Abstract
Several immune regulatory cell types participate in the protection against autoimmune diseases such as autoimmune diabetes. Of these immunoregulatory cells, we and others have shown that peripheral CD4−CD8− double negative (DN) T cells can induce antigen-specific immune tolerance. Particularly, we have described that diabetes-prone mice exhibit a lower number of peripheral DN T cells compared to diabetes-resistant mice. Identifying the molecular pathways that influence the size of the DN T cell pool in peripheral lymphoid organs may thus be of interest for maintaining antigen-specific immune tolerance. Hence, through immunogenetic approaches, we found that two genetic loci linked to autoimmune diabetes susceptibility, namely Idd2 and Idd13, independently contribute to the partial restoration of DN T cell proportion in secondary lymphoid organs. We now extend these findings to show an interaction between the Idd2 and Idd13 loci in determining the number of DN T cells in secondary lymphoid organs. Using bioinformatics tools, we link potential biological pathways arising from interactions of genes encoded within the two loci. By focusing on cell cycle, we validate that both the Idd2 and Idd13 loci influence RAD51 expression as well as DN T cell progression through the cell cycle. Altogether, we find that genetic interactions between Idd2 and Idd13 loci modulate cell cycle progression, which contributes, at least in part, to defining the proportion of DN T cells in secondary lymphoid organs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A prominent role in the prevention of autoimmunity has been credited to various regulatory cell subsets, including peripheral immunoregulatory CD4−CD8− double negative (DN) T cells. Indeed, DN T cells have been shown to inhibit immune responses in an antigen-specific manner (Zhang et al. 2006; Zhang et al. 2000). This has major implications since the use of DN T cells in immunotherapy should therefore lead to fewer side effects and a decreased risk of infection, which remains a major concern in the application of broad immunosuppressive regimens (Bluestone et al. 2010a). As a result, DN T cells have been studied in various disease models, revealing that DN T cells display a promising therapeutic potential in the induction of allo- and xenograft tolerance (Chen et al. 2003; Ma et al. 2008; Young et al. 2002; Zhang et al. 2000) and in the prevention of both graft-vs-host disease (He et al. 2007; McIver et al. 2008; Young et al. 2003; Young et al. 2002) and autoimmune diabetes (Dugas et al. 2010; Duncan et al. 2010; Ford et al. 2007; Zhang et al. 2011).
Using TCR transgenic models, our work has supported the contribution of DN T cells towards immune tolerance in autoimmune diabetes (Dugas et al. 2010). Specifically, we have demonstrated that TCR transgenic DN T cell proportion is low in autoimmune diabetes-susceptible mice (Dugas et al. 2010). However, the immunoregulatory function of TCR transgenic DN T cells remains unaltered in these mice (Hillhouse et al. 2010), demonstrating that a deficiency in DN T cell number, rather than function, contributes to autoimmune diabetes susceptibility. Correspondingly, the injection of DN T cells is sufficient to reduce the incidence of autoimmune diabetes in autoimmune diabetes-susceptible mice in the TCR transgenic setting (Dugas et al. 2010). These results highlight the importance of identifying the genetic determinants controlling DN T cell number as they may reveal key molecular targets to promote DN T cell development, thereby supporting immune tolerance.
As for most autoimmune diseases, susceptibility to autoimmune diabetes is a complex genetic trait (Bluestone et al. 2010b; Concannon et al. 2009). NOD (non-obese diabetic) mice, which spontaneously develop autoimmune diabetes, have facilitated the study of genetic factors underlying autoimmune diabetes susceptibility (Anderson and Bluestone 2005; Wicker et al. 2005). To date, more than 20 Idd (Insulin-dependent diabetes) loci linked to diabetes predisposition have been identified in NOD mice (www.T1Dbase.org), and the concordance with human diabetes susceptibility has been demonstrated for some of these loci (Driver et al. 2011; Wicker et al. 2005). Subsequent analysis of NOD.Idd congenic mice has revealed Idd regions which are sufficient to reduce insulitis and/or diabetes incidence (Ghosh et al. 1993; Lyons et al. 2000; McAleer et al. 1995; Serreze et al. 1994). Interestingly, we have also linked certain Idd regions to the regulation of DN T cell number (Collin et al. 2014; Dugas et al. 2014), where increased DN T cell number is associated with autoimmune diabetes resistance (Collin et al. 2014; Dugas et al. 2010). More specifically, using a linkage analysis approach and congenic strain validation, we have shown that DN T cell proportion in peripheral lymphoid organs is a multigenic trait and that both Idd2 and Idd13 loci each contribute to the partial restoration of DN T cell numbers in the spleen and lymph nodes (Collin et al. 2014; Dugas et al. 2014). These results suggest that Idd2 and Idd13 loci modulate diabetes development, at least in part, by influencing DN T cell number.
Idd2 and Idd13, located on chromosome 9 and 2, respectively, are two loci linked to autoimmune diabetes susceptibility, while Idd2 is also linked to insulitis (Ghosh et al. 1993; Prochazka et al. 1987; Serreze et al. 1998; Serreze et al. 1994; Wicker et al. 1995). Recently, our group has shown that, in contrast to Idd13 (Hillhouse et al. 2016), the Idd2 locus also protects NOD.H2k mice from diabetes development and insulitis in the 3A9 TCR:insHEL transgenic model (Collin et al. 2014). In addition to their role in DN T cell number modulation in the non-transgenic and/or TCR transgenic setting, both Idd2 and Idd13 loci are thought to play a role in the selection of diabetogenic T cells (Collin et al. 2014; Dugas et al. 2014; Pearce et al. 1995; Serreze et al. 1998). Accordingly, the Idd13 interval encodes for several candidate genes implicated in the negative selection of autoreactive T cells including the Mer tyrosine kinase (Mertk) gene as well as multiple apoptosis-associated genes (Liston et al. 2007; Liston et al. 2004; Serreze et al. 1998; Wallet et al. 2009; Zucchelli et al. 2005), and Idd2 encodes for Thy1, Cbl, and Il10ra, all likely contributors to T cell biology (Lang and Bellgrau 2004; Prochazka et al. 1987). In addition to DN T cell numbers, the Idd13 locus also influences the proportion of merocytic dendritic cells, another cell population associated with autoimmune diabetes susceptibility (Pelletier and Lesage 2013). Altogether, the Idd2 and Idd13 loci clearly play a role in autoimmune diabetes progression via different mechanisms, including the modulation of DN T cell number.
For our studies, we take advantage of a spontaneous autoimmune diabetes model, namely the 3A9 TCR:insHEL transgenic mouse model. In this model, the insHEL transgene promotes hen egg lysosyme (HEL) expression under the rat insulin promoter, thus inducing pancreatic β cell specific expression, while the 3A9 TCR transgene yields T cells recognizing a HEL peptide presented in the context of I-Ak (Akkaraju et al. 1997; Ho et al. 1994). To allow for effective HEL peptide presentation on I-Ak, this pair of insHEL and 3A9 TCR transgenes was introduced onto B10.BR and NOD.H2k genetic backgrounds, resulting in the diabetes-resistant 3A9 TCR:insHEL B10.BR and diabetes-susceptible 3A9 TCR:insHEL NOD.H2k mouse strains (Lesage et al. 2002). Importantly, 3A9 DN T cell proportion is decreased in secondary lymphoid organs of mice from the diabetes-susceptible NOD.H2k background in comparison to the diabetes-resistant B10.BR background (Dugas et al. 2010), where this difference is partially restored by the presence of the Idd2 or Idd13 locus from the B10.BR background in NOD.H2k congenic mice (Collin et al. 2014; Dugas et al. 2014).
Herein, we investigated the interactions between genetic determinants encoded within the Idd2 and Idd13 loci and how they influence DN T cell proportion. To do so, we generated 3A9 TCR and 3A9 TCR:insHEL NOD.H2k-Idd2/13 double congenic mice. Using the double congenic mice, we demonstrate an interaction between Idd2 and Idd13 loci in determining the number of DN T cells in secondary lymphoid organs, where DN T cell proportion is fully restored in the lymph nodes of 3A9 TCR:insHEL NOD.H2k-Idd2/13 mice. By exploiting DAVID and STRING bioinformatics databases, we link potential biological pathways arising from interactions of genes encoded within the Idd2 and Idd13 loci. This in silico approach suggested that the Idd2 and Idd13 loci may regulate cell cycle progression. Using congenic mice, we validate that the Idd2 and Idd13 loci influence progression through S-phase and DN T cell proliferation. Moreover, we find that both the Idd2 and Idd13 loci modulate RAD51 expression. Together, the bioinformatics data, the cell cycle analysis, and the regulation of RAD51 expression suggests that Idd2 and Idd13 loci modulate cell cycle progression, which likely contributes to defining the proportion of DN T cells in secondary lymphoid organs.
Results
The proportion of 3A9 DN T cells is significantly lower in the lymph nodes of 3A9 TCR NOD.H2k mice in comparison to 3A9 TCR B10.BR mice, even in the absence of cognate antigen (Dugas et al. 2010). This suggests that the NOD genetic background, independent of the autoimmune response, defines the low number of DN T cells in secondary lymphoid organs. To identify these genetic determinants, we previously performed a linkage analysis on an F2 outcross from these two parental strains and found that immunoregulatory DN T cell proportion is regulated by a multigenic trait in secondary lymphoid organs (Collin et al. 2014). By exploiting the 3A9 TCR transgenic mouse model, we subsequently demonstrated that both Idd2 and Idd13 loci independently contribute to defining the proportion and number of DN T cells in the spleen and lymph nodes (Collin et al. 2014; Dugas et al. 2014). Still, 3A9 TCR NOD mice congenic for either of these loci presented with a modest increase in DN T cell number relative to mice bearing the B10.BR genetic background. As a result, we proposed that interactions between genetic determinants encoded within the Idd2 and Idd13 loci cooperate to define the proportion and number of immunoregulatory DN T cells. We segregated the F2 mice from our aforementioned linkage analysis (Collin et al. 2014) according to their genotype at the SNP with the highest LOD score on both chromosome 2 (Idd13) and chromosome 9 (Idd2), namely rs6193859 and D9Mit232, respectively, and plotted the proportion of 3A9 DN T cells from the skin-draining lymph nodes (Fig. 1a, Table 1). F2 mice bearing NOD homozygous alleles at the Idd2 locus present with a low proportion of DN T cells, which is not significantly influenced by the presence of either NOD or B10.BR alleles at the Idd13 locus (Fig. 1a, white circles). However, we find a significant increase in the proportion of DN T cells in F2 mice bearing homozygous B10.BR alleles at both loci (19.34 ± 1.42) when compared to mice bearing homozygous NOD alleles at Idd2 (13.60 ± 1.10), at Idd13 (13.65 ± 1.36), or at both loci (11.82 ± 2.05) (Table 1). This suggests that B10.BR alleles from both Idd2 and Idd13 loci may have an additive or synergistic effect in regulating DN T cell proportion.
Interactions between chromosome 2 and chromosome 9 in determining 3A9 DN T cell proportion in 3A9 TCR F2 mice. a The proportion of 3A9 DN T cells in the skin-draining lymph nodes of 3A9 TCR F2 mice is represented based on their genotype on chromosome 2 at rs6193859 (x axis) and chromosome 9 at D9Mit232 (colored circles). N/N = homozygous NOD.H2k alleles, B/N = heterozygous alleles, B/B = B10.BR homozygous alleles. Black circles, homozygous B10.BR alleles at D9Mit232; gray circles, heterozygous alleles at D9Mit232; white circles, homozygous NOD.H2k alleles at D9Mit232. Mean ± SEM are represented. b Schematic representation of mouse chromosome 2 and chromosome 9 from NOD.H2k-Idd2/13 congenic mice. The delimitation of the congenic interval as defined by Illumina medium density or PCR reaction. B10 alleles are shown in black, NOD alleles in white, and alleles of undetermined origin in gray. The markers are positioned according to assembly NCBI m37 coordinates
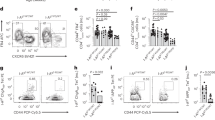
To test this potential interaction between Idd2 and Idd13 loci, we generated 3A9 TCR NOD.H2k-Idd2/13 as well as 3A9 TCR:insHEL NOD.H2k-Idd2/13 double congenic mice, where the limits of the intervals were determined by genotyping and medium density SNP Illumina platform (Fig. 1b). As previously reported, B10.BR mice present with a higher proportion of 3A9 DN T cells than NOD.H2k mice in both the spleen and lymph nodes of either the 3A9 TCR single transgenic or 3A9 TCR:insHEL double transgenic mice (Fig. 2) (Collin et al. 2014; Dugas et al. 2010; Dugas et al. 2013; Dugas et al. 2014; Hillhouse et al. 2010). In addition, both the NOD.H2k-Idd2 and NOD.H2k-Idd13 single congenic mice show an increase in the proportion of 3A9 DN T cells relative to NOD.H2k mice (Fig. 2) (Collin et al. 2014; Dugas et al. 2014). Although the proportion of 3A9 DN T cells was increased in the spleen of NOD.H2k-Idd2/13 double congenic mice relative to the parental NOD.H2k strain, the proportion of 3A9 DN T cells of the double congenic mice did not exceed that of NOD.H2k-Idd13 single congenic mice in the spleen (Fig. 2a). In contrast to the spleen, and in agreement with the F2 linkage data of 3A9 DN T cells in the lymph nodes (Fig. 1), we observe a statistically significant increase in the proportion of 3A9 DN T cells of NOD.H2k-Idd2/13 double congenic mice compared to single congenic mice in the lymph nodes (Fig. 2b). Importantly, the proportion of 3A9 DN T cells in the lymph nodes of 3A9 TCR:insHEL NOD.H2k-Idd2/13 double congenic mice was comparable to that of the 3A9 TCR:insHEL B10.BR parental strain (Fig. 2b). Notably, the increased proportion of 3A9 DN T cells in the lymph nodes of NOD.H2k-Idd2/13 double congenic mice relative to NOD.H2k, although not as striking, is corroborated in absolute numbers (Fig. 3), and is not the result of a decrease in the proportion of either CD4 or CD8 T cells (Table 2). Together, these data suggest that an interaction between Idd2 and Idd13 loci increases the proportion of 3A9 DN T cells in the NOD.H2k transgenic mice.
The Idd2 and Idd13 loci cooperate to define the proportion of 3A9 DN T cells in lymph nodes. The proportion of 3A9 DN T cells is shown for 3A9 TCR and 3A9 TCR:insHEL transgenic mice in the spleen (a) and the lymph nodes (b). Each dot represents data from one mouse. Mean ± SEM are shown. p value * < 0.05, ** < 0.01, *** < 0.001 according to one-way ANOVA with Bonferroni post-hoc test. ns, non-significant
The Idd2 and Idd13 loci cooperate to define the absolute number of 3A9 DN T cells in lymph nodes. The absolute number of 3A9 DN T cells is shown for 3A9 TCR and 3A9 TCR:insHEL transgenic mice in the spleen (a) and the lymph nodes (b). Each dot represents data from one mouse. Mean ± SEM are shown. p value * < 0.05, ** < 0.01, *** < 0.001 according to one-way ANOVA with Bonferroni post-hoc test. ns, non-significant
The increased proportion of 3A9 DN T cells in secondary lymphoid organs of NOD.H2k-Idd2/13 double congenic mice relative to that of NOD.H2k mice may potentially result from an increase in thymic selection in the 3A9 TCR:insHEL NOD.H2k-Idd2/13 mice. We thus quantified the proportion of 3A9 DN T cells in the thymus of the various 3A9 TCR single transgenic and 3A9 TCR:insHEL double transgenic mice. As for the spleen and lymph nodes, we find a higher proportion and absolute number of 3A9 DN T cells in the thymus of B10.BR mice relative to NOD.H2k mice, in both the 3A9 TCR single transgenic and 3A9 TCR:insHEL double transgenic mice (Fig. 4). In addition, the NOD.H2k-Idd13 single congenic mice show a modest increase in the proportion of 3A9 DN T cells relative to NOD.H2k mice, whereas the increase in 3A9 DN T cells for NOD.H2k-Idd2 is only observed in the 3A9 TCR:insHEL double transgenic mice (Fig. 4). Notably, although the proportion and absolute number of 3A9 DN T cells was modestly increased in the thymus of NOD.H2k-Idd2/13 double congenic mice relative to NOD.H2k mice, this increase was comparable to that observed in the NOD.H2k-Idd13 single congenic mice (Fig. 4). This suggests the absence of an additive effect of the Idd2 and Idd13 loci in defining DN T cell number in the thymus. In line with our previous work showing that thymic DN T cell proportion in 3A9 TCR mice does not correlate with 3A9 DN T cell proportion in secondary lymphoid organs (Dugas et al. 2013), we find that the increased proportion of 3A9 DN T cells in the secondary lymphoid organs of NOD.H2k-Idd2/13 double congenic mice relative to NOD.H2k mice is not a consequence of increased thymic differentiation towards the DN T cell lineage.
The impact of Idd2 and Idd13 loci on the proportion and number of 3A9 DN T cells in secondary lymphoid organs in not due to increased thymic differentiation. The proportion (a) and absolute number (b) of 3A9 DN T cells in the thymus is shown for 3A9 TCR and 3A9 TCR:insHEL transgenic mice. Each dot represents data from one mouse. Mean ± SEM are shown. p value * < 0.05, ** < 0.01 according to one-way ANOVA with Bonferroni post-hoc test. ns, non-significant
In the 3A9 TCR:insHEL B10.BR, BALB.K, and NOD.H2k mice, we have previously shown that elevated numbers 3A9 DN T cells in secondary lymphoid organs correlate with a decrease in diabetes incidence and that injection of 3A9 DN T cells in the BALB.K setting is sufficient to prevent diabetes onset (Dugas et al. 2010). We have also shown that 3A9 TCR:insHEL NOD.H2k-Idd2 mice exhibit an increase in 3A9 DN T cells and a lower incidence of diabetes relative to 3A9 TCR:insHEL NOD.H2k mice (Collin et al. 2014). However, we recently found that 3A9 TCR:insHEL NOD.H2k-Idd13 mice, which present an increase in 3A9 DN T cells relative to their NOD.H2k counterpart (Fig. 2) (Dugas et al. 2014), exhibit an increase in diabetes incidence (Hillhouse et al. 2016). This result suggests that, in contrast to Idd2, the increase in DN T cell number conferred by the Idd13 locus is not sufficient to control insulitis present in the 3A9 TCR:insHEL mouse model (as detailed in (Hillhouse et al. 2016)). Because of the contrasting phenotypes of Idd2 and Idd13 in controlling diabetes incidence in 3A9 TCR:insHEL NOD.H2k mice, we decided to study the impact of the combination of both Idd2 and Idd13 loci. Expectedly, 3A9 TCR:insHEL NOD.H2k-Idd2/13 double congenic mice presented a level of insulitis that was comparable to both 3A9 TCR:insHEL B10.BR and NOD.H2k mice (Fig. 5). Still, all of the nine 3A9 TCR:insHEL NOD.H2k-Idd2/13 mice that were aged to over 28 weeks remained diabetes-free and never exhibited glucosuria nor glycemia (Table 3). The combination of the Idd2 and Idd13 loci in the 3A9 TCR:insHEL NOD.H2k mice also significantly reduced the serum level of HEL-specific IgG autoantibodies (Table 3). Taken together, these data show that the combined presence of Idd2 and Idd13 loci in 3A9 TCR:insHEL NOD.H2k mice enhances the proportion of 3A9 DN T cells in secondary lymphoid organs and decreases the level of autoantibodies, as well as prevents diabetes onset.
We next sought to define the biological process related to the increase in DN T cell proportion which is mediated by the combination of the Idd2 and Idd13 congenic intervals. We considered all of the genes encoded within the Idd2 and Idd13 loci. We then queried the DAVID bioinformatics database based on the gene list from the Idd2 and Idd13 loci. The Idd2 and Idd13 gene lists independently revealed nine and five biological processes respectively, while 11 biological processes were identified by the combination of the two gene lists (Table 4). Using the combined Idd2 and Idd13 gene lists obtained by the DAVID analysis, we next independently queried the STRING database for each of these 11 biological processes. We selectively considered associations from genes in Idd2 and Idd13 that showed a confidence score above 0.7 for the given biological process (Table 5). We find strong interactions between genes in all biological processes, except for lipid transport where the four links between genes from Idd2 and Idd13 all presented with a confidence below 0.7. Based on this observation, we decided to further restrict our attention to biological processes where a confidence score above 0.7 was given to most of the gene interactions (i.e., more than 50% of the interactions). Out of the 11 biological processes, only 3 fit these criteria, namely phospholipid catabolism, T cell, and cell cycle (Table 5, highlighted in gray).
The DAVID analysis grouped 7 genes from the Idd2 locus and 28 genes from the Idd13 locus under phospholipid catabolism. STRING revealed that only two genes from the Idd2 locus, namely Arhgef12 and Pafah1b2, were associated with genes from the Idd13 locus (Fig. 6a). Accordingly, Arhgef12 was linked to Adra1d while Pafah1b2 was linked to four genes of the Pla2g4 family, specifically Pla2g4b, Pla2g4d, Pla2g4e, and Pla2g4f. Based on ImmGen, Arhgef12 is strongly expressed in germinal center B cells and Pafah1b2 is expressed in many immune cell populations (immgen.org). However, neither Adra1d nor any of the four Pla2g4 genes from the Idd13 locus are expressed in hematopoietic cells (immgen.org). As we have previously shown that DN T cell proportion is regulated at least in part by hematopoietic-intrinsic factors (Dugas et al. 2014), it is unclear how the Arhgef12 and Pafah1b2 genes from the Idd2 locus interact with Adra1d or any of the four Pla2g4 genes from the Idd13 locus to influence the proportion of DN T cells, as none of the potentially interacting genes in Idd13 are expressed in the hematopoietic system.
The T cell biological process showed interactions between genes from the CD3 complex encoded in the Idd2 locus and B2m from the Idd13 locus (Fig. 6b). This interaction stems from the antigen presentation of peptides to CD8+ T cells, where the chains of the CD3 complex are necessary for TCR expression which recognizes peptides in the context of MHC class I molecules that are associated with β2m. However, we have previously demonstrated that B2m deficiency does not decrease the proportion of 3A9 DN T cells in both the spleen and lymph nodes of 3A9 TCR transgenic mice, as shown in B2m-deficient mice from both B10.BR and NOD.H2k genetic backgrounds (Dugas et al. 2014). The T cell biological process is thus unlikely to explain the additive interaction between Idd2 and Idd13 loci in defining the proportion of DN T cells.
Finally, STRING analysis of the cell cycle progression reveals multiple relevant interactions between genes from the Idd2 and Idd13 loci (Fig. 6c). For one, Chek1 in Idd2 partners with Rad51, Mcm8, and Cdc25b in Idd13 to regulate cell cycle progression (Kramer et al. 2004; Lutzmann et al. 2012; Patil et al. 2013; Sorensen et al. 2005). In addition, Zw10 in Idd2 and Bub1, Bub1b, and Casc5 in Idd13 all localize to the kinetochore and facilitate mitotic progression (Anderson et al. 2002; Genin et al. 2012; Taylor and McKeon 1997; Vallee et al. 2006; Varma et al. 2006). Importantly, all of these genes are ubiquitously expressed.
To determine if cell cycle progression is possibly impaired in NOD.H2k and could therefore define DN T cell number, we performed cell cycle analysis using propidium iodide staining in total spleen cells from B10.BR, NOD.H2k and NOD.H2k-Idd2/13 mice. A greater proportion of cells from NOD.H2k mice were actively engaged in S-phase (Fig. 7a). Interestingly, cells from NOD.H2k-Idd2/13 mice exhibited a comparable cell cycle profile to that of B10.BR mice with a similar proportion of cells in S-phase (Fig. 7a). This finding suggests that the Idd2 and Idd13 loci on the NOD.H2k background are sufficient to modulate the proportion of cells in S-phase. We confirm that this is not a particularity of the MHC haplotype, as we find that spleen cells from NOD mice also have a higher proportion of cells in S-phase relative to those from C57BL/6 mice (Fig. 7b). More importantly, we find that, among T cell subsets, the impact of the Idd2 and Idd13 loci on proliferation is specific to DN T cells (Fig. 7c). Indeed, when comparing T cell subsets from both NOD.H2k and NOD.H2k-Idd2/13 mice, we find a similar proportion of either CD4+ or CD8+ T cells that express Ki-67. However, a lower proportion of DN T cells from NOD.H2k-Idd2/13 mice stained positive for Ki-67 relative to DN T cells from NOD.H2k mice (Fig. 7c). As for S-phase measurements, the fraction of Ki-67+ DN T cells from NOD.H2k-Idd2/13 mice is comparable to that of B10.BR mice. Altogether, these data suggest that the low proportion of DN T cells in the NOD background is influenced by allelic variants of genes encoded within the Idd2 and Idd13 loci, which together influence the cell cycle of immunoregulatory DN T cells.
Allelic variants in the Idd2 and Idd13 loci influence cell cycle. Total spleen cells were stained with propidium iodide and the percentage of cells in S-phase was quantified by flow cytometry, for a B10.BR, NOD.H2k and NOD.H2k-Idd2/13 mice (n = 5) and b C57BL/6 (B6) and NOD mice (n = 7). Mean ± SEM are shown. c Spleen cells from B10.BR and NOD.H2k-Idd2/13 mice were stained with antibodies to CD3, CD4, CD8, and Ki-67. The percentage of Ki-67+ cells was quantified for CD4+ (CD3+CD4+CD8−), CD8+ (CD3+CD4−CD8+), and DN (CD3+CD4−CD8−) T cells. n = 5. Mean ± SEM are shown. Student t test, p value * < 0.05, ** < 0.01, *** < 0.001, ns = non-significant
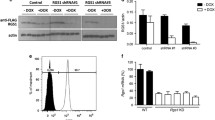
Progression of DN T cells through the cell cycle is influenced by the Idd2 and Idd13 loci. Of the candidate genes encoded within Idd13, our attention turned to Rad51, as inhibition of RAD51 has recently been shown to inhibit diabetes development (Ratiu et al. 2017). RAD51 is a recombinase involved in DNA repair, an essential step to allow efficient progression through the cell cycle. RAD51 acts downstream of γ-H2AX and CHEK1, both of which are encoded within Idd2. Interestingly, we found that RAD51, but not γ-H2AX, was expressed more abundantly in spleen cells from NOD.H2k mice relative to B10.BR mice (Fig. 8 and data not shown). We also found that NOD.H2k mice carrying either the Idd2 or the Idd13 locus both showed a reduction in RAD51 expression, which was not enhanced in NOD.H2k-Idd2/13 mice (Fig. 8). This result suggests that polymorphisms within the Idd13 locus, which encodes for Rad51, effectively modulate RAD51 expression. Importantly, it also suggests that polymorphisms in the Idd2 locus, likely in either H2afx or Chek1, also impact RAD51 expression. Yet, the polymorphism in both Idd2 and Idd13 did not cooperate to restore RAD51 expression to the levels observed in B10.BR mice, suggesting that although Idd2 and Idd13 clearly regulate RAD51 expression, other pathways are also affected by the two loci.
RAD51 expression is modulated by polymorphisms on both the Idd2 and Idd13 loci. One representative Western blot (a) and data compilation (b) of relative RAD51 protein levels in total spleen cells from B10.BR, NOD.H2k, NOD.H2k-Idd2, NOD.H2k-Idd13, and NOD.H2k-Idd2/13 (n = 3). alpha-tubulin is used as a loading control.
Discussion
We have previously shown that autoimmune diabetes susceptibility is associated with decreased DN T cell proportion (Collin et al. 2014; Dugas et al. 2010). In order to identify the genetic determinants defining DN T cell number, we used a linkage analysis approach and congenic strain validation and demonstrated that DN T cell proportion is influenced by Idd2 and Idd13 (Collin et al. 2014; Dugas et al. 2014). However, as the contribution of each locus was modest, we sought to investigate the interactions between the genetic determinants encoded within Idd2 and Idd13 loci and how they cooperate to define DN T cell numbers. Here, we analyzed our linkage analysis data (Collin et al. 2014) and revealed that F2 mice bearing homozygous B10.BR alleles at both the Idd2 and Idd13 loci exhibit a significant increase in DN T cell proportion when compared to mice bearing homozygous NOD alleles at either or both loci. In addition, we show that Idd2 and Idd13 together increase the proportion of DN T cells in both 3A9 TCR and 3A9 TCR:insHEL NOD.H2k mice. Moreover, in a cohort of nine female mice, none of the 3A9 TCR:insHEL NOD.H2k-Idd2/13 mice develop autoimmune diabetes. Together, these data suggest an additive effect of the mouse Idd2 and Idd13 loci in the restoration of DN T cells, a trait associated with autoimmune diabetes resistance. Lastly, we demonstrated that cell cycle and DN T cell proliferation are regulated by the Idd2 and Idd13 loci. Together with our bioinformatics analysis, this suggests that cell cycle progression, by modulating RAD51 expression, is likely contributing to defining the proportion of DN T cells in secondary lymphoid organs.
Of interest, RAD51 has recently been linked to type 1 diabetes (Ratiu et al. 2017). In this study, they find that inhibiting RAD51 protects from diabetes development by enhancing the proportion of immunoregulatory B cells. Notably, DN T cells are able to eliminate B cells and in vivo injection of DN T cells leads to a decrease in autoantibody levels (Dugas et al. 2010; Hillhouse et al. 2010; Ma et al. 2008). It is thus tempting to suggest that inhibiting RAD51 leads to a specific increase in DN T cells which would eliminate autoantibody-producing B cells, thereby resulting in a proportional increase in immunoregulatory B cells, altogether preventing autoimmune diabetes onset. Alternatively, as we have shown that RAD51 is generally increased in the spleen of NOD mice, inhibiting RAD51 may independently affect many cell types, all actively participating in the induction or maintenance of immune tolerance.
Identifying the molecular pathways that influence the size of the DN T cell pool in peripheral lymphoid organs is of interest as it will facilitate the identification of molecular targets that regulate DN T cell numbers. During our analysis of DAVID and STRING bioinformatics databases exploiting the Idd2 and Idd13 gene lists, we were able to limit our focus on three biological processes mediated by strong interactions of genes within the two loci, namely phospholipid catabolism, T cell biology, and cell cycle progression. However, concerning phospholipid catabolism, the five genes located within the Idd13 locus that were found to interact with genes from the Idd2 locus, specifically Adra1d, Pla2g4b, Pla2g4d, Pla2g4e, and Pla2g4f, are not expressed in hematopoietic cells (immgen.org). As we have previously shown that DN T cell proportion is regulated at least in part by hematopoietic-intrinsic factors (Dugas et al. 2014), we found it unlikely that the phospholipid catabolism is the biological process influencing DN T cell numbers. Likewise, the T cell biological process was highlighted by interactions between genes from the CD3 complex encoded in the Idd2 locus and B2m from the Idd13 locus, where we have previously shown that B2m-deficiency does not influence 3A9 DN T cell proportion in secondary lymphoid organs (Dugas et al. 2014). Therefore, the T cell biological process is not likely influencing DN T cell numbers. As a result, we shifted our focus to the third biological process highlighted by our bioinformatics analyses, cell cycle. The STRING analysis of the cell cycle progression revealed several relevant interactions between genes from the Idd2 and Idd13 loci, with various genes of interest being ubiquitously expressed. We further validate that cell cycle and proliferation are regulated by the Idd2 and Idd13 loci as a greater proportion of lymphocytes from NOD.H2k mice are in S-phase. Moreover, the proportion of Ki-67+ DN T cells is also increased in NOD.H2k mice in comparison to both B10.BR and NOD.H2k-Idd2/13 mice. The greater proportion of proliferating DN T cells in NOD.H2k mice, which exhibit a lower proportion of DN T cells in lymphoid organs, relative to NOD.H2k-Idd2/13 mice is counterintuitive. However, the increased proportion of cells in S-phase may suggest a block in cell cycle progression, yielding lower numbers of DN T cells. Altogether, our data thus suggest that DN T cell proportion is influenced by allelic variants of genes encoded within the Idd2 and Idd13 loci, which together influence cell cycle progression.
The cell cycle consists of a series of sequential phases, specifically the G0/G1, S, G2, and M (mitosis) phases, leading to cell division. During the cell cycle, multiple checkpoints are encountered prior to entry into the subsequent phase, which provides an important surveillance mechanism for faithful replication and division of cells (Elledge 1996). The STRING analysis of the cell cycle progression revealed that Chek1 in Idd2 partners with Rad51, Mcm8, and Cdc25b from the Idd13 locus. Indeed, each of these four genes play an important role during S-phase by controlling DNA replication and the DNA damage response to replicative stress. For instance, the DNA helicase MCM8 is a significant player in the elongation phase of DNA replication (Maiorano et al. 2005), it promotes RAD51 recruitment to DNA damage sites and it facilitates homologous recombination-mediated repair of DNA double-strand breaks (Park et al. 2013). As RAD51 is central in replication fork progression (Berti and Vindigni 2016), MCM8 may participate in this pathway during S-phase. In addition to MCM8, Cdc25b, which encodes a phosphatase that is essential for cell cycle transition by promoting the dephosphorylation of key substrates (Lammer et al. 1998), is also a key player during S-phase. For instance, down-regulation of Cdc25 mRNA levels in HeLa cells delays S-phase and leads to a proliferation defect (Garner-Hamrick and Fisher 1998). Furthermore, CDC25B has been involved in centrosome duplication, a process that is initiated in S-phase (Boutros et al. 2007). Interestingly, unscheduled expression of CDC25B during DNA replication has been linked to replicative stress and the formation of DNA lesions (Bugler et al. 2010). In this regard, CHEK1 also plays a dual role during DNA replication. Indeed, CHEK1 is linked to fork progression during S-phase by controlling the initiation of DNA replication (Petermann et al. 2010), and is also critical in the response to single-stranded DNA lesions by phosphorylating key substrates, including the CDC25 phosphatases, and promoting the activation of DNA damage checkpoints (Patil et al. 2013). Interestingly, the conditional deletion, knockdown, or inhibition of Chek1 is associated with an accumulation of cells in S-phase cells as well as increased cell death (Lam et al. 2004; Sorensen et al. 2005), which mirrors our observations for DN T cells from mice exhibiting NOD alleles for Idd2 and Idd13. Indeed, our findings suggest that the Idd2 and Idd13 loci regulate the proportion of cells accumulating in S-phase and of Ki-67+ DN T cells. Moreover, we find that NOD.H2k mice carry fewer DN T cells than both B10.BR and NOD.H2k-Idd2/13 mice in the transgenic setting and that this difference in number may be regulated, at least in part, by apoptosis (Hillhouse et al. 2010). Interestingly, CHEK1 phosphorylates RAD51 at serine-309 and recruits it to DNA repair foci in order to promote homologous recombination repair and cell survival (Sorensen et al. 2005). The fact that we observe increased RAD51 levels in the NOD.H2k mice and that RAD51 is central during S-phase argues for a model where dysregulation of RAD51 levels impairs progression of DN T cells through S-phase resulting in a block in their cell cycle progression. Consequently, these results suggests that potential genetic polymorphisms influencing the interactions between genes encoded within Idd2, such as Chek1, and those encoded within Idd13, such as Rad51, Mcm8, and Cdc25b, modulate DN T cell number.
In summary, our data clearly demonstrates an interaction between the Idd2 and Idd13 loci in determining the number of DN T cells in secondary lymphoid organs, where DN T cell proportion and number are increased in the 3A9 TCR:insHEL NOD.H2k-Idd2/13 mice relative to both single congenic mice and to 3A9 TCR:insHEL NOD.H2k. Our findings also suggest that the S-phase is likely contributing to defining the proportion of DN T cells in secondary lymphoid organs. Although cell cycle progression was the biological process that we identified through our bioinformatics approach, we cannot exclude that other pathways contribute to defining the proportion of DN T cells in secondary lymphoid organs. Further investigation of NOD.H2k-Idd2/13 double congenic mice may reveal other relevant pathways as a target for potentiating the number of immunoregulatory DN T cells, which may be of clinical use for the antigen-specific treatment of various autoimmune pathologies.
Materials and methods
Mice
3A9 TCR transgenic and HEL transgenic mice, where HEL is expressed under the rat insulin promoter (insHEL), on B10.BR and NOD.H2k backgrounds have been previously described (Lesage et al. 2002). The 3A9 TCR NOD.H2k-Idd2 congenic strain bears B10.BR alleles on chromosome 9 between 29.4 and 50.0 Mb (corresponding to markers rs6385855 and rs13480186) at a locus included within Idd2 and was described elsewhere (Collin et al. 2014). Idd13 congenic mice were a gift of D.V. Serreze and were crossed to NOD.H2k 3A9 TCR:insHEL background to introduce the transgenes (Dugas et al. 2014). The NOD.H2k-Idd2/Idd13 strain was obtained by an intercross between 3A9 TCR:insHEL NOD.H2k-Idd2 mice and 3A9 TCR:insHEL NOD.H2k-Idd13 mice. Offspring were genotyped using the markers D9Mit129, D9Mit328, D2Mit132, D2Mit48, and D2Mit338. Mice bearing B10.BR alleles at all of these markers were selected for further intercrosses. After two generations of homozygous mating, an Illumina medium density linkage, serviced through The Centre for Applied Genomics at the Hospital for Sick Children, was performed on the tail DNA from congenic mice to delimit the boundaries of both the Idd2 and Idd13 loci in the NOD.H2k-Idd2/dd13 mice; Idd2 (chromosome 9:29.6–61.4 Mb) and Idd13 (chromosome 2:114.0–156.1 Mb) where positions are based on NCBI Build m38. All of the mouse strains were maintained at the Maisonneuve-Rosemont Hospital animal house facility (Montreal, Canada).
Flow cytometry
Eight- to 12-week-old non-diabetic mice (diastix negative) were analyzed. Spleen, lymph nodes, and thymus were pressed through a 70-μm cell strainer (Thermo Fisher Scientific, Ottawa, Ontario, Canada). Spleen cell suspensions were treated with NH4Cl to lyse red blood cells. Single cell suspensions were stained with the following antibodies to quantify 3A9 DN T cells: CD3 (17A2; Biolegend), TCRβ (H57-597, Biolegend), CD4 (GK1.5; Biolegend and eBioscience), CD8α (53-6.7; Biolegend), CD19 (6D5; Biolegend), and anti-clonotypic 1G12 (homemade) (Peterson et al. 1999) antibody followed by detection with IgG1 (RMG1-1; Biolegend). For Ki-67, cells were subsequently permeabilized using the eBioscience FoxP3 staining kit according to the manufacturer’s protocol. Following permeabilization, cells were incubated with either Ki-67 (B56, BD) or its isotype control (IgG1 κ; BD) for 30 min at 4 °C. For propidium iodide, single cell suspensions from the spleen were washed in PBS and thoroughly resuspended in cold 70% ethanol for at least 1 h at 4 °C. Cells were resuspended in 250 μl of propidium iodide solution (40 μg/ml propidium iodide, Invitrogen; 4 μg/ml RNase, USBiological life sciences; diluted in PBS) per 1 million cells. Stained cells were kept on ice and rapidly processed on a flow cytometer. All data were collected either on a FACSCanto, LSRII, Fortessa x-20, or a BD FACSCalibur (BD Biosciences, Mississauga, Ontario, Canada). All data was analyzed with FlowJo software (Treestar, Ashland, OR, USA).
Histology
H&E staining was performed on 5 to 7 μm pancreas cuts from paraffin blocks, for two to three non-successive cuts for each mouse. H&E slides were scored for infiltration as previously described (Hillhouse et al. 2013), and according to the following scale: 0 = no infiltration, 1 = peri-insulitis, 2 = infiltration < 50%, 3 = infiltration > 50%, 4 = complete infiltration.
ELISA
Total IgG was measured by ELISA according to the manufacturer’s protocol (Bethyl Laboratories Inc., Montgomery, TX, USA). Serum anti-HEL IgG and IgG1 levels were measured by ELISA on Nunc Maxisorp plates (Thermo Fisher Scientific) coated with 100 μg/ml HEL prepared in NaHCO3 at pH 9.5, developed with goat anti-mouse IgG-HRP (Biolegend, clone poly4053) or rat anti-mouse IgG1-biotin (Biolegend, clone RMG1-1) followed by avidin-HRP (Biolegend). A reference pool of sera from diabetic and non-diabetic TCR:iHEL mice was set to contain 100 arbitrary units for anti-HEL antibodies or 10 arbitrary units for total IgG antibodies.
Diabetes incidence study
Diabetes incidence was monitored daily in female 3A9 TCR:iHEL mice for overt signs of diabetes (wet cage, hunched posture) and every 2 weeks for urine glucose levels using Diastix (Bayer, Toronto, Ontario, Canada) starting at 8 to 12 weeks of age. After two successive positive Diastix tests, overt diabetes is confirmed by blood glucose levels higher than 17 mmol/l. The age of diabetes onset is set at the first detection of elevated urine glucose levels. The mice were sacrificed within 1 week of detection of high blood glucose or when they reached more than 28 weeks of age. At culling, tail DNA was collected to verify the genotype of the mouse. The serum was collected and the pancreas was conserved in formalin for at least 48 h at room temperature before being sent for paraffin inclusion.
Pathway analysis
Genes lists (and MGI ID) from Idd2 (chromosome 9:29.6–50.3 Mb) and Idd13 (chromosome 2:114.0–156.1 Mb) were retrieved by the Genes and Markers Query search tool from MGI (Eppig et al. 2015). MGI ID were submitted to DAVID bioinformatics database (Huang et al. 2009a, b). To rescue some unmapped genes (via MGI ID) that did not match with the DAVID database, we sent two lists of unmapped genes to DAVID to get their DAVID ID. MGI and DAVID ID lists were submitted to DAVID and were merged for each locus. Mus musculus genome was specified as background. A third list was built combining all genes from Idd2 and Idd13 (Idd2/13) loci. All olfactory genes (beginning with “Olfr”) were removed from gene lists as they are expected to have little impact on immunological processes. Olfactory genes represent 21% of genes from Idd2 locus. For these three gene lists (Idd2 only, Idd13 only, Idd2 and Idd13 combined), all categories from “disease,” “functional categories,” and “pathways” were selected as well as the subcategories containing “all” from the “gene ontology” category. Functional annotation clustering was performed with default options except for three options: “similarity threshold,” “final group membership,” and “EASE.” They were set to 0.4, 0.6, and 0.05, respectively. Each cluster made by each analysis was named based on the biological processes present in that cluster and compared to the other two analysis results. Highly similar clusters, in the same and/or in different analysis, were grouped under the same biological process. Genes from each biological process produced with the Idd2/13 genes list were compiled separately and identified as located on chromosome 2 (Idd13) or 9 (Idd2). DAVID revealed a total of 9 pathways for Idd2, 5 for Idd13, and 15 for the combined list of Idd2/13. One biological process (transferase) present in Idd2/13 contained only genes from Idd13 and was excluded from further analysis. The metabolic process was too broad as it comprised almost all of the genes from both loci and this pathway was also discarded for STRING analysis.
STRING analysis
Gene (protein) lists of 13 of the 15 identified biological processes for Idd2/13 were submitted to STRING analysis individually (Szklarczyk et al. 2015). All associations made by STRING between the input list and their database were manually validated. Only “co-expression,” “experiments,” and “databases” categories were selected for the active prediction methods. Confidence score was set to 0.15 to retrieve text file results and was set to 0.7 to retrieve pathway interactions images. Only links between genes from Idd2 and Idd13 loci were selected for further analysis. At least 50% of all links (between genes from the two loci) had to have a confidence score of at least 0.7 to be selected as a final biological process of interest (Table 5). Two different biological processes (sodium transport and peptides) did not show any interaction between genes from Idd2 and Idd13 and were excluded from further investigations.
Western blots
Dry pellets of homogenized spleen from B10.BR, NOD.H2k, NOD.H2k-Idd2, NOD.H2k-Idd13, and NOD.H2k-Idd2/13 were resuspended in NuPAGE® LDS Sample Buffer (106 mM Tris-HCl, 141 mM Tris Base, 2% lithium dodecyl sulfate, 10% Glycerol, 0.51 mM EDTA, 0.22 mM SERVA Blue G250, 0.175 mM Phenol Red) and sonicated at 30% intensity for 10 s three times. Protein concentrations were acquired by NanoDrop Microvolume Spectrophotometer. The proteins were loaded on 12% NuPAGE® gels at 200 V. Migrated proteins were transferred to PVDF membranes, blocked with 5% milk and 3% BSA in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h at room temperature. Separate membranes were incubated overnight at 4 °C with mouse anti-αTubulin (Abcam, ab7291), rabbit anti-RAD51 (BioAcademia, 70-002), and anti-H2AX (Bethyl, A300-082A) antibodies. Following washes with TBS-T, the membranes were incubated with either anti-rabbit or anti-mouse-Horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Chemiluminescence was acquired on an Azure c600 (Azure Biosystems). Quantification and normalization of the protein bands were performed on ImageJ.
Statistical analysis
Statistical significance was determined using one-way ANOVA with Bonferroni post-HOC test for differences between more than two groups and a paired or unpaired Student t test was used when testing for significant difference between two groups, as appropriate.
References
Akkaraju S, Ho WY, Leong D, Canaan K, Davis MM, Goodnow CC (1997) A range of CD4 T cell tolerance: partial inactivation to organ-specific antigen allows nondestructive thyroiditis or insulitis. Immunity 7:255–271
Anderson MS, Bluestone JA (2005) THE NOD MOUSE: a model of immune dysregulation. Annu Rev Immunol 23:447–485
Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D (2002) Projection of an immunological self shadow within the thymus by the aire protein. Science 298:1395–1401
Berti M, Vindigni A (2016) Replication stress: getting back on track. Nat Struct Mol Biol 23:103–109
Bluestone JA, Auchincloss H, Nepom GT, Rotrosen D, St Clair EW, Turka LA (2010a) The immune tolerance network at 10 years: tolerance research at the bedside. Nat Rev Immunol 10:797–803
Bluestone JA, Herold K, Eisenbarth G (2010b) Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464:1293–1300
Boutros R, Lobjois V, Ducommun B (2007) CDC25B involvement in the centrosome duplication cycle and in microtubule nucleation. Cancer Res 67:11557–11564
Bugler B, Schmitt E, Aressy B, Ducommun B (2010) Unscheduled expression of CDC25B in S-phase leads to replicative stress and DNA damage. Mol Cancer 9:29
Chen W, Ford MS, Young KJ, Zhang L (2003) Infusion of in vitro-generated DN T regulatory cells induces permanent cardiac allograft survival in mice. Transplant Proc 35:2479–2480
Collin R, Dugas V, Pelletier AN, Chabot-Roy G, Lesage S (2014) The mouse idd2 locus is linked to the proportion of immunoregulatory double-negative T cells, a trait associated with autoimmune diabetes resistance. J Immunol 193:3503–3512
Concannon P, Rich SS, Nepom GT (2009) Genetics of type 1A diabetes. N Engl J Med 360:1646–1654
Driver JP, Serreze DV, Chen YG (2011) Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Semin Immunopathol 33:67–87
Dugas V, Beauchamp C, Chabot-Roy G, Hillhouse EE, Lesage S (2010) Implication of the CD47 pathway in autoimmune diabetes. J Autoimmun 35:23–32
Dugas V, Chabot-Roy G, Beauchamp C, Guimont-Desrochers F, Hillhouse EE, Liston A, Lesage S (2013) Unusual selection and peripheral homeostasis for immunoregulatory CD4 (−) CD8 (−) T cells. Immunology 139:129–139
Dugas V, Liston A, Hillhouse EE, Collin R, Chabot-Roy G, Pelletier AN, Beauchamp C, Hardy K, Lesage S (2014) Idd13 is involved in determining immunoregulatory DN T-cell number in NOD mice. Genes Immun 15:82–87
Duncan B, Nazarov-Stoica C, Surls J, Kehl M, Bona C, Casares S, Brumeanu TD (2010) Double negative (CD3+ 4− 8−) TCR alphabeta splenic cells from young NOD mice provide long-lasting protection against type 1 diabetes. PLoS One 5:e11427
Elledge SJ (1996) Cell cycle checkpoints: preventing an identity crisis. Science 274:1664–1672
Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE (2015) The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res 43:D726–D736
Ford MS, Chen W, Wong S, Li C, Vanama R, Elford AR, Asa SL, Ohashi PS, Zhang L (2007) Peptide-activated double-negative T cells can prevent autoimmune type-1 diabetes development. Eur J Immunol 37:2234–2241
Garner-Hamrick PA, Fisher C (1998) Antisense phosphorothioate oligonucleotides specifically down-regulate cdc25B causing S-phase delay and persistent antiproliferative effects. Int J Cancer 76:720–728
Genin A, Desir J, Lambert N, Biervliet M, Van Der Aa N, Pierquin G, Killian A, Tosi M, Urbina M, Lefort A, Libert F, Pirson I, Abramowicz M (2012) Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum Mol Genet 21:5306–5317
Ghosh S, Palmer SM, Rodrigues NR, Cordell HJ, Hearne CM, Cornall RJ, Prins JB, McShane P, Lathrop GM, Peterson LB, Wicker LS, Todd JA (1993) Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat Genet 4:404–409
He KM, Ma Y, Wang S, Min WP, Zhong R, Jevnikar A, Zhang ZX (2007) Donor double-negative Treg promote allogeneic mixed chimerism and tolerance. Eur J Immunol 37:3455–3466
Hillhouse EE, Beauchamp C, Chabot-Roy G, Dugas V, Lesage S (2010) Interleukin-10 limits the expansion of immunoregulatory CD4−CD8− T cells in autoimmune-prone non-obese diabetic mice. Immunol Cell Biol 88:771–780
Hillhouse EE, Collin R, Chabot-Roy G, Guyon MJ, Tessier N, Boulay M, Liscourt P, Lesage S (2013) Nearby construction impedes the progression to overt autoimmune diabetes in NOD mice. J Diabetes Res 2013:620313
Hillhouse EE, Liston A, Collin R, Desautels E, Goodnow CC, Lesage S (2016) TCR transgenic mice reveal the impact of type 1 diabetes loci on early and late disease checkpoints. Immunol Cell Biol 94:709–713
Ho WY, Cooke MP, Goodnow CC, Davis MM (1994) Resting and anergic B cells are defective in CD28-dependent costimulation of naive CD4+ T cells. J Exp Med 179:1539–1549
Huang d W, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13
Huang d W, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J (2004) Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol 6:884–891
Lam MH, Liu Q, Elledge SJ, Rosen JM (2004) Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell 6:45–59
Lammer C, Wagerer S, Saffrich R, Mertens D, Ansorge W, Hoffmann I (1998) The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci 111(Pt 16):2445–2453
Lang J, Bellgrau D (2004) Animal models of type 1 diabetes: genetics and immunological function. Adv Exp Med Biol 552:91–116
Lesage S, Hartley SB, Akkaraju S, Wilson J, Townsend M, Goodnow CC (2002) Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med 196:1175–1188
Liston A, Hardy K, Pittelkow Y, Wilson SR, Makaroff LE, Fahrer AM, Goodnow CC (2007) Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling. Genome Biol 8:R12
Liston A, Lesage S, Gray DHD, O' Reilly LA, Strasser a, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM. (2004) Generalized resistance to thymic deletion in the NOD MouseA polygenic trait characterized by defective induction of Bim. Immunity 21:817–830
Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, Forichon L, de Massy B, Mechali M (2012) MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell 47:523–534
Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS (2000) The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity 13:107–115
Ma Y, He KM, Garcia B, Min W, Jevnikar A, Zhang ZX (2008) Adoptive transfer of double negative T regulatory cells induces B-cell death in vivo and alters rejection pattern of rat-to-mouse heart transplantation. Xenotransplantation 15:56–63
Maiorano D, Cuvier O, Danis E, Mechali M (2005) MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120:315–328
McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, Rodrigues NR, Prins JB, Serreze DV et al (1995) Crosses of NOD mice with the related NON strain. A polygenic model for IDDM. Diabetes 44:1186–1195
McIver Z, Serio B, Dunbar A, O'Keefe CL, Powers J, Wlodarski M, Jin T, Sobecks R, Bolwell B, Maciejewski JP (2008) Double-negative regulatory T cells induce allotolerance when expanded after allogeneic haematopoietic stem cell transplantation. Br J Haematol 141:170–178
Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, Luo Y, Schimenti JC, Gambus A, Walter JC, Dutta A (2013) The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol 33:1632–1644
Patil M, Pabla N, Dong Z (2013) Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci 70(21):4009–4021
Pearce RB, Formby B, Healy K, Peterson CM (1995) Association of an androgen-responsive T cell phenotype with murine diabetes and Idd2. Autoimmunity 20:247–258
Pelletier AN, Lesage S (2013) The Idd13 congenic interval defines the number of merocytic dendritic cells, a novel trait associated with autoimmune diabetes susceptibility. J Autoimmun 43:70–77
Petermann E, Woodcock M, Helleday T (2010) Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A 107:16090–16095
Peterson DA, DiPaolo RJ, Kanagawa O, Unanue ER (1999) Quantitative analysis of the T cell repertoire that escapes negative selection. Immunity 11:453–462
Prochazka M, Leiter EH, Serreze DV, Coleman DL (1987) Three recessive loci required for insulin-dependent diabetes in nonobese diabetic mice. Science 237:286–289
Ratiu JJ, Racine JJ, Hasham MG, Wang Q, Branca JA, Chapman HD, Zhu J, Donghia N, Philip V, Schott WH, Wasserfall C, Atkinson MA, Mills KD, Leeth CM, Serreze DV (2017) Genetic and small molecule disruption of the AID/RAD51 axis similarly protects nonobese diabetic mice from type 1 diabetes through expansion of regulatory B lymphocytes. J Immunol 198:4255–4267
Serreze DV, Bridgett M, Chapman HD, Chen E, Richard SD, Leiter EH (1998) Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J Immunol 160:1472–1478
Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH (1994) Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med 180:1553–1558
Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T (2005) The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7:195–201
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Taylor SS, McKeon F (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89:727–735
Vallee RB, Varma D, Dujardin DL (2006) ZW10 function in mitotic checkpoint control, dynein targeting and membrane trafficking: is dynein the unifying theme? Cell Cycle 5:2447–2451
Varma D, Dujardin DL, Stehman SA, Vallee RB (2006) Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J Cell Biol 172:655–662
Wallet MA, Flores RR, Wang Y, Yi Z, Kroger CJ, Mathews CE, Earp HS, Matsushima G, Wang B, Tisch R (2009) MerTK regulates thymic selection of autoreactive T cells. Proc Natl Acad Sci U S A 106:4810–4815
Wicker LS, Clark J, Fraser HI, Garner VE, Gonzalez-Munoz A, Healy B, Howlett S, Hunter K, Rainbow D, Rosa RL, Smink LJ, Todd JA, Peterson LB (2005) Type 1 diabetes genes and pathways shared by humans and NOD mice. J Autoimmun 25(Suppl):29–33
Wicker LS, Todd JA, Peterson LB (1995) Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol 13:179–200
Young KJ, DuTemple B, Phillips MJ, Zhang L (2003) Inhibition of graft-versus-host disease by double-negative regulatory T cells. J Immunol 171:134–141
Young KJ, Yang L, Phillips MJ, Zhang L (2002) Donor-lymphocyte infusion induces transplantation tolerance by activating systemic and graft-infiltrating double-negative regulatory T cells. Blood 100:3408–3414
Zhang D, Zhang W, Ng TW, Wang Y, Liu Q, Gorantla V, Lakkis F, Zheng XX (2011) Adoptive cell therapy using antigen-specific CD4−CD8− T regulatory cells to prevent autoimmune diabetes and promote islet allograft survival in NOD mice. Diabetologia 54:2082–2092
Zhang ZX, Ma Y, Wang H, Arp J, Jiang J, Huang X, He KM, Garcia B, Madrenas J, Zhong R (2006) Double-negative T cells, activated by xenoantigen, lyse autologous B and T cells using a perforin/granzyme-dependent, fas-fas ligand-independent pathway. J Immunol 177:6920–6929
Zhang ZX, Yang L, Young KJ, DuTemple B, Zhang L (2000) Identification of a previously unknown antigen-specific regulatory T cell and its mechanism of suppression. Nat Med 6:782–789
Zucchelli S, Holler P, Yamagata T, Roy M, Benoist C, Mathis D (2005) Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 22:385–396
Funding
This work was supported by a grant from the Canadian Diabetes Association (OG-3-13-4018) to SL. SL holds a Senior scholarship and RC holds studentship both from Fonds de recherche du Québec-Santé, and VMD holds a studentship from l’Université de Montréal. MK received a doctoral fellowship from the Cole Foundation. AO is the Canada Research Chair (Tier 2) in Genome Stability and Hematological Malignancies. Work in the AO laboratory was supported by a Transition Grant from the Cole Foundation and an internal Operating Fund from the Sir Mortimer B. Davis Foundation of the Jewish General Hospital.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Collin, R., Doyon, K., Mullins-Dansereau, V. et al. Genetic interaction between two insulin-dependent diabetes susceptibility loci, Idd2 and Idd13, in determining immunoregulatory DN T cell proportion. Immunogenetics 70, 495–509 (2018). https://doi.org/10.1007/s00251-018-1060-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-018-1060-8