer (pH 8.0)] and 0.5 ml phenol:chloroform:isoamyl alcohol mixture (25:24:1) were homogenized (3×30 s at 5,000 rpm) in a Beadbeater (Braun, Melsungen, Germany) with 10 s between shakings. Following centrifugation (5 min, 3,000 g), 300 μl supernatant was transferred to RNase-free Eppendorf tubes. Another 0.5 ml CTAB buffer was added to the remaining sludge suspension and the extraction procedure was repeated, giving a final volume of 600 μl. The phenol was removed by mixing with an equal volume of chloroform:isoamyl alcohol (24:1) and centrifuging (3,000 g, 10 s). The aqueous phase was transferred to a new Eppendorf tube and nucleic acids were precipitated with two volumes of 30% (w/v) polyethylene glycol 6000/1.6 M NaCl for 2 h at room temperature. Subsequently the Eppendorf tube was centrifuged (18,000 g, 4°C, 10 min). Finally, the nucleic acid pellet was washed in ice-cold 70% (v/v) ethanol and dried under vacuum for 10 min prior to re-suspension in 100 μl RNase-free water.
Polymerase chain reaction and denaturing gradient gel electrophoresis of the 16S rRNA genes
The polymerase chain reaction (PCR) mastermix contained 500 nM of each primer, 200 µM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 µl thermophilic DNA polymerase 10× reaction buffer (MgCl2-free), 2.5 U Taq DNA Polymerase (Promega, Madison, Wis.), 400 ng/µl bovine serum albumin (Boehringer, Mannheim, Germany), and DNase- and RNase-free filter-sterile water (Sigma-Aldrich, Steinheim, Germany) to a final volume of 100 µl (Boon et al. 2000). DNA template (1 μl) was added to 24 µl of mastermix.
The 16S rRNA gene fragments of the isolates were amplified by PCR using the forward primer P338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and the reverse primer P518r (5′-ATTACCGCGGCTGCTGG-3′). A GC-clamp of 40 bp (Muyzer et al. 1993) was added to the forward primer to make denaturing gradient gel electrophoresis (DGGE) analysis possible. The PCR program consisted of 10 min at 95°C; 30 cycles of 1 min at 94°C, 1 min at 53°C, 2 min at 72°C; and a final elongation for 10 min at 72°C.
DGGE was performed using the Bio-Rad D Gene System (Hercules, Calif.). PCR samples were loaded onto 7% (w/v) polyacrylamide gels in 1× TAE (20 mM Tris, 10 mM acetate, 0.5 mM EDTA, pH 7.4). The polyacrylamide gels were cast with a denaturing gradient ranging from 50 to 65%, using 7 M urea and 40% formamide as 100% denaturant (Boon et al. 2002). The electrophoresis was run for 16 h at 60°C and 40 V. The gels were stained with SYBR GreenI nucleic acid gel stain (1:10,000 dilution; FMC BioProducts, Rockland, Me.) and photographed (Boon et al. 2000).
PCR-DGGE of the ureC gene
The PCR-mastermix composition was identical to that described above. To amplify the genes encoding the ureC subunit of the urease enzyme, PCR was performed with primers UreC-F (5′-TGGGCCTTAAAATHCAYGARGAYTGGG-3′) and UreC-R (5′-GGTGGTGGCACACCATNANCATRTC-3′) as previously described by Reed (2001). A GC clamp (Muyzer et al. 1993), was attached to the 5′ end of the UreC-F primer. The length of the expected amplified fragment with the GC clamp was 422 bp. PCR was performed in a 9600 thermal cycler (Perkin-Elmer, Norwalk, Conn.) as follows: 94°C for 5 min, followed by 35 cycles of 92°C for 1 min, 50°C for 1 min, and 72°C for 2 min. A final extension was carried out at 72°C for 10 min. DGGE analysis was performed as described above using polyacrylamide gels with a denaturing gradient ranging from 40 to 60%. The electrophoresis was run for 16 h at 60°C, and 45 V.
DNA sequencing
Putative ureC gene fragments and 16S rRNA gene fragments were excised from DGGE gels, reamplified by PCR and sequenced. DNA sequencing of PCR fragments was carried out by ITT Biotech-Bioservice (Bielefeld, Germany). Analysis of DNA sequences and homology searches were completed with standard DNA sequencing programs and the BLAST server of the National Center for Biotechnology Information using the BLAST algorithm (Altschul et al. 1997). Phylogenetic trees were constructed using the RDP Phylip Interface (Maidak et al. 2001)
Nucleotide sequence accession numbers
Nucleotide sequences for 16S rRNA gene fragments have been deposited with the GenBank database under accession numbers AY157610–AY157613. Nucleotide sequences for UreC gene fragments have accession numbers AY167743 and AY167744.
Sludge analysis
Calcareous sludge was sampled from both reactors after 4 weeks and 6 weeks. These samples were analysed for total and volatile suspended solids (TSS and VSS) (Greenberg et al. 1992), while the sludge settling rate (SSR) and the sludge volume index (SVI) values were also determined. This is done by allowing 1 l sludge to settle for 30 min, measuring the rate at which it settled, and then measuring the final sludge volume. The latter was subsequently related to the TSS value. Sludge samples were subsequently dried (50°C, 48 h) and pulverised. The dried sludge was examined by X-ray diffraction (XRD powder diagrams) using a Phillips PW 1140 diffractometer equipped with a Ni filter and a Cu Kα radiation source; compounds were identified according to criteria set by the Joint Committee on Powder Diffraction Standards and the American Society for Testing Materials.
Epifluorescence and scanning electron microscopy
Fresh sludge samples (50 µl) were stained with Live/Dead (Backlight, L-13152, Molecular Probes, Leiden, The Netherlands) staining solution for 15–30 min, and subsequently analysed with epifluorescence microscopy using an Axioskop II plus microscope (Zeiss, Germany) equipped with FLUO3 and CY3 filters to selectively visualise live (green) and dead (red) stained bacteria, respectively. Digital images were captured with a 1-CCD camera using HPDC software (Hamamatsu Photonics, Herssching, Germany), and separate "live" and "dead" images from identical positions were merged using the capturing software. Images were further analysed for particle size distribution using digital image analysis (DIA) software (Olympus MicroImage 4.0; Olympus Optical, Hamburg, Germany). Dried sludge (28°C, 14 days) samples were gold-coated and analysed by scanning electron microscopy (SEM) using a JSM 840 instrument (JEOL, Tokyo, Japan).
Results
Reactor performance
After a 2-week start-up period, a further 2 weeks were required for the establishment of optimal reactor performance, defined by the removal of about 80% of the soluble Ca2+ from the wastewater, within 8 h HRT. Anaerobic effluent from a full-scale UASB reactor was used as influent, with considerable variations in most of the wastewater parameters (Table 1). Table 1 also shows the weekly average effluent data for the two BCC reactors from start-up over a 6-week period. Three operational periods are distinguished (week 1, week 2 and weeks 3–6), based on the applied HRT and urea addition. The first week and second week show high calcium removal efficiencies, which can be attributed to the high urea load and long HRT used during this period. After normalising the HRT (week 3), a decrease in reactor efficiency (calcium removal) was initially observed, partly due to an accidental under-dosage of urea (Table 1). However, the overall performance evolved positively during the subsequent 4 weeks until a period of optimal precipitation efficiency was established and maintained (week 6, Table 1). The total COD decreased with respect to the influent, and this decrease was similar for all three periods. This is, however, ascribed to the sedimentation and co-precipitation of organic mater (e.g. paper fibres) in the BCC reactors, rather than to microbial degradation. Nitrogen (Kjeldahl and ammonium) increased proportional to addition of urea during each period, while nearly no soluble phosphate could be detected in the effluent. Soluble magnesium removal was not detected in any of the time periods. Week 6 represents the data for optimal performance of the BCC reactors, showing a near neutral pH, and high calcium removal, both total and soluble. The evolution in reactor performance was also evident from the evolution of urease activity in the reactors (Fig. 1). The repetitive addition of urea to the inoculum sludge resulted in about a 10-fold increase in urease activity. At week 1, the urease activity was about 7.2 mg urea l−1 reactor h−1, meaning that at least 70 h would be required for the hydrolysis of 500 mg urea l−1 reactor. At week 6, a urease activity of about 100 mg (=1.68 mmol) urea l−1 reactor h−1 was detected, meaning that less than 5 h would be required for complete urea hydrolysis. Whilst numerous bacteria have the urease gene, it is clear that the expression thereof in the activated sludge inoculum was insufficient for BCC requirements, and that stimulation of expression is essential to optimal operation of these reactors.
Single reactor cycle analyses
CaCO3 precipitation from the wastewater used here is directly proportional to urea hydrolysis, but not dependant on the pH. Figure 2 shows the evolution of pH, Ca2+, and TAN within a single reactor cycle during a period of optimal performance (week 6; Table 1), with urea (Fig. 2A) and without urea addition as a negative control (Fig. 2B). Ca2+ precipitation in the absence of any urea addition is limited to about 15%, with hardly any pH shift or TAN evolution evident in the control cycle (Fig. 2B). In the reactor cycle with urea addition (Fig. 2A), visible pH fluctuations are apparent, but it should be noted that the pH of the bulk solution did not exceed 7.5 during the entire precipitation reaction. \( {\rm{NH}}_4^ + - {\rm{N}} \) evolution proceeds rapidly (3 h) to at least 95% relative to the initial urea addition, and Ca2+ precipitation is roughly directly proportional to urea hydrolysis.
DGGE analysis and sequencing
The essence of BCC reactor performance is the combined ureolytic activity of the mixed microbial populations in the sludge. We attempted to characterise this community using genetic tools. The microbial community of the reactors stabilised within a 3-week period to a community with about 15 dominant species detectable by DGGE. A clear development of bands can be seen, especially in the upper half of the gel, representing bacteria that become dominant over time. This development was rapid and persistent from the first week onwards; very few changes occurred in the composition of the dominant bacteria of both reactors. The changes in the bottom of the gel were not further analysed since the appearing and disappearing bands could not be linked to the performance of the reactor. This trend was evident in both reactors. The five major bands were excised but clean sequences were obtained for only four of these (Fig. 3A). The closest relatives of cultured bacteria to the excised sequences were confirmed with BLAST software to be Porphyromonas endodontalis L16491 (Paster et al. 1994) (BCC 2), Porphyromonas cangingivalis g577600 (Collins et al. 1994) (BCC 3), Arcobacter skirrowii L14625 (Stanley et al. 1993) (BCC 4), and Bacteroides stercoris g1325946 (Ruimy et al. 1996) (BCC 5) (Fig. 3B). The urease-PCR-DGGE analysis detected the amplification of only two bands in the inoculum sludge, with one of these subsequently disappearing, leaving only a single dominant band for all the remaining time points in both reactors. Both these bands were excised and sequenced. The corresponding predicted amino-acid sequences were compared with one another and with protein sequences from known isolates. The two bands were clearly different from one another, showing 77% nucleotide alignment, and only 61% alignment (76% positives) in their amino acid sequences. Both bands corresponded to previously sequenced urease genes. One displayed maximum homology (88% identity; 92% positive) with urease genes from the cyanobacteria Synechococcus sp. AF035751 (Sakamoto et al. 1998) and Anabaena variabilis AF065139 (Collier et al. 1999). The dominant band displayed maximum similarity (64% identity; 78% positive) with urease genes from Sinohizobium meliloti S69145 (Miksch et al. 1994) and Yersinia pestis AF095636 (Sebanne et al. 2001).
Denaturing gradient gel electrophoresis (DGGE) separation of 16S rDNA amplified from the replicate reactors; excised and sequenced bands are indicated (2–5). Sequencing of band 1 was unsuccessful. B A phylogenetic tree constructed with partial 16S rDNA sequences (BCC 2–BCC 5). Sequence analysis is described in the text; only 94 base positions were included in the calculations
Sludge analyses
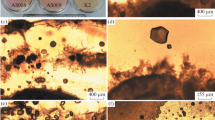
A calcareous sludge, which functioned both as a niche for the microbial species, and as crystal nucleation sites for CaCO3 precipitation, developed in the reactor. A significant increase in TSS was seen, while VSS remained more or less stable (Table 2). The inoculum sludge had an SSR of about 2 m h−1 and the calcareous sludge displayed an SSR increase up to 9 m h−1 after 6 weeks. The SVI portrays the compactability of this calcareous sludge (Table 2) and, together with the SSR, shows that the calcareous sludge has excellent settling properties. XRD analysis of the dried sludge revealed a homogenous mineral fraction comprised almost entirely of calcite, with only trace amounts of fluorite (CaF2) (Fig. 4). Figure 5 shows the transformation of the inoculum sludge into calcareous sludge over the experimental period, with the crystalline nature of the BCC sludge being evident from the light microscopy image (Fig. 5B). DIA of particle size distribution showed approximately 65% of the sludge particles having a radius of less than 100 µm (data not shown). Commercial Live/Dead reagents stain bacteria with intact cellular membranes (thus alive) green, and cells with damaged membranes red. The Live/Dead staining shows that the surface of the sludge particles is colonised with living bacteria, while dead bacteria can also be seen (Fig. 5C, D). SEM images of the calcareous sludge revealed sludge flocs that are clearly crystalline in nature, yet not entirely dense or solid (Fig. 6A). In fact, a complex floc structure is evident, including multiple channels and pores. The fossilised remains of bacterial cells, mostly rod-shaped, can be seen as well as large angular crystals (Fig. 6B).
Light microscopy images showing the evolution of the organic inoculum sludge (A) to a homogenous calcareous sludge (B). (C) A merged Live/Dead stained epifluorescence image of the image in (B) together with enlargement of the marked section (D) of the calcareous sludge flocs colonised with living bacteria (green) as well as the remains of dead bacteria (red)
Discussion
The focus of this work was the characterisation of the bio-catalyst in a BCC reactor in terms of composition and function. A detailed description of BCC reactor performance operational parameters and economic perspectives is presented elsewhere (Hammes et al. 2003a, 2003b). In these previous studies it was established that the two main parameters of critical importance for optimal working of BCC reactors are: (1) a high actual ureolytic capacity, and (2) the presence of crystal nucleation sites. It was opted to inoculate the reactors with activated sludge directly from the industrial wastewater plant, to represent organisms autochthonous to the wastewater being used. Moreover, the sludge already contained some CaCO3 particles as such (data not shown), these being a possible starting point for nucleation sites to develop. Although the urease enzyme is common to many micro-organisms (Mobley and Hausinger 1989; Reed et al. 2001), Table 1 shows that several weeks were still required before optimal calcium removal under standard operational conditions could be achieved. This evolution was directly ascribed to the evolution of the two main parameters mentioned above: urease activity and the presence of crystal nucleation sites.
Crystal nucleation site development was not specifically quantified, but the definitive increase in TSS (Table 2), coupled with the identification of this inorganic component as CaCO3 (specifically calcite) showed that this process was indeed occurring. Considering the wastewater composition, mineral phases that could be expected include struvite (NH4MgPO4), magnesium-calcite and CaCO3 (Doyle and Parsons 2002). Table 1 shows no significant decrease in soluble Mg2+, and only small amounts of P being removed (about 5 mg l−1), in contrast to the large amounts of soluble Ca2+ (>500 mg l−1), thus probably explaining why only CaCO3 as mineral phase was detected with XRD. Of the five known CaCO3 polymorphs, only the stable forms (aragonite and calcite) could reasonably be expected to form. While aragonite is often detected during biological precipitation, previous studies concerning ureolytic MCP also identified the main crystal type as calcite (Stocks-Fischer et al. 1999; Bang et al. 2001; Bachmeier et al. 2002; Kawaguchi and Decho 2002). Fluorite precipitation has been reported previously in association with microbial urea hydrolysis in dental plaque biofilms (Wong et al. 2002). The presence of trace amounts of fluorite in the BCC sludge is ascribed to impurities in the wastewater. The fact that the BCC reactor system utilises self-developed crystal nucleation sites, rather than commercial alternatives (e.g. silver sand), saves significantly on operational (purchasing) and transport costs. It does, however, also require that this process be studied in detail, so that it can be properly controlled under operational conditions.
Urease activity within the BCC reactors is of vital importance for the process. The evolution of urease activity was specifically measured, and displayed a 10-fold increase over the experimental period (Fig. 1). Since the sludge volume was more or less maintained, this increase can be ascribed to the evolution and domination of urease-positive micro-organisms in the reactors. Bacteria are known to hydrolyse urea for the purposes of: (1) increasing the ambient pH (Stocks-Fischer et al. 1999; Burne and Marquis 2000), (2) utilising it as a nitrogen source (Burne and Chen 2001), and (3) using it as a source of energy (Mobley and Hausinger 1989). The pH increase in our experiments is not likely to act as a physiological incentive for the bacteria. The use of urea as nitrogen source is possible, but does not explain the excess amounts hydrolysed. Indeed, Table 1 clearly shows that most of the hydrolysed urea is converted and released as TAN. This extended hydrolysis thus may relate to the fact that certain organisms utilise urea hydrolysis as a mechanism of generating ATP (Mobley and Hausinger 1989). In this reaction, ammonia from urea becomes protonated intracellularly, yielding ammonium in the cytoplasm. This results in formation of a pH gradient, energising of the cell membrane, and ATP synthesis. This unique mechanism has been described for a variety of urease-positive micro-organsims including Ureaplasma urealyticum and Bacillus pasteurii (Mobley and Hausinger 1989; Smith et al. 1993; Jahns 1996). This mechanism would not only be ecologically beneficial to the microbial population, but may also be a key factor in BCC reactor operation and functioning. This mechanism will provide certain bacteria with the ability to survive and rapidly proliferate, thereby becoming the selective factor in the development of the BCC reactor microbial population.
A dynamic evolution in the BCC reactor population was indeed observed. Both the Bacteroides sp. and the Porphyromonas sp. belong to the Bacteroidales order, while Arcobacter sp. are members of the epsilon subdivision of Proteobacteria. These all represent anaerobic bacterial species, typical to dental cavities and the gastro-intestinal tract (Stanley et al. 1993; Paster et al. 1994). It is not surprising that anaerobic bacteria dominate in the BCC reactors, since anaerobic (UASB) effluent was used as wastewater and the reactors were not specifically aerated. Though these species represent some of the most dominant bacterial species in the BCC reactors, this does not necessarily imply that these are the causative organisms of the hydrolysis/precipitation process. In fact, a comprehensive literature search could not specifically link any of these species with urease activity, ATP generation through urease activity, or calcium carbonate precipitation. Concerning the latter point, the view is taken in this study that ureolytic MCP apparently does not involve specific calcium precipitating bacteria, but rather micro-organisms that selectively express their urease genes under specific environmental conditions.
Bacterial urease genes are known to differ considerably in genetic composition, thereby facilitating their molecular analysis (Mobley and Hausinger 1989). We attempted to use a new DGGE-based protocol to detect urease gene diversity in the microbial population of the reactors. The UreC PCR reaction was previously used to differentiate between urease genes from different microbial species (Reed 2001). Differences in the protein sequences of the detected urease PCR-DGGE bands did show that it was possible to distinguish between different urease genes from environmental samples. The low diversity of the urease gene fragment was striking. Apparently only one type of urease dominated the microbial community, with this type probably being responsible for the conversion of urea. DGGE of the 16S rDNA showed several new bands, while the calcium precipitation rate was enhanced, so it is possible that different bacteria posses the same urease gene, or that only one of these strains was responsible for the hydrolysis-precipitation reaction. It is also known that urease genes can be plasmid-encoded (Dupuy et al. 1997), and that horizontal gene transfer between the different sludge species could have occurred.
The basic biochemical mechanism of ureolytic MCP is that bacterial urea hydrolysis increases both the pH and the DIC of an environment, thus favouring precipitation (Douglas and Beveridge 1998; Castanier et al. 1999). Considering the initial pH (7.35), alkalinity (35 meq l−1), and Ca2+ concentration (617.2 mg l−1 =15.4 mM) of the anaerobic effluent used in these experiments, over-saturation of CaCO3 can be roughly calculated as follows (Eqs. 6, 7; Van Haandel and Lettinga 1994):
This 149 times super-saturation in the raw wastewater theoretically suggests that about 99% of Ca2+ will precipitate as CaCO3, without any additives of treatments. However, as Fig. 2B indicates, Ca2+ precipitation in the absence of any urea addition is only about 15%. This is partially explained by the complex nature of real wastewater, for instance the binding of calcium ions to organic matter. Van Haandel and Lettinga (1994) reported experimental findings showing that the K s values for CaCO3 in anaerobic effluent could be as low as 2.0×10−7. Using this value in the calculation above, the corresponding over-saturation is only 2.8 times, suggesting at least 5.44 mM Ca2+ remaining in solution before any treatment or additives (Eq. 8).
Hardly any pH shift is evident in the control cycle with no urea addition (Fig. 2B). In the reactor cycle with urea addition (Fig. 2A), pH evolution is governed by the buffer capacity of the wastewater (primarily bicarbonate), as well as by a possible pH increase from urea hydrolysis (Eqs. 3, 4, 5), and by a possible pH decrease from CaCO3 precipitation (Eqs. 1, 2). Although visible pH fluctuations are apparent, it should be noted that the bulk solution pH barely exceeds 7.5 during the entire precipitation reaction. \( {\rm{NH}}_4^ + - {\rm{N}} \)evolution confirms the high ureolytic activity in the reactors: almost all of the added urea is converted completely to \( {\rm{NH}}_4^ + \) within about 3 h. Typical Michaelis-Menten enzyme kinetics suggest a very high substrate affinity. About 8 mM urea was hydrolysed, while about 12.5 mM Ca2+ was precipitated (Fig. 2A), of which at least 3 mM could be attributed to chemical precipitation (Fig. 2B). It is thus apparent that Ca2+ precipitation is directly proportional to urea hydrolysis, even though little correlation between the pH and Ca2+ evolution could be detected.
Apart from this "link-in-time" of CaCO3 precipitation with the bacterial mechanisms (urea hydrolysis), a "link-in-space" was also established with microscopy techniques. The Live/Dead staining shows that the surface of the sludge particles, which represent crystal nucleation sites, is colonised with living bacteria, as well as showing some remains of dead bacteria (Fig. 5C, D), thereby showing that the calcareous sludge provides a niche for bacteria to attach and proliferate. This was confirmed by SEM analysis. Although intact bacterial cells could not be detected because the dry sludge was viewed without any additional ethanol critical point drying, the fossilised remains of bacterial cells, mostly rod-shaped, could be seen (Fig. 6B). These SEM images displayed quite some resemblance to SEM images of microbial CaCO3 precipitation from natural environments (Wright 1999). Cellular incrustation could have involved specific precipitation mechanisms on the cellular level, such as the functioning of bacteria as crystal nucleation sites (Schultze-Lam et al. 1996; Stocks-Fischer et al. 1999), and calcium concentration by bacteria (McConnaughey and Whelan 1997). The SEM analysis also suggested that the precipitation event might well lead to physical capturing and eventual "die-off" of the organisms. Southam (2000) suggested that cell-surface mineralisation leads to "die-off" of bacteria, due to limitations of nutrient and proton transport. It is well documented that bacteria are often captured and fossilised during the precipitation reaction (Trewin and Knoll 1999). Exactly this phenomenon sparked a debate as to whether mineral precipitates on the ALH84001 meteorite could represent past life in outer space (McKay et al. 1996).
The above results showed that BCC reactors require about 4–6 weeks start-up before optimal performance is reached, mainly depending on the development of a high urease activity and sufficient crystal nucleation sites. During optimal performance, CaCO3 precipitation is directly proportional to urea hydrolysis, which is an intracellular biochemical process. Furthermore, it was shown that viable micro-organisms inhabit the surface of the calcareous sludge flocs, which in this case serves as crystal nucleation sites in the precipitation process. It is proposed that this co-localisation of the biochemical catalysts and the crystal nucleation sites results in the formation of a microenvironment favouring CaCO3 precipitation (Fig. 7). As a result, localised elevated pH levels would cause a shift in the bicarbonate equilibrium, resulting in \( {\rm{CO}}_3^{2 - } \) formation at the cellular level, which immediately precipitates as CaCO3 due to the presence of crystal nucleation sites. This would explain the near absence of any significant bulk solution pH increase (Fig. 2). Calcium required for precipitation would be in constant supply as a result of the continuous concentration gradient that results from the precipitation reaction.
In conclusion, BCC reactors, combining autochthonous ureolytic bacteria on calcareous sludge flocs, represent an efficient process for calcium removal from industrial wastewater. The localisation of the bio-catalyst on the crystal nucleation sites allows for the possibility of micro-environment chemistry, which could be a key factor in future implementations of this process. The high degree of adaptability of this process makes BCC reactors an attractive technology for wastewaters with elevated levels of heavy metals, phosphate and radionuclides.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bachmeier KL, Williams AE, Warmington JR, Bang SS (2002) Urease activity in microbiologically-induced calcite precipitation. J Biotechnol 93:171–181
Bang SS, Galinat JK, Ramakrishnan V (2001) Calcite precipitation induced by polyurethane-immobilized Bacillus pasteurii. Enzyme Microb Technol 28:404–409
Boon N, Goris J, De Vos P, Verstraete W, Top EM (2000) Bioaugmentation of activated sludge by an indigenous 3-chloroaniline degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913
Boon N, De Windt W, Verstraete W, Top EM (2002) Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol Ecol 39:101–112
Burne RA, Chen RE (2001) Bacterial ureases in infectious diseases. Microbes Infect 2:533–542
Burne RA, Marquis RE (2000) Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193:1–6
Castanier S, Le Métayer-Levrel G, Perthuisot JP (1999) Ca-carbonates precipitation and limestone genesis—the microbiologist point of view. Sediment Geol 126:9–23
Collier JL, Brahamsha B, Palenik B (1999) The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447–459
Collins MD, Love DN, Karjalainen J, Kanervo A, Forsblom B, Willems A, Stubbs S, Sarkiala E, Bailey GD, Wigney DI, Jousimies-Somer H (1994) Phylogenetic analysis of members of the genus Porphyromonas and description of Porphyromonas cangingivalis sp. nov. and Porphyromonas cansulci sp. nov. Int J Syst Bacteriol 44:674–679
Douglas S, Beveridge TJ (1998) Mineral formation by bacteria in natural microbial communities. FEMS Microbiol Ecol 26:79–88
Doyle JD, Parsons SA (2002) Struvite formation, control and recovery. Water Res 36:3925–3940
Dupuy B, Daube G, Popoff MR, Cole ST (1997) Clostridium perfringens urease genes are plasmid borne. Infect Immun 65:2313–2320
Fujita Y, Ferris EG, Lawson RD, Colwell FS, Smith RW (2000) Calcium carbonate precipitation by ureolytic subsurface bacteria. Geomicrobiol J 17:305–318
Greenberg AE, Clesceri LS, Eaton AD (eds) (1992) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, Washington, D.C.
Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491
Hammes F, Seka A, De Knijf S, Verstraete W (2003a) A novel approach to calcium removal from calcium-rich industrial wastewater. Water Res 37:699–704
Hammes F, Seka A, Van Hege K, Van de Wiele T, Vanderdeelen J, Siciliano SD, Verstraete W (2003b) Calcium removal from industrial wastewater by bio-catalytic CaCO3 precipitation. J Chem Technol Biotechnol (in press)
Jahns T (1996) Ammonium/urea-dependent generation of a proton electrochemical potential and synthesis of ATP in Bacillus pasteurii. J Bacteriol 178:403–409
Kawaguchi T, Decho AW (2002) A laboratory investigation of cyanobacterial extracellular polymeric secretions (EPS) in influencing CaCO3 polymorphism. J Cryst Growth 240:230–235
Kowalchuk GA, Bodelier PLE, Heilig GHJ, Stephen JR, Laanbroek HJ (1998) Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol Ecol 27:339–350
Lloyd JR, Lovley DR (2001) Microbial detoxification of metals and radionuclides. Curr Opin Biotechnol 12:248–253
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (ribosomal database project). Nucleic Acids Res 29:173–174
McConnaughey TA, Whelan FF (1997) Calcification generates protons for nutrient and bicarbonate uptake. Earth Sci Rev 42:95–117
McKay DS, Gibson EK, Thomas-Keprta K, Vali H, Romanek CS, Clemett SJ, Chillier XDF, Maechling CR, Zare RN (1996) Search for past life on Mars: possible relic biogenic activity in Martian meteorite ALH84001. Science 273:924–930
Miksch G, Arnold W, Lentzsch P, Priefer UB, Puhler A (1994) A 4.6 kb DNA region of Rhizobium meliloti involved in determining urease and hydrogenase activities carries the structural genes for urease (ureA, ureB, ureC) interrupted by other open reading frames. Mol Gen Genet 242:539–550
Mobley HLT, Hausinger RP (1989) Microbial ureases: significance, regulation and molecular characterisation. Microbiol Rev 53:85–108
Muyzer G, De Waal EC, Uitterlinden A (1993) Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Paster BJ, Dewhirst FE, Olsen I, Fraser GJ (1994) Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J Bacteriol 176:725–732
Reed KE (2001) Restriction enzyme mapping of bacterial urease genes: using degenerate primers to expand experimental outcomes. Biochem Mol Biol Educ 29:239–244
Ruimy R, Podglajen I, Breuil J, Christen R, Collatz E (1996) A recent fixation of cfiA genes in a monophyletic cluster of Bacteroides fragilis is correlated with the presence of multiple insertion elements J Bacteriol 178:1914–1918
Sakamoto T, Delgaizo VB, Bryant DA (1998) Growth on urea can trigger death and peroxidation of the cyanobacterium Synechococcus sp. strain PCC 7002. Appl Environ Microbiol 64:2361–2366
Schultze-Lam S, Fortin D, Davis BS, Beveridge TJ (1996) Mineralisation of bacterial surfaces. Chem Geol 132:171–181
Sebbane F, Devalckenaere A, Foulon J, Carniel E, Simonet M (2001) Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect Immun 69:170–176
Smith DGE, Russel WC, Ingledew WJ, Thirkell D (1993) Hydrolysis of urea by Ureaplasma urealyticum generates a transmembrane potential with resultant ATP synthesis. J Bacteriol 175:3253–3258
Southam G (2000) Bacteria surface-mediated mineral formation. In: Lovley DR (ed) Environmental microbe-metal interactions, ASM Press, Washington D.C.
Stanley J, Linton D, Burnens AP, Dewhirst FE, Owen RJ, Porter A, On SL, Costas M (1993) Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol 139:2495–2504
Stocks-Fisher S, Galinat JK, Bang SS (1999) Microbiological precipitation of CaCO3. Soil Biol Biochem 3:1563–1571
Stumm W, Morgan JJ (1981) Aquatic chemistry, 2nd edn. Wiley, New York
Trewin NH, Knoll AH (1999) Preservation of Devonian chemotrophic filamentous bacteria in calcite veins. Palaios 14:288–294
Van Haandel AC, Lettinga GL (1994). Anaerobic sewage treatment. A practical guide for regions with a hot climate. Wiley, Chichester, UK
Warren LA, Maurice PA, Parmar N, Ferris FG (2001) Microbially mediated calcium carbonate precipitation: implications for interpreting calcite precipitation and for solid-phase capture of inorganic contaminants. Geomicrobiol J 18:93–115
Wong L, Sissons CH, Pearce EIF, Cutress TW (2002) Calcium phosphate deposition in human dental plaque microcosm biofilms induced by a ureolytic pH-rise procedure. Arch Oral Biol 47:779–790
Wright DT (1999) The role of sulphate-reducing bacteria and cyanobacteria in dolomite formation in distal ephemeral lakes of the Coorong region, South Australia. Sediment Geol 126:147–157
Acknowledgements
This research was sponsored by VPK (Oudegem, Belgium), AVECOM nv (Belgium) and by the project grant G.O.A. (1997–2002) of the "Ministerie van de Vlaamse GemeenschapVlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek" (Belgium), and by an equipment grant (epifluorescence microscope) from the Flemish Fund for Scientific Research (FWO-Vlaanderen). Special thanks to Els Jolie and Greet Van De Velde for analytical measurements, and to Sofie Dobbelaere, and Thomas Vande Sijpe for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hammes, F., Boon, N., Clement, G. et al. Molecular, biochemical and ecological characterisation of a bio-catalytic calcification reactor. Appl Microbiol Biotechnol 62, 191–201 (2003). https://doi.org/10.1007/s00253-003-1287-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1287-6